Abstract
We have developed a robust cytopathic effect-based high-throughput screening assay to identify inhibitors of dengue virus (DENV) infection. Screening of a small natural product library yielded 11 hits. Four of these were found to be potent inhibitors of DENV, although serotype differences were noted. Taken together, these data suggest that screening of larger and more complex molecule libraries may result in the identification of more potent and specific DENV inhibitors.
TEXT
Two billion people in over 100 countries are at risk of infection from one of the four dengue virus serotypes (DENV-1 to -4) (5, 9). At present, there is no effective vaccine available against DENV (4), and no antiviral drugs have been approved for its treatment. As such, there is a major unmet medical need to identify and develop effective anti-DENV drugs.
While propagating DENV-2 in human hepatoma-derived Huh7.5.1 cells, we noticed a viral-induced cytopathic effect (CPE) (Fig. 1A). A similar but less-pronounced phenotype was also observed in Vero and A549 cells (see Fig. S1A in the supplemental material). Based on this finding, we hypothesized that we could develop a DENV-induced CPE high-throughput screening (HTS) assay in which inhibitors of virus replication would prevent cell death (Fig. 1B). In this assay, toxic compounds would not score as positive hits as they would further increase cell death (Fig. 1B).
Fig 1.
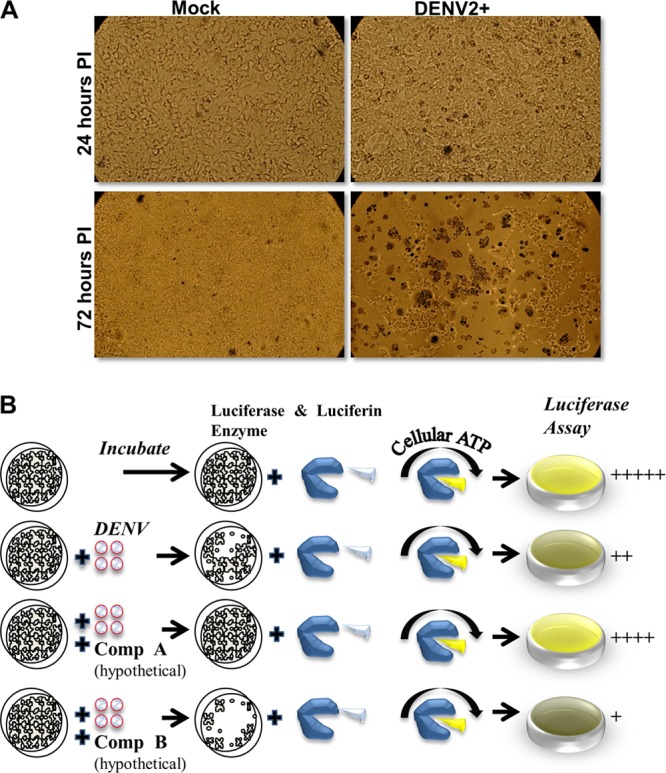
Cytopathic effect of dengue virus on Huh7.5.1. (A) DENV-2 infection of human hepatoma cell line Huh7.5.1 (MOI = 1) induced massive cell death at 72 h postinfection (PI). Huh7.5.1 were seeded in a 96-well format at a density of 1.8 × 104/well and stained with 1% trypan blue after the indicated times. Eighty to 90% cell death in infected wells was observed compared to uninfected wells. (B) Overall design of the screening assay. The cell death resulting from dengue infection causes a loss of cellular ATP, and hence the ATP levels reflect the levels of viral infection and replication before and after drug treatments. In the assay, Huh7.5.1 cells are exposed to dengue virus in the presence of candidate inhibitors. After a period of incubation the cells are lysed in a buffer containing both the luciferase enzyme and luciferin substrate. Any remaining ATP in the cell culture will drive the oxidation of luciferin, resulting in the emission of light that is quantitated using a luminometer. Compound A and B are hypothetical and do not denote specific molecules.
