Abstract
We generated and characterized a series of spontaneous mutants of Lactococcus lactis IL1403 with average 6- to 11-fold-lowered sensitivities to the circular bacteriocin garvicin ML (GarML). Carbohydrate fermentation assays highlighted changes in carbohydrate metabolism, specifically loss of the ability to metabolize starch and maltose, in these mutants. PCR and sequencing showed that a 13.5-kb chromosomal deletion encompassing 12 open reading frames, mainly involved in starch and maltose utilization, had spontaneously occurred in the GarML-resistant mutants. Growth experiments revealed a correlation between sensitivity to GarML and carbon catabolite repression (CCR); i.e., sensitivity to GarML increased significantly when wild-type cells were grown on maltose and galactose as sole carbohydrates, an effect which was alleviated by the presence of glucose. Among the genes deleted in the mutants were malEFG, which encode a CCR-regulated membrane-bound maltose ABC transporter. The complementation of mutants with these three genes recovered normal sensitivity to the bacteriocin, suggesting an essential role of the maltose ABC transporter in the antimicrobial activity of GarML. This notion was supported by the fact that the level of sensitivity to GarML was dose dependent, increasing with higher expression levels of malEFG over a 50-fold range. To our knowledge, this is the first time a specific protein complex has been demonstrated to be involved in sensitivity to a circular bacteriocin.
INTRODUCTION
Bacteriocins are ribosomally synthesized peptides or proteins of bacterial origin which display antimicrobial activity, most often against strains closely related to the producer (45, 54). Bacteriocins are frequently active at nanomolar concentrations and in general act by pore formation or disruption of the integrity of the target cell membrane (31). The bacteriocin-producing strain as a rule has one or, occasionally among lantibiotics, two immunity determinants, which render(s) the producer immune to the deleterious effects of the respective bacteriocin (17, 20). Specific bacteriocins generally display well-defined (broad or narrow) inhibitory spectra; i.e., they are active only against selected genera or species while having no antagonistic effects on others. This phenomenon is consistent with the theory that bacteriocins utilize a specific receptor molecules on target cells to exert their effects (18, 24, 45). However, only two target molecules are hitherto known. The class I bacteriocin nisin and some closely related lantibiotics have all been shown to employ lipid II, a cell wall precursor molecule, as a docking site. Dependent on their concentration, these bacteriocins can either inhibit peptidoglycan biosynthesis (at low bacteriocin concentrations) or form lethal pores in the cytoplasmic membrane (at high bacteriocin concentrations) (8, 57, 58). Among class II bacteriocins, the pediocin-like bacteriocins (class IIa) and lactococcin A have been demonstrated to target the membrane-located components of the mannose phosphotransferase system (man-PTS) of sensitive cells (15). The efficiency of the man-PTS as a receptor for class IIa bacteriocins was also shown to depend on specific sequence regions of these man-PTS subunits (32, 33).
Circular bacteriocins form a separate class of bacteriocins (56), characterized by their N- to C-terminal covalent link forming a circular backbone. Circular bacteriocins are synthesized as linear precursor proteins, containing a signal peptide (2 to 35 amino acid residues) which is cleaved off during the maturation process. The linear peptides (58 to 70 amino acid residues) are cyclized by the formation of an amide bond between the N- and C-terminal residues, before being exported out of the cell. The details of these mechanisms, the potential coupling of the three processes, and the enzymes responsible are still unclear (10, 39). Circular bacteriocins are subdivided into two classes (11, 41): subclass i includes cationic peptides with limited sequence identity and a high isoelectric point (pI ∼10), whereas subclass ii circular bacteriocins share high sequence identity, with more acidic residues, and have a lower isoelectric point (pI ∼5). Characterizations of the three-dimensional structures of several circular bacteriocins have revealed that they share a compact globular structure consisting of repeated α-helical motifs surrounding a hydrophobic core (41). This highly stable circular structure makes the circular bacteriocins particularly resilient, with characteristic traits such as high thermo-, pH-, and proteolytic stability. These traits make the circular bacteriocins especially interesting for potential industrial applications. For in-depth reviews on circular bacteriocins, we refer the reader to van Belkum et al. (56) and Maqueda et al. (39).
