Abstract
Integrons are natural expression vectors due to the presence of an intrinsic promoter (Pc). Although rare, gene cassettes can harbor their own promoter. This study determined the functionality of an internal promoter in the qnrVC1 cassette whose presence was suggested by a level of transcription similar to that of the preceding cassette (aadA2) and confirmed by in silico analysis. Its functionality was determined by 5′ rapid amplification of cDNA ends (RACE) and cloning into promoter-probe vectors. PqnrVC was found in the qnrVC cassette family, stressing its role in contributing to resistance manifestation.
TEXT
The integron is a genetic element which produces an integrase capable of inserting, excising, and rearranging gene cassettes by site-specific recombination. This element is also considered a natural expression system due to the presence of a promoter (Pc) that controls the transcription of gene cassettes inserted in the array (12). Although gene cassettes are generally promoterless functional units, internal cassette-specific promoters have been identified by in silico analyses and by the resistance phenotype changes of transformed Escherichia coli (1, 2, 9, 11, 13). In general, the expression level of cassette-associated genes is influenced by their relative position in the array (cassette position effect), where genes closer to Pc are more expressed than distal ones (3), and by the presence of cassette-specific promoters.
The qnrVC1 cassette was previously found in a class 1 integron, preceded by the aadA2 cassette, from a clinical Brazilian Vibrio cholerae strain (4). Different from other gene cassettes, which have a specific attC recombination site, qnrVC1 was associated with an attC typically found in Vibrio chromosomal integrons (CIs). More precisely, its attC corresponded to a Vibrio parahaemolyticus repeat (VPR), which is one of the signatures of the V. parahaemolyticus CI. Moreover, although qnrVC1 has been found in a class 1 integron, it is more similar to the chromosomal qnr genes from Vibrionaceae than to the plasmid-borne determinants, such as qnrA1.
This work fully characterized and determined the functionality of an internal cassette promoter distributed among the qnrVC alleles that was present in several genetic elements, ensuring their expression and the emergence of the resistance phenotype.
The transcription of the aadA2-qnrVC1 array was assessed by real-time reverse transcription (RT)-PCR using Power SYBR green PCR master mix (Applied Biosystems). The transcription of gene cassettes was measured in both the wild V. cholerae strain (VC627) and in an E. coli DH5α strain transformed with the pGEM-T Easy vector (Promega) harboring the aadA2-qnrVC1 insert. The single-copy housekeeping rpoA gene from V. cholerae and the β-lactamase (bla) gene, present in the pGEM vector as a selective marker, were used for normalizing the transcription of wild-type and cloned genes, respectively (Table 1). The use of the bla gene eliminates any interference of plasmid copy number on relative quantification of cloned genes. The criteria to interpret the results are described elsewhere (5). Similar relative quantification values were obtained for the aadA2 (2.68 ± 0.04 for the wild-type strain and 2.77 ± 0.09 for recombinant strains) and qnrVC1 (2.51 ± 0.1 for the wild-type strain and 2.60 ± 0.07 for recombinant strains) genes, indicating similar transcript amounts. These results may be explained by (i) the occurrence of a read-through of the RNA polymerase along this array producing a full-length transcript (3) or (ii) the presence of an internal specific cassette promoter controlling qnrVC1 transcription.
Table 1.