During assay development, we chose to measure DENV-induced CPE by monitoring cellular ATP levels, which positively correlate with cellular viability. Cellular ATP levels are measured by a sensitive luciferase assay that is based on luciferase's requirement for ATP in producing light. To optimize the signal/background (S/B) ratio, we compared the effect of the multiplicity of infection (MOI) and the assay time (see Fig. S1B in the supplemental material). We found that the MOI of 1 and an assay time of 72 h provided an S/B ratio of >16. Using this assay, we found that mycophenolic acid (MPA), a known inhibitor of DENV, yielded a 50% effective concentration (EC50) value of 0.8 μM (see Fig. S2A in the supplemental material), which is similar to what others have reported (3), and a 50% cytotoxic concentration (CC50) of greater than 10 μM (see Fig. S2B). Furthermore, we determined that the Z-factor for the assay was 0.78. The Z-factor is a measure of statistical effect size and is used to judge whether the response in a particular HTS assay is large enough to warrant further attention. A Z-factor between 0.5 and 1 suggests an excellent and robust HTS assay. Taken together, these data demonstrate the efficiency of our assay to rapidly assess inhibitors of DENV infection and to calculate EC50 and CC50 values.
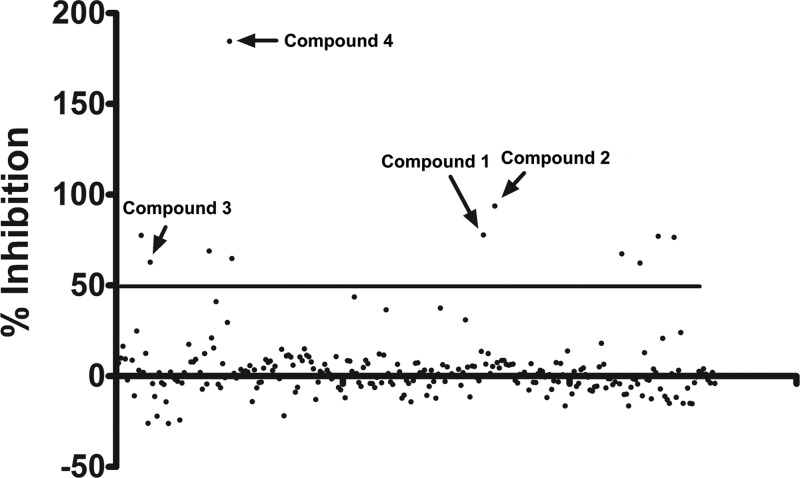
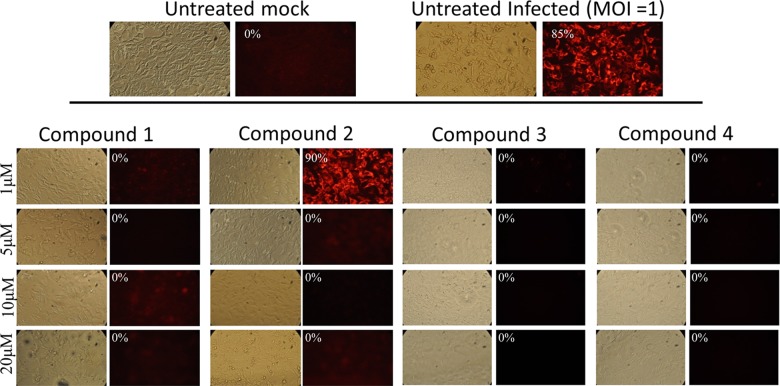
We next screened a library of 235 natural compounds from the National Cancer Institute (NCI) to identify novel inhibitors of DENV-2. The library was screened using a single concentration of 20 μM. We identified 11 compounds that inhibited DENV replication by more than 50% (Fig. 2). To further validate their antiviral activity, we performed immunofluorescence assay (IFA) staining to determine the percentage of viral infection in the presence of inhibitors. Six of the 11 compounds were subsequently shown to inhibit DENV infection at concentrations equal to or less than 2 μM by IFA (Fig. 3 and data not shown). Four of these compounds were subjected to further analyses, including EC50 and CC50 determination (Table 1). Compounds 1, 3, and 4 were found to be potent inhibitors of DENV-2 infection with EC50s less than 1 μM. Compound 2 was somewhat less active and displayed an EC50 of 2.4 μM. Importantly, none of the compounds were toxic at the highest concentration tested (10 μM).