Enterocin AS-48, produced by Enterococcus faecalis subsp. liquefaciens, is the first-discovered and most studied circular bacteriocin (38). Other circular bacteriocins include gassericin A (3, 27, 28), circularin A (27), carnocyclin A (42), subtilosin (4, 29), butyrivibriocin AR10 (26), uberolysin (59), lactocyclicin Q (52), and leucocyclicin (43). A recently identified circular bacteriocin is garvicin ML (GarML) (6). GarML is a 60-amino-acid subclass i circular bacteriocin produced by Lactococcus garvieae DCC43, which was isolated from mallard ducks (Anas platyrhynchos) (51). Protein structure modeling suggested that GarML, similar to other circular bacteriocins, folds into a compact globular bundle comprised of conserved α-helices enclosing a hydrophobic core. A cluster of basic amino acid residues imparts a positive charge on the surface of the peptide, which is thought to attract the peptide to the negatively charged surface of the target cell (6). GarML also shares the common characteristics of circular bacteriocins, being resistant to high temperatures, alkaline or acidic pH, and proteolytic enzymes. The antimicrobial spectrum of GarML is relatively broad; the most sensitive species are those most closely related to the producer, Lactococcus garvieae and Lactococcus lactis. However, strains of Lactobacillus, Pediococcus, Streptococcus, Enterococcus, Propionibacterium, Clostridium, and Listeria have all been shown to be highly or moderately sensitive (6). In a continued effort to unveil the mode of action of bacteriocins, an important prerequisite for the use of bacteriocins in potential future food safety or medical applications (11), in the present study, we present the identification a maltose ATP binding cassette (ABC) transporter which is required for the antimicrobial activity of this novel circular bacteriocin.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and their derivatives used in this study are listed in Table 1. Lactococcal strains were routinely grown at 30°C in M17 medium (Oxoid, Hampshire, United Kingdom) supplemented with 0.4% (wt/vol) glucose, galactose, or maltose. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C with shaking at 225 rpm. Erythromycin or tetracycline was added at a final concentration of 5 μg ml−1, and chloramphenicol was added at a final concentration of 10 μg ml−1 for selection in lactococci. Ampicillin was added at a final concentration of 100 μg ml−1, chloramphenicol was added at a final concentration of 30 μg ml−1, and tetracycline was added at a final concentration of 12.5 μg ml−1 for selection in E. coli.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. lactis | ||
| IL1403 | Indicator strain for GarML | 9a |
| IL1403 200B1 | GarML-resistant isolate | This study |
| IL1403 200C1 | GarML-resistant isolate | This study |
| IL1403 150G1 | GarML-resistant isolate | This study |
| IL1403 150G2 | GarML-resistant isolate | This study |
| IL1403 150H3 | GarML-resistant isolate | This study |
| IL1403 150H4 | GarML-resistant isolate | This study |
| Lactococcus garvieae DCC43 | Producer strain of GarML | 51 |
| Plasmids | ||
| pNZ8037 | Expression vector containing the nisin-inducible promoter PnisA; MCS; Camr | 12 |
| pNZ9530 | Lactococcal expression vector of nisRK; Ermr | 34 |
| pCG11 | Expression vector with malEFG fusion under the nisin-inducible promoter PnisA; Camr | This study |
Camr, chloramphenicol resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance; Ermr, erythromycin resistance; MCS, multiple-cloning site.
Bacteriocin preparation and assays.
The bacteriocin GarML was concentrated from the supernatant of the producing strain L. garvieae DCC43 by precipitation with 45% (wt/vol) ammonium sulfate. Bacteriocin sensitivity was determined by using microtiter plate assays (MPAs), where 100-fold dilutions of the indicator strains were exposed to 2-fold serial dilutions of bacteriocin. Bacteriocin sensitivity was assessed by the minimal concentration of bacteriocin producing 50% growth inhibition, i.e., the MIC50, of the indicator strain. One bacteriocin unit (BU) was defined as the amount of bacteriocin required to produce 50% growth inhibition in 200 μl of culture. Alternatively, bacteriocin sensitivity was determined by a spot-on-lawn soft-agar assay, where the concentrated supernatant was spotted directly onto a soft-agar plate containing a 100-fold dilution of the indicator strain. Bacteriocin activity was seen as clear zones of growth inhibition of the indicator strain. The proteinaceous nature of the antimicrobial substance was confirmed by the addition of proteinase K.
Isolation of L. lactis IL1403 GarML-resistant mutants.
Bacterial cultures grown overnight were 100-fold diluted and added to soft agar containing bacteriocin at concentrations ranging from 150 to 300 BU ml−1. The solution was plated onto agar, and incubation was carried out until colonies appeared. The bacteriocin sensitivity phenotypes of the produced mutants were confirmed and quantified by microtiter plate assays. The clonal identity of the mutant isolates as L. lactis IL1403 was confirmed by multilocus sequence typing (MLST) of four partial genes: pheS, encoding phenylalanine tRNA synthetase; rpoA, encoding the RNA polymerase alpha chain; pepX, encoding X-prolyl dipeptidyl peptidase); as well as the 16S rRNA gene (L200142), according to the scheme described by Rademaker et al. (49). The primers used for MLST are given in Table S1 in the supplemental material.
Growth and phenotypic assays.
Growth assays were performed in microtiter plates with 100-fold-diluted cultures grown overnight using Bioscreen C (Oy Growth Curves Ab Ltd., Helsinki, Finland), measuring the optical density at 600 nm (OD600) in intervals. An API 50 CH kit (bioMérieux, Marcy l'Etoile, France) was used for determining carbohydrate metabolism patterns of bacterial cultures. The stability of the bacteriocin-resistant phenotype was assessed during >50 generations of growth without selective pressure (bacteriocin) in liquid cultures. At regular intervals, 10 colonies from each culture were replica plated onto plates containing 0, 25, 50, and 100 BU ml−1 bacteriocin to determine the sensitivity level of the cell population.
Genetic techniques.
Transformation of lactococci was performed as described previously (25). Isolation of total DNA was performed using a Thermo Savant FastPrep FP120 cell disrupter (Qbiogene Inc., CA) and an E.Z.N.A. plasmid miniprep kit (Omega Bio-tek Inc., GA). Plasmid isolation was carried out by using an E.Z.N.A. plasmid miniprep kit or a Qiagen plasmid midi kit (Qiagen, Düsseldorf, Germany) following enzymatic lysis with lysozyme (4 mg ml−1) and mutanolysin (100 U ml−1) at 37°C for 30 min when required. Cloning techniques were in general performed according to methods described by Sambrook and Russell (50). Restriction enzymes, calf intestinal alkaline phosphatase, and T4 DNA ligase (New England BioLabs Inc., MA) were used according to the supplier's instructions. Oligonucleotides were supplied by Invitrogen Life Technologies (Scotland, United Kingdom). PCR was performed using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, MA), and PCR products were purified using NucleoSpin Extract II (Macherey-Nagel, Düren, Germany). Sequencing was performed by using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, CA). The primers used in this study are given in Table S1 in the supplemental material.