Primers used in this study
| Primers | Sequence (5′–3′) | Target |
|---|---|---|
| Real-time RT-PCR | ||
| TR AADA2 F | GCGATGAGCGAAATGTAGTG | aadA2 transcription |
| TR AADA2 R | GGCAGGTAGGCGTTTTATTG | |
| TR QNRVC F | CTTCTCACATCAGGACTTGCA | qnrVC1 transcription |
| TR QNRVC R | AATCGCACCCTTCCAATG | |
| TR RPOA F | GAACAAATCAGCACGACACA | V. cholerae rpoA transcription |
| TR RPOA R | CACAACCTGGCATTGAAGA | |
| TR AMP F | TTTATCCGCCTCCATCCA | bla transcription from the pGEM vector |
| TR AMP R | AGCCATACCAAACGACGAG | |
| Conventional PCR and sequencing | ||
| INT1P F | AAACCTTGCGCTCGTTC | Class 1 integron variable region |
| INB | AAGCAGACTTGACCT | |
| INF | GGCATCCAAGCAGCAAG | |
| M13 F | CGCCAGGGTTTTCCCAGTCACGAC | Cloned insert into pGEM vector |
| M13 R | TCACACAGGAAACAGCTATGAC | |
| T7 GFP Reverse | TAATACGACTCACTATAGGG GGGTAAGCTTTCCGTATGTAGC | Cloned insert into pGLOW |
| 5′ RACE | ||
| GSP1—QNRVC | CACAGCCTTGTACTCTAAAC | First-strand cDNA synthesis |
| GSP2—QNRVC | ACACCACGGCTTAAATCTGA | PCR for accessing the TSS |
| AUAP | GGCCACGCGTCGACTAGTAC | |
| Promoter regions (promoterless probe-vector) | ||
| pQL896-Pc F | GAGCTCGAATTCAAACCTTGCGCTCGTTC | Pc promoter region |
| pQL896-Pc R | CTGCAGAAGCTTGTTGCTGCTCCATAACATCA | |
| pGLOW-Pc F | AAACCTTGCGCTCGTTC | |
| pGLOW-Pc R | GCATACTGCAATCATCCTGTTCGGTCAAGGTTCTGGA | |
| pQL896-QNR F | GAGCTCGAATTCCCGGCGTTATGTGCTTTCT | Putative qnrVC promoter region |
| pQL896-QNR R | CTGCAGAAGCTTTGCAAGTCCTGATGTGAGAAG | |
| pGLOW-QNR F | TTGGCTAAAACGGGGTGT | |
| pGLOW-QNR R | GCATACTGCAATCATCCTCATGCTGTGGCTCCAAAA |
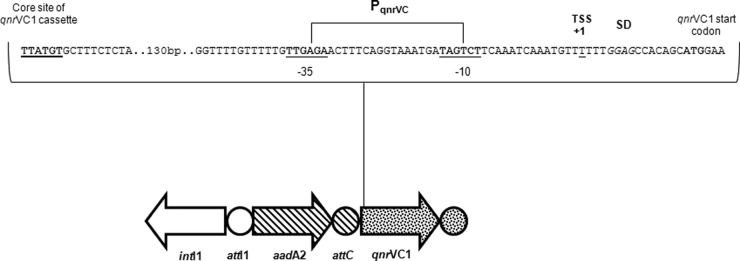
Analyses in silico were performed to verify the presence of internal cassette-specific promoters of aadA2 and qnrVC1 using four promoter prediction programs: Neural Network for Promoter Prediction (NNPP) version 2.2 (Berkeley Drosophila Genome Project, http://www.fruitfly.org/index.html), BPROM (SoftBerry, http://linux1.softberry.com/berry.phtml), prokaryotic promoter analysis using SAK (6), and Prokaryotic Promoter Prediction (PPP) (http://bioinformatics.biol.rug.nl/websoftware/ppp/ppp_start.php). No reliable promoter was found for aadA2; however, a putative internal promoter for qnrVC1 was determined with high scores by all programs. This promoter (−35 TTGAGA |16 bp| −10 TAGTCT) (Fig. 1), named PqnrVC here, presented high similarity with E. coli σ70-dependent promoters (−35 TTGACA |16 to 18 bp| −10 TATAAT) once it preserved 8 (underlined above) of the 12 canonical nucleotides of the −35 and −10 hexamers (10). Most gene cassettes feature a short 5′ untranslated region (UTR); for example, this region in the aadA2 gene cassette has only 9 bp. The qnrVC1 cassette presented an unusual 5′ UTR of 216 bp, supporting the presence of expression signals such as a promoter and a translation initiation region (TIR). A Shine-Dalgarno sequence was identified seven nucleotides upstream from the qnrVC1 start codon (Fig. 1).
Fig 1.
Schematic representation of the transcription/translation motifs found in the class 1 integron harboring the qnrVC1 gene cassette. The recombination core site representing the beginning of the qnrVC1 cassette is highlighted in bold and thickly underlined. The PqnrVC and the first base (+1) of its corresponding TSS (underlined base) are shown. The Shine-Dalgarno (SD) sequence and the start codon of qnrVC1 are indicated in italics and in boldface, respectively.
Interestingly, PqnrVC had been found in qnrVC2, qnrVC3, and qnrVC4 (4, 7, 14). The 5′ UTRs of these alleles are very similar in sequence and length to that of qnrVC1. Moreover, not only qnrVC alleles, but all BLAST hits obtained in January 2012, showed conservation of PqnrVC. These results demonstrate that PqnrVC is a general property of these qnr determinants.
The functionality of PqnrVC was experimentally assessed by cloning into promoterless vector-probe plasmids (pLQ and pGlow) to test its ability in driving expression of a resistance gene (cat) and a reporter gene (gfp). The strong Pc version from the class 1 integron was also cloned as a positive control. These promoters were amplified with modified primers (Table 1) and were cloned into the polylinker region of the pLQ896 and pLQ897 vectors (2), kindly provided by Paul H. Roy. Recombinants transformed with both pLQ896-Pc and pLQ896-PqnrVC were able to grow in 25 mg/liter of chloramphenicol and presented MIC values of ≥256 mg/liter, while E. coli DH5α, used as a control strain, had a MIC of 8 mg/liter.