Fig 2.
Screening of a small compound library. The initial high-throughput screen of the library of 235 small compounds yielded 11 positive hits (over 50% inhibition). The virus used was DENV-2, and the concentration of each compound was 20 μM. The four compounds that were found to inhibit DENV-2 in the immunostaining assays shown in Fig. 3 are indicated with arrows.
Fig 3.
IFA of four compounds from the CPE assay. All four compounds inhibited DENV-2 infection of Huh7.5.1 cells albeit with different efficacies. Shown are light microscopy images of cells and IFA staining of DENV prM (MOI = 1; 48 h postinfection) using the 2H2 antibody (red). The number in each IFA panel indicates the estimated percentage of positively stained cells (infected by DENV).
Table 1.
Chemical names, structures, EC50s, and CC50s of four inhibitors identified in the preliminary screening
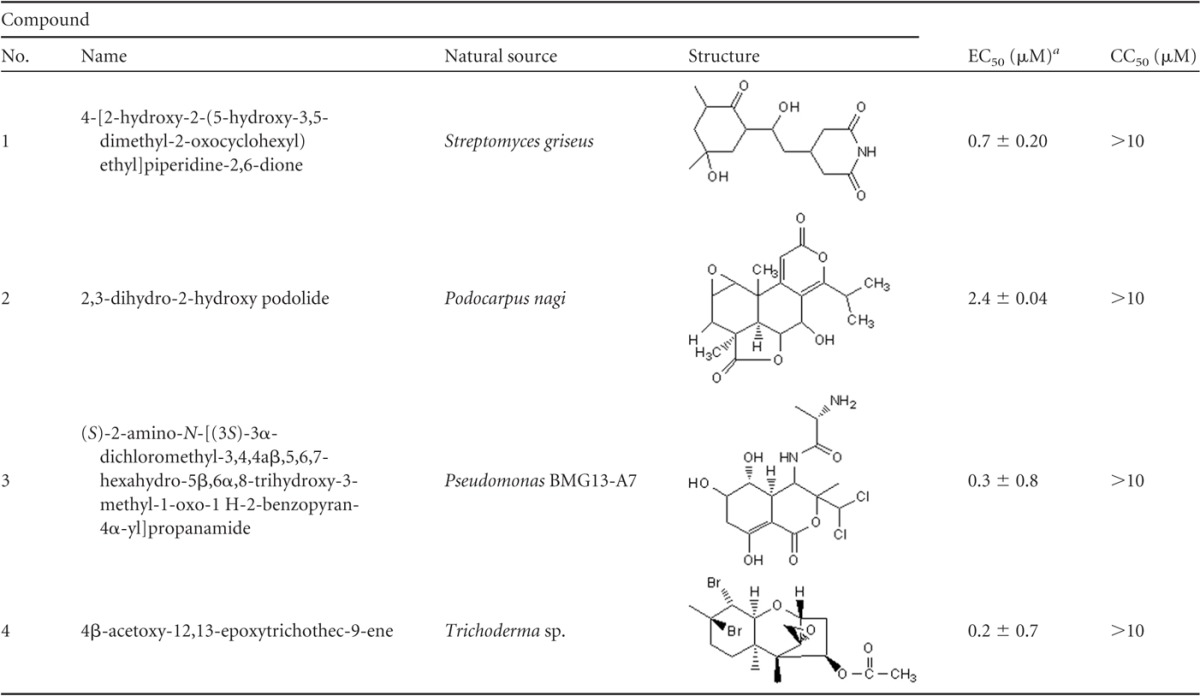
Mean and standard error of the mean for two independent experiments performed in duplicate. EC50 was determined against DENV-2.