Complementation of GarML-resistant mutants with the L. lactis maltose ABC transporter.
The open reading frames (ORFs) of malE and malFG were amplified from L. lactis IL1403 genomic DNA by PCR, using primer pairs malEFG-F/malE-R2 and malFG-F2/malEFG-R, respectively. The PCR products were then spliced by overlap extension (SOE) PCR using primers malEFG-F and malEFG-R. The malEFG fusion product was confirmed by sequencing and subsequently ligated as a transcriptional fusion into the NcoI/SmaI site of pNZ8037 (12), creating pCG11. In this expression vector, the transcription of malEFG is under the control of the nisin-inducible promoter PnisA. The vectors pCG11and pNZ9530 (34) were introduced into wild-type L. lactis IL1403 and GarML-resistant mutant isolates 200B3, 200C1, and 150G1 by electroporation. The transcription of malEFG was induced by the addition of nisin at concentrations ranging from 0.1 to 10 ng ml−1, and sensitivity to bacteriocin was assayed by MPAs with standard GM17 medium as described above. The ability of the malEFG-complemented GarML-resistant mutants to ferment maltose was assessed by growth assays with M17 medium supplemented with maltose (0.4%, wt vol-1).
RESULTS
Generation and characterization of GarML-resistant mutants.
Upon exposure to GarML at concentrations ranging from 150 to 300 BU ml−1, L. lactis IL1403 colonies arose at an average frequency from 10−7 at the lower concentrations (150 BU ml−1) to 10−8 at the higher concentrations (200 to 250 BU ml−1) of bacteriocin. No colonies were obtained at concentrations above 250 BU ml−1. Colonies were subcultured in nonselective medium, and the sensitivity level of these cultures was determined by microtiter plate assay (MPA). Three phenotypes were recognized: (i) the majority of colonies (68% from three biological replicas) showed wild-type sensitivity (results not shown), indicating that tolerance to the bacteriocin was due to a transient adaptation; (ii) a few colonies (approximately 20%) showed a slight increase in the MIC50 (2 to 3 times more tolerant to GarML than the wild type), and we defined these isolates as low-level-tolerant isolates; and (iii) a total of six isolates from four different biological replicates showed a consistent average 6- to 11-fold increase in the MIC50 against GarML, corresponding to 30 to 55 BU ml−1, compared to wild-type L. lactis IL1403 (5 BU ml−1). High-level resistance to the bacteriocin, as is often observed for class IIa bacteriocins (23, 47) and lantibiotics (35), was not observed. The isolates showed slight variations in MIC50 values between measurements but no consistent internal variation; i.e., all six isolates appeared to be within the same level of sensitivity to GarML. These isolates, denoted 200B3, 200C1, 150G1, 150G2, 150H3, and 150H4 (with the main number indicating the concentration of bacteriocin exposure and the letter identifying the biological replicate), were defined as being GarML resistant and were selected for further studies. The GarML-resistant phenotype was found to be specific; i.e., the isolates were not affected in their sensitivity to other bacteriocins targeting lactococci: lactococcin A, lactococcin G, and nisin (data not shown). The GarML-resistant phenotype was shown to persist for more than 50 generations of growth without selective pressure in liquid medium (data not shown), thus indicating that that the phenotype resulted from a stable genetic change. The clonal identity of these isolates as Lactococcus lactis IL1403 was confirmed by multilocus sequence typing (MLST) of four genetic loci: pheS, rpoA, pepX, and the 16S rRNA gene (data not shown).
GarML-resistant mutants are defective in maltose and starch catabolism.
The L. lactis IL1403 GarML-resistant mutants were further phenotypically characterized. Carbohydrate fermentation assays highlighted two specific differences that distinguished all six mutant isolates from the wild type: while wild-type L. lactis IL1403 ferments starch and maltose, the mutant isolates did not grow on either carbohydrate. Growth experiments confirmed these observations (Fig. 1). Growth of the wild type and mutants in standard GM17 medium (containing 0.4% glucose) did not show any significant differences in the growth rate or in the maximal optical density at the stationary growth phase of the cultures, indicating that the GarML-resistant phenotype does not cause changes in the growth performance with glucose as the main energy source. However, clear differences were observed when maltose was used as the sole carbohydrate source. Consistent with data from previous reports, wild-type L. lactis IL1403 grows at a low rate to a maximum absorbance (OD600) of approximately 0.5 in maltose-containing M17 medium (53). The GarML-resistant mutants, however, grew only to an absorbance of 0.2, corresponding to the residual growth in M17 medium without added sugar. These findings thus confirmed that the GarML-resistant mutants had lost the ability to utilize maltose.
Fig 1.