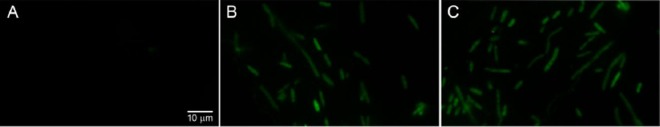
The PqnrVC and Pc were amplified with primers modified as recommended (Table 1) and cloned into the pGlow-TOPO TA expression kit (Invitrogen). After fixation, ampicillin-resistant fluorescent clones harboring the constructions pGlow-PqnrVC and pGlow-Pc were visualized in an Axio Imager M1 fluorescence microscope (Carl Zeiss) with a 60× objective lens by using a fluorescein isothiocyanate (FITC) filter (excitation/emission peaks of 489 nm/509 nm). Only background fluorescence was observed in the negative-control recombinant (E. coli transformed with recircularized pGlow vector) (Fig. 2). This in vitro analysis is one more demonstration of PqnrVC functionality.
Fig 2.
In vitro PqnrVC promoter activity in the qnrVC1 gene cassette. Panel A shows an image of E. coli::pGlow-TOPO TA, corresponding to the recircularized vector used as a negative control for background fluorescence. Panels B and C show the green fluorescent protein (GFP)-fluorescent E. coli::pGlow-Pc and E. coli::pGlow-PqnrVC, respectively, resulting from transformation of E. coli TOP10 cells with the pGlow-TOPO TA containing the promoter regions fused into the ATG codon of gfp.
In order to determine whether qnrVC1 transcription begins under the control of its putative internal promoter, its transcription start site (TSS) was assessed by the 5′ rapid amplification of cDNA ends (RACE) strategy as previously described (5). Sequence analysis revealed a TSS with the +1 position located 14 bp upstream from the initiation codon of qnrVC1. PqnrVC was placed 14 bp upstream from this TSS (Fig. 1), an interval which is considered an acceptable distance between a promoter and its start site (8).
The lack of a promoter region is a remarkable characteristic of gene cassettes, and the identification of structures harboring such motifs is, so far, rare. The possibility of internal cassette promoters is, in fact, an extra element in the evolution of the antimicrobial resistance phenotype, minimizing the position effect, since the cassette-specific promoter guarantees the transcription of even genes that are distal relative to Pc.
ACKNOWLEDGMENTS
We thank the Cellular Ultrastructure Laboratory (IOC/FIOCRUZ)/Equipamento solidário Project (FAPERJ) for fluorescence microscopy technical assistance.
This work was supported by FAPERJ, CNPq, and an Oswaldo Cruz Institute grant.
Footnotes
Published ahead of print 5 March 2012
REFERENCES
- 1. Biskri L, Mazel D. 2003. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 47:3326–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bissonnette L, Champetier S, Buisson JP, Roy PH. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173:4493–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collis CM, Hall RM. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fonseca EL, Dos Santos Freitas F, Vieira VV, Vicente AC. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 14:1129–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fonseca EL, dos Santos Freitas F, Vicente AC. 2011. Pc promoter from class 2 integrons and the cassette transcription pattern it evokes. J. Antimicrob. Chemother. 66:797–801 [DOI] [PubMed] [Google Scholar]
- 6. Gordon L, Chervonenkis AY, Gammerman AJ, Shahmuradov IA, Solovyev VV. 2003. Sequence alignment kernel for recognition of promoter regions. Bioinformatics 19:1964–1971 [DOI] [PubMed] [Google Scholar]
- 7. Kim HB, et al. 2010. Transferable quinolone resistance in Vibrio cholerae. Antimicrob. Agents Chemother. 54:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendoza-Vargas A, et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naas T, Mikami Y, Imai T, Poirel L, Nordmann P. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozoline ON, Deev AA, Arkhipova MV. 1997. Non-canonical sequence elements in the promoter structure. Cluster analysis of promoters recognized by Escherichia coli RNA polymerase. Nucleic Acids Res. 25:4703–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ploy M-C, Courvalin P, Lambert T. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob. Agents Chemother. 42:2557–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stokes HW, Hall RM. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669–1683 [DOI] [PubMed] [Google Scholar]
- 13. Stokes HW, Hall RM. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26:10–19 [DOI] [PubMed] [Google Scholar]
- 14. Xia R, Guo X, Zhang Y, Xu H. 2010. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob. Agents Chemother. 54:3471–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]