We next evaluated the activity of the 4 compounds against different DENV serotypes (see Fig. S3 in the supplemental material). At a 2 μM concentration, compound 1 completely inhibited the DENV-2, -3, and -4 serotypes but had displayed no activity against DENV-1. Similarly, compound 2 also exhibited more potent activity against the DENV-2, -3, and -4 serotypes compared to DENV-1. In contrast, compounds 3 and 4 and MPA inhibited the different DENV serotypes to the same extent. We also tested the inhibitory activity of the compounds against a related Flaviviridae virus, hepatitis C virus (HCV), and against the unrelated Paramyxoviridae virus, Newcastle disease virus (NDV) (see Fig. S4 in the supplemental material). All four compounds showed inhibitory activity toward HCV, albeit that compound 2 and compound 4 were somewhat less potent. Interestingly, compounds 1 and 3 also inhibited NDV replication. However, compounds 2 and 4 and MPA had little to no effect on NDV replication. Therefore, these compounds appear to preferentially inhibit viruses from the Flaviviridae family. The mechanisms by which these compounds inhibit virus replication still need to be determined, and it is unknown if they target viral or cellular enzymes/proteins. Of note, all four of these compounds have been extensively studied for their use as possible antitumor reagents because of their limited cytotoxicity (1, 2, 6–8, 10, 11).
In summary, we have successfully established a robust technological platform that can be used to screen for novel inhibitors of DENV replication. Importantly, the assay can be readily improved to facilitate HTS. One limitation of the assay is that it cannot distinguish between direct-acting antiviral drugs and compounds that inhibit cell death pathways. As such, subsequent experiments are required.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the central development research fund (CRDF) from the University of Pittsburgh.
We thank F. Chisari and C. Basler for providing the Huh7.5.1 cell line and NDV-GFP. We are grateful to Aram Lee for critical reading of the manuscript.
Footnotes
Published ahead of print 5 March 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Chan J, Khan SN, Harvey I, Merrick W, Pelletier J. 2004. Eukaryotic protein synthesis inhibitors identified by comparison of cytotoxicity profiles. RNA 10:528–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dales S. 1965. Effects of streptovitacin A on the initial events in the replication of vaccinia and reovirus. Proc. Natl. Acad. Sci. U. S. A. 54:462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diamond MS, Zachariah M, Harris E. 2002. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304:211–221 [DOI] [PubMed] [Google Scholar]
- 4. Durbin AP, Whitehead SS. 2011. Next-generation dengue vaccines: novel strategies currently under development. Viruses 3:1800–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gubler DJ. 2006. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found. Symp. 277:3–16 [DOI] [PubMed] [Google Scholar]
- 6. Kondo S, Horiuchi Y, Hamada M, Takeuchi T, Umezawa H. 1979. A new antitumor antibiotic, bactobolin produced by Pseudomonas. J. Antibiot. 32:1069–1071 [DOI] [PubMed] [Google Scholar]
- 7. Kubo I, Muroi H, Himejima M. 1993. Combination effects of antifungal nagilactones against Candida albicans and two other fungi with phenylpropanoids. J. Nat. Prod. 56:220–226 [DOI] [PubMed] [Google Scholar]
- 8. Levitt NH, Crowell RL. 1967. Comparative studies of the regeneration of HeLa cell receptors for poliovirus T1 and coxsackievirus B3. J. Virol. 1:693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodenhuis-Zybert IA, Wilschut J, Smit JM. 2010. Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci. 67:2773–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabin AB. 1966. Different effects of chloramphenicol, dactinomycin, and streptovitacin A on synthesis of tumor and virion antigens in SV40 virus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 55:1141–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stafford ME, McLaughlin CS. 1973. Trichodermin, a possible inhibitor of the termination process of protein synthesis. J. Cell. Physiol. 82:121–128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.