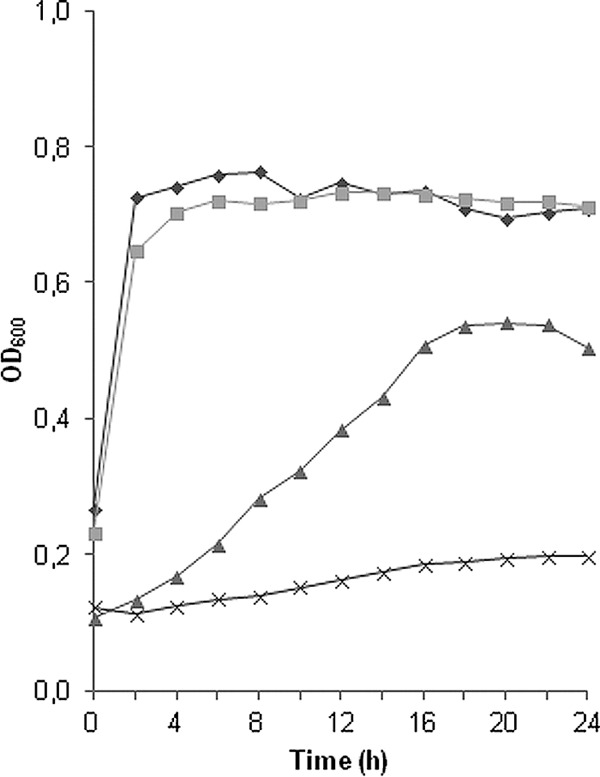
Growth of wild-type L. lactis IL1403 and spontaneous GarML-resistant mutants in glucose- and maltose-containing medium. Growth was measured by OD600 in M17 medium with glucose (0.4%) or maltose (0.4%). Graphs show the growth of wild-type L. lactis IL1403 in glucose (♦) and maltose (▲) and the growth of L. lactis IL1403 GarML-resistant mutants in glucose (■) and maltose (×). The average growth for all six mutant isolates 200B3, 200C1, 150G1, 150G2, 150H3, and 150H4 is shown as single graphs for simplification.
Carbon catabolite repression has an effect on sensitivity to GarML.
Sugar metabolism in most bacteria is normally regulated through a hierarchical mechanism known as carbon catabolite repression (CCR), in which the presence of a preferred sugar (e.g., glucose) represses the expression of genes involved in the metabolism of sugars with less energy output (e.g., galactose and maltose) (13). In L. lactis, several genes involved in maltose utilization (encoding maltose phosphorylase, the maltose ATP binding cassette [ABC] transporter, and β-phosphoglucomutase) are regulated by CCR (1, 46, 48) via catabolite-responsive elements (cre) (Fig. 2) that are bound by the serine-phosphorylated HPr/CcpA repression protein complex in response to high glucose availability in the cell (44). To examine whether sensitivity to GarML is linked to CCR regulation, bacteriocin sensitivity assays were performed on wild-type and GarML-resistant mutant isolates grown in medium with a single sugar or a mixture of different sugars; glucose (CCR inducing), maltose (non-CCR inducing), and galactose (non-CCR inducing). Galactose was selected as a control, as genes involved in the utilization of this carbohydrate are also regulated by CCR in L. lactis (37). The results showed that wild-type L. lactis IL1403 indeed became more sensitive to GarML when grown on maltose and/or galactose than when grown on glucose (Fig. 3). The effect was most pronounced on maltose, where the wild type displayed an average 4-fold reduction in the MIC50 (increased sensitivity to GarML) relative to growth on glucose, corresponding to approximately 1.25 BU ml−1. Growth on glucose alone or in combination with maltose or galactose, however, alleviated the effect and restored a normal level of sensitivity to GarML, which is consistent with a response to a carbon catabolite repressive situation. These results demonstrate that relief of CCR in general, and growth on maltose specifically, has a distinct effect on the sensitivity of L. lactis IL1403 to GarML. The L. lactis IL1403 GarML-resistant mutants, however, did not display any significant changes in sensitivity to the bacteriocin regardless of the type of sugar(s) added to the growth medium (data not shown).
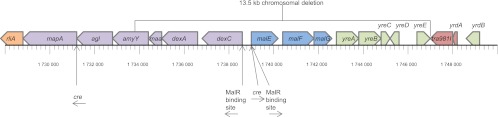
Fig 2.
Chromosomal region encoding functions involved in starch/maltose breakdown, uptake, and conversion in L. lactis IL1403. The rliA gene encodes a transcriptional regulator; mapA encodes maltose phosphorylase; agl encodes α-glucosidase; amyY encodes α-amylase; maa encodes maltose O-acetyltransferase; dexA encodes oligo-1,6-glucosidase; dexC encodes neopullulanase; malEFG encode the maltose ABC transporter substrate binding protein and two permease proteins, respectively; yreABCDE encode hypothetical proteins; L200065 encodes a hypothetical protein; tra981I encodes a transposase of IS981I; yrdA encodes a transposase; and yrdB encodes a hypothetical protein. The 13.3-kb deletion characterized for GarML-resistant mutants is indicated. Genes shown in purple are involved in sugar utilization, blue genes are components of the maltose ABC transporter, green genes are hypothetical proteins with unknown functions, and IS981I-related genes are marked in red. The MalR transcriptional activator (encoded by rliA) is indicated in orange, and its putative binding sites are indicated with arrows (2). Catabolite-responsive elements (cre) subject to regulation by carbon catabolite repression (CCR) are also indicated by arrows (1).
Fig 3.
