Abstract
Pseudomonas aeruginosa PAO1 lon mutants have phenotypes of deficiencies in cell division, swarming, twitching, and biofilm formation as well as a phenotype of ciprofloxacin supersusceptibility. In this study, we demonstrated that a lon mutant was also supersensitive to the DNA-damaging agent UV light. To understand the influence of lon in causing these phenotypes, global gene expression was characterized by performing microarrays on the lon mutant and the PAO1 wild type grown in the presence of subinhibitory concentrations of ciprofloxacin. This revealed major differences in the expression of genes involved in the SOS response and DNA repair. Real-time quantitative PCR confirmed that these genes were highly upregulated upon ciprofloxacin exposure in the wild type but were significantly less induced in the lon mutant, indicating that Lon modulates the SOS response and consequentially ciprofloxacin susceptibility. As the known Lon target SulA is a member of the SOS response regulon, the influence of mutating or overexpressing this gene, and the