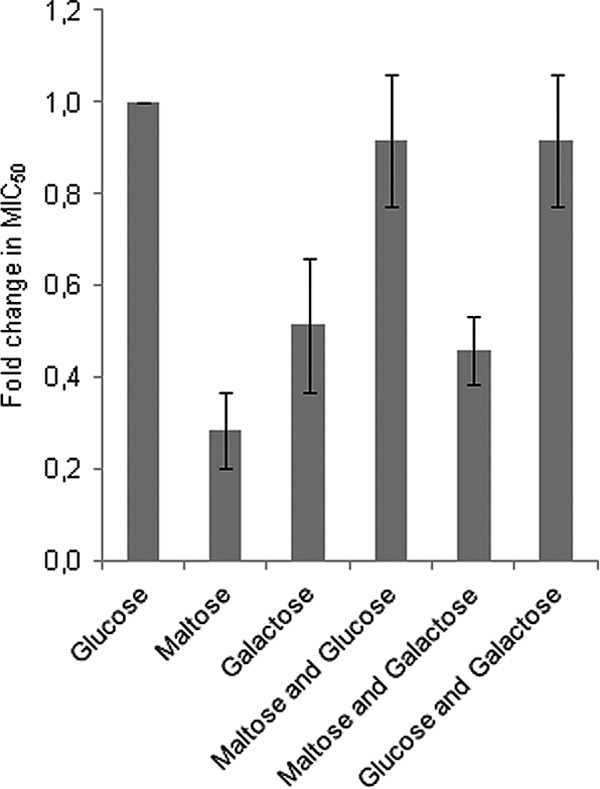
Sensitivity of wild-type L. lactis IL1403 to garvicin ML in different media. Sensitivity to garvicin ML is given as average fold change in the MIC50 in M17 medium containing 0.4% sugar (glucose, maltose, galactose, glucose and maltose, and glucose and galactose) relative to growth on glucose (set to 1). Error bars show standard deviations from three biological replicates.
GarML-resistant mutants have a large deletion in a chromosomal region involved in carbohydrate metabolism.
In L. lactis IL1403, a chromosomal region of approximately 15 kb located at kb 1727 to 1742 in the genome (GenBank accession no. AE005176) contains most of the genes involved in the breakdown, uptake, and conversion of starch and maltose (Fig. 2). An α-amylase (encoded by amyY) located in this region is responsible for the hydrolysis of starch into maltose, and a dedicated ABC transporter (encoded by malEFG) is the sole uptake system for maltose in L. lactis (36). By PCR analysis, it was found that the malE, malF, and malG structural genes were present in wild-type L. lactis strain IL1403, as expected from the genome sequence, but to our surprise, these genes were absent in all six of the GarML-resistant mutant isolates. Further investigation revealed that a minimum 12.6-kb region encompassing 12 ORFs was absent in all six GarML-resistant mutants, indicating that a chromosomal deletion had occurred in these mutants. The putative deletion was subsequently confirmed by sequencing of mutant isolates 150G2 and 150H4, showing an identical 13.5-kb deletion (region from nucleotide [nt] 1733559 to nt 1747095) in both mutants (Fig. 2). The fact that these two independently selected mutants contain the same deletion in their genomes indicates that a sequence-specific DNA rearrangement had occurred. The chromosomal deletion, comprising an α-amylase (amyY), a maltose O-acetyltransferase (maa), an oligo-1,6-glucosidase (dexA), a neopullulanase (dexC), and the maltose ABC transporter substrate binding and permease components (malEFG) as well as additional hypothetical proteins (yreA, yreB, yreC, yreD, yreE, and L200065), thus explains the inability of the GarML-resistant mutants to ferment starch and maltose.
Directly adjacent to the deleted chromosomal region are two genes (tra981I and yrdA) encoding transposon-related functions for insertion sequence (IS) element IS981I (Fig. 2), which provide the most likely explanation for this deletion event. This multicopy IS element was previously shown to be active under laboratory conditions and to have caused similar large chromosomal deletions by replicative transposition in L. lactis IL1403 (14).
The presence of msmK (encoding the multiple-sugar ABC transporter ATP-binding protein) and pgmB (encoding β-phosphoglucomutase), involved in maltose transport and breakdown, respectively, was also assessed by PCR. These genes are implicated in the metabolism of other sugars and are located separately from the starch/maltose gene cluster. However, none of these genes were found to be affected in the six GarML-resistant mutants.
Complementation of mutants with the maltose ABC transporter restores sensitivity to GarML.
The deletion characterized for GarML-resistant mutants led us to hypothesize that the deletion of a potential receptor molecule for the bacteriocin could link the observed genotype to the bacteriocin-resistant phenotype. Most bacteriocins of lactic acid bacteria (LAB) affect the permeability of the membrane, and therefore, it is generally believed that potential bacteriocin receptors on target cells are likely to be membrane located in order to allow a specific interaction. This is true for the receptor of class IIa bacteriocins, lactococcin A, and lipid II-targeting lantibiotics (8, 15, 57). Genes within the deleted region of the GarML-resistant mutants, however, code for either hypothetical proteins with an unknown function or secreted or intracellular enzymes (Fig. 2). The only exception is the maltose ABC transporter encoded by an operon comprising three genes, malE, malF, and malG, encoding the substrate binding and two permease components of the permease, respectively. As indicated previously, the malEFG operon is regulated at the transcriptional level by CCR but also by the transcriptional activator MalR (encoded by the proximately located rliA gene) in the presence of maltose (Fig. 2) (2, 44). This corresponded very well with our observation that growth on maltose and non-CCR-inducing sugars affected sensitivity to GarML. Together, these findings suggested that the maltose ABC transporter is implicated in the sensitivity to GarML.
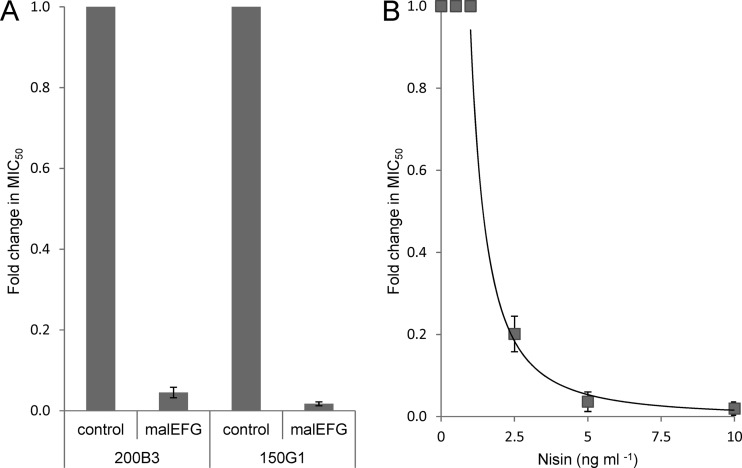
To test whether this membrane-located protein complex is involved in sensitivity to GarML, the L. lactis IL1403 GarML-resistant mutants (lacking malEFG as well as other genes) were complemented with malEFG using a two-plasmid system comprising pNZ9530 and pCG11, with the latter expressing the malEFG genes from a nisin-inducible promoter. Indeed, the complementation of the resistant mutants with these three genes rendered resistant cells highly sensitive to GarML. The complemented mutant isolates 200B3 and 150G1 showed average 22- and 58-fold reductions in the MIC50, respectively (Fig. 4A), relative to that of the control (empty vector). This corresponds not only to a restoration of the wild-type level of sensitivity (5 BU ml−1) but to an even further reduction in the MIC50: 0.3 BU ml−1 for mutant isolate 200B3 and 0.7 BU ml−1 for mutant isolate 150G1. Furthermore, it was shown that the sensitivity of the complemented clones responded to the inducer nisin in a dose-dependent manner. The graded expression of malEFG by induction with 0.1 to 10 ng ml−1 nisin yielded a power correlation (y = 0.942 x−1.783; R2 = 0.978) between the expression level and sensitivity to GarML for the complemented mutant L. lactis IL1403 200B3 (Fig. 4B). The level of sensitivity to GarML for mutant isolate 200B3 thus ranges approximately 50-fold (1 to 50 BU ml−1), increasing with the expression level of malEFG. The sensitivity level of the control (empty vector) remained unchanged irrespective of the induction level. At these induction levels (<10 ng ml−1 nisin), no detrimental effects on the growth of the clones were observed. However, at a higher concentration of the inducer (≥10 ng ml−1 nisin), in a few cases, we observed lethality for the complemented mutant but not for the control, even in the absence of bacteriocin. It should be noted that at 10 ng ml−1 nisin, the concentration of nisin should be sublethal to L. lactis (34), and the observed lethality is more likely caused by detrimental effects commonly resulting from the overexpression of membrane proteins (40). Growth assays of the complemented mutants on medium containing maltose showed that functional complementation was not achieved, as the mutants did not regain the capacity to metabolize maltose (data not shown). This could reflect a possible requirement for one or several of the additional deleted genes and/or regulatory sequences for maltose utilization in L. lactis.
Fig 4.
Sensitivity of mutant isolates complemented with the maltose ABC transporter (malEFG) to GarML. Sensitivity is given as average fold changes in the MIC50 relative to the respective controls (set to 1). The expression of malEFG was induced by the addition of nisin. Error bars show standard deviations from three biological replicates. (A) Sensitivity of GarML-resistant mutant isolates 200B1 and 150G1 complemented with the maltose ABC transporter (malEFG) at an induction level of 1 ng ml−1 nisin relative to controls (empty vector). (B) Correlation between sensitivity to GarML and expression levels of the maltose ABC transporter in GarML-resistant mutant strain 200B3, with the induction level ranging from 0.1 to 10 ng ml−1 relative to the control (no added nisin). No change in the MIC50 was observed at concentrations below 1 ng ml−1 nisin. A sharp decrease in the MIC50 from 1 to 10 ng ml−1 nisin is indicated by a power trend line (y = 0.942 x−1.783; R2 = 0.978).
DISCUSSION
Most bacteriocins act by disruption of the integrity of the target cell membrane (24, 31). The mechanisms of action of most circular bacteriocins have not yet been determined, but for two of the most studied cases, enterocin AS-48 and carnocyclin A, it was previously shown that the bacteriocins can permeabilize liposomes and/or lipid bilayers (21, 22). Also, previous studies of the circular bacteriocins gassericin A and subtilosin A indicated that the bacteriocins do not require a target receptor (28, 55). Thus, it has been suggested that circular bacteriocins in general exert their activity independently of any target molecule in the cell membrane of sensitive cells. For the circular bacteriocin GarML, we observed a relatively broad but defined inhibition spectrum and large interspecies variation in sensitivity to GarML, which led us to hypothesize that a specific target molecule might be involved in the recognition of this bacteriocin. As detailed further below, this study provides evidence that GarML requires the maltose ABC transporter on target cells for antimicrobial activity.
Spontaneous GarML-resistant mutants with 6- to 11-fold-increased sensitivities to the bacteriocin were shown to be defective in maltose utilization (Fig. 1), caused by a large deletion of the chromosomal region encoding these functions (Fig. 2). Furthermore, growth experiments showed a clear correlation between sensitivity to GarML and carbon catabolite repression (CCR) (Fig. 3), where growth on the non-CCR-inducing sugars maltose and/or galactose (in the absence of glucose) increased the sensitivity to GarML markedly (Fig. 3). Growth in maltose-containing medium thus renders wild-type L. lactis IL1403 even more sensitive to GarML, up to approximately 44-fold compared to the resistant mutants (grown on glucose). These findings thus pointed to a possible role of the maltose- and CCR-regulated maltose ABC transporter in sensitivity to GarML, and the subsequent complementation of GarML-resistant mutants with this permease confirmed this hypothesis by effectively restoring the sensitivity to the bacteriocin (Fig. 4A). Moreover, the response to the expression of the maltose ABC transporter appeared to be dose dependent, as higher expression levels of these genes increased the sensitivity of cells to GarML (Fig. 4B), implying a direct correlation between the potency of the bacteriocin and the expression level of the maltose transporter. These results support the notion that the maltose ABC transporter plays an essential role in sensitivity to this bacteriocin. The maltose ABC transporter may potentially function as a target receptor for the bacteriocin, rendering the permease open for the efflux of intracellular solutes, eventually leading to cell death. However, we cannot rule out the possibility that the ABC transporter may be used as a docking molecule, in a manner similar to that of lipid II for the class I lantibiotic nisin (8, 58), or that it transports the bacteriocin to another intracellular target. Continued research efforts may shed further light on the details of the mode of action of GarML.
To our knowledge, this is the first time that a specific protein complex has been demonstrated to be involved in the sensitivity to a circular bacteriocin and also the first example of an ABC transporter acting as a cellular target for bacteriocins in Gram-positive bacteria. However, the finding corroborates recent discoveries in the bacteriocin field. For class IIa bacteriocins and lactococcin A, the sugar transporter man-PTS has been shown to function as a target receptor on sensitive cells (15), and for another bacteriocin, lactococcin 972, the sugar transporter CelB has been implicated in sensitivity (9). Pending future discoveries, it is therefore tempting to predict that sugar uptake systems may emerge as a common theme in bacteriocin target recognition among class II bacteriocins of Gram-positive bacteria and probably also among some microcins of Gram-negative bacteria (5). In this context, bacteriocin production can be viewed as a competitive mechanism (16, 31) targeting competitors for the (primary) food source of the producer strain. In the case of GarML, it is feasible that the producer strain L. garvieae DCC43 antagonizes other lactococci, generally believed to be of plant origin (30), competing for the plant-derived sugar maltose by targeting the maltose uptake system. Maltose utilization is indeed a widespread trait among lactococci (7, 19), and a protein BLAST search revealed that the L. lactis IL1403 maltose ABC transporter subunits have high sequence identity to the homologous proteins in the sequenced strain L. garvieae ATCC 49156 (71%, 67%, and 77% identities to MalE, MalF, and MalG, respectively, with 100% coverage for all), lending further support to this notion. Future work should therefore be directed at elucidating any potential common mechanisms in related strains containing homologous maltose ABC transporters and whether this may reflect the activity spectrum of GarML.
GarML has a predicted structure homologous to other class ii circular bacteriocins, where basic amino acid residues in patches on the surface of the compact hydrophobic globular structure are thought to play a role in interactions with the negatively charged membrane on target cells (6, 41). The fact that several circular bacteriocins have been reported to act on lipid bilayers/liposomes may support the notion that such unspecific surface interactions are sufficient for antimicrobial activity. However, several of these studies were performed with bacteriocin concentrations significantly above the levels required for antimicrobial activity in vivo. For GarML, we have demonstrated that the maltose ABC transporter facilitates high-level sensitivity to this bacteriocin. However, consistent with other circular bacteriocins, we observed receptor-independent killing at higher concentrations of GarML. Our results therefore suggest that this class of bacteriocins may indeed require a specific interaction with a target receptor/mediator for antimicrobial activity at low concentrations. Such a dual concentration-dependent mode of action was demonstrated previously for the lantibiotic nisin (58) and can also be supported by data from previous studies of pore formation with the circular bacteriocin subtilosin A (55). The finding that an ABC transporter is involved in bacteriocin sensitivity is unprecedented for Gram-positive bacteria, and the identification of a novel type of target molecule for antimicrobial peptides could potentially be of importance both for future applications of circular bacteriocins specifically and for drug design and delivery in general.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Research Council of Norway.
We thank Zhian Salehian and Mari Christine Brekke for technical assistance.
Footnotes
Published ahead of print 12 March 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Andersson U, Molenaar D, Radstrom P, de Vos WM. 2005. Unity in organisation and regulation of catabolic operons in Lactobacillus plantarum, Lactococcus lactis and Listeria monocytogenes. Syst. Appl. Microbiol. 28:187–195 [DOI] [PubMed] [Google Scholar]
- 2. Andersson U, Radstrom P. 2002. Physiological function of the maltose operon regulator, MalR, in Lactococcus lactis. BMC Microbiol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arakawa K, et al. 2010. HPLC purification and re-evaluation of chemical identity of two circular bacteriocins, gassericin A and reutericin 6. Lett. Appl. Microbiol. 50:406–411 [DOI] [PubMed] [Google Scholar]
- 4. Babasaki K, Takao T, Shimonishi Y, Kurahashi K. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98:585–603 [DOI] [PubMed] [Google Scholar]
- 5. Bieler S, Silva F, Belin D. 2010. The polypeptide core of microcin E492 stably associates with the mannose permease and interferes with mannose metabolism. Res. Microbiol. 161:706–710 [DOI] [PubMed] [Google Scholar]
- 6. Borrero J, et al. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 77:369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bounaix S, Benachour A, Novel G. 1996. Presence of lactose genes and insertion sequences in plasmids of minor species of the genus Lactococcus. Appl. Environ. Microbiol. 62:1112–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brotz H, et al. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 9. Campelo AB, et al. 2011. The Lcn972 bacteriocin-encoding plasmid pBL1 impairs cellobiose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 77:7576–7585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a. Chopin A, Chopin MC, Moillo-Batt A, Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263 [DOI] [PubMed] [Google Scholar]
- 10. Conlan BF, Gillon AD, Craik DJ, Anderson MA. 2010. Circular proteins and mechanisms of cyclization. Biopolymers 94:573–583 [DOI] [PubMed] [Google Scholar]
- 11. Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 12. de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 14. de Visser JA, Akkermans AD, Hoekstra RF, de Vos WM. 2004. Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics 168:1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Draper LA, Ross RP, Hill C, Cotter PD. 2008. Lantibiotic immunity. Curr. Protein Pept. Sci. 9:39–49 [DOI] [PubMed] [Google Scholar]
- 18. Drider D, Fimland G, Hechard Y, McMullen LM, Prevost H. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elliott JA, Collins MD, Pigott NE, Facklam RR. 1991. Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole-cell protein patterns. J. Clin. Microbiol. 29:2731–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fimland G, Johnsen L, Dalhus B, Nissen-Meyer J. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J. Pept. Sci. 11:688–696 [DOI] [PubMed] [Google Scholar]
- 21. Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. 1991. Permeation of bacterial cells, permeation of cytoplasmic and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J. Bacteriol. 173:886–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gong X, Martin-Visscher LA, Nahirney D, Vederas JC, Duszyk M. 2009. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim. Biophys. Acta 1788:1797–1803 [DOI] [PubMed] [Google Scholar]
- 23. Gravesen A, et al. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361–2369 [DOI] [PubMed] [Google Scholar]
- 24. Hechard Y, Sahl HG. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545–557 [DOI] [PubMed] [Google Scholar]
- 25. Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kalmokoff ML, Cyr TD, Hefford MA, Whitford MF, Teather RM. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can. J. Microbiol. 49:763–773 [DOI] [PubMed] [Google Scholar]
- 27. Kawai Y, Kemperman R, Kok J, Saito T. 2004. The circular bacteriocins gassericin A and circularin A. Curr. Protein Pept. Sci. 5:393–398 [DOI] [PubMed] [Google Scholar]
- 28. Kawai Y, Saito T, Kitazawa H, Itoh T. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62:2438–2440 [DOI] [PubMed] [Google Scholar]
- 29. Kawulka K, et al. 2003. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125:4726–4727 [DOI] [PubMed] [Google Scholar]
- 30. Kelly WJ, Ward LJ, Leahy SC. 2010. Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2:729–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kjos M, et al. 2011. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157:3256–3267 [DOI] [PubMed] [Google Scholar]
- 32. Kjos M, Nes IF, Diep DB. 2009. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology 155:2949–2961 [DOI] [PubMed] [Google Scholar]
- 33. Kjos M, Salehian Z, Nes IF, Diep DB. 2010. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 192:5906–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kramer NE, Van Hijum SAFT, Knol J, Kok J, Kuipers OP. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents. Chemother. 50:1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Law J, et al. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luesink EJ, van Herpen RE, Grossiord BP, Kuipers OP, de Vos WM. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789–798 [DOI] [PubMed] [Google Scholar]
- 38. Maqueda M, et al. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399–416 [DOI] [PubMed] [Google Scholar]
- 39. Maqueda M, et al. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2–22 [DOI] [PubMed] [Google Scholar]
- 40. Marreddy RK, et al. 2011. The response of Lactococcus lactis to membrane protein production. PLoS One 6:e24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin-Visscher LA, Gong X, Duszyk M, Vederas JC. 2009. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 284:28674–28681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin-Visscher LA, et al. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Masuda Y, et al. 2011. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 77:8164–8170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Monedero V, Kuipers OP, Jamet E, Deutscher J. 2001. Regulatory functions of serine-46-phosphorylated HPr in Lactococcus lactis. J. Bacteriol. 183:3391–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nes IF, Yoon SS, Diep DB. 2007. Ribosomally synthesized antimicrobial peptides (bacteriocins) in lactic acid bacteria: a review. Food Sci. Biotechnol. 16:675–690 [Google Scholar]
- 46. Nilsson U, Radstrom P. 2001. Genetic localization and regulation of the maltose phosphorylase gene, malP, in Lactococcus lactis. Microbiology 147:1565–1573 [DOI] [PubMed] [Google Scholar]
- 47. Opsata M, Nes IF, Holo H. 2010. Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qian N, Stanley GA, Bunte A, Radstrom P. 1997. Product formation and phosphoglucomutase activities in Lactococcus lactis: cloning and characterization of a novel phosphoglucomutase gene. Microbiology 143(Pt 3):855–865 [DOI] [PubMed] [Google Scholar]
- 49. Rademaker JL, et al. 2007. Diversity analysis of dairy and nondairy Lactococcus lactis isolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 73:7128–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Sanchez J, et al. 2007. Antimicrobial and safety aspects, and biotechnological potential of bacteriocinogenic enterococci isolated from mallard ducks (Anas platyrhynchos). Int. J. Food Microbiol. 117:295–305 [DOI] [PubMed] [Google Scholar]
- 52. Sawa N, et al. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl. Environ. Microbiol. 75:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sjoberg A, Hahn-Hagerdal B. 1989. Beta-glucose-1-phosphate, a possible mediator for polysaccharide formation in maltose-assimilating Lactococcus lactis. Appl. Environ. Microbiol. 55:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tagg JR, Dajani AS, Wannamaker LW. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thennarasu S, et al. 2005. Membrane permeabilization, orientation, and antimicrobial mechanism of subtilosin A. Chem. Phys. Lipids 137:38–51 [DOI] [PubMed] [Google Scholar]
- 56. van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol. 19:411–418 [DOI] [PubMed] [Google Scholar]
- 57. Wiedemann I, et al. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285–296 [DOI] [PubMed] [Google Scholar]
- 58. Wiedemann I, et al. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772–1779 [DOI] [PubMed] [Google Scholar]
- 59. Wirawan RE, Swanson KM, Kleffmann T, Jack RW, Tagg JR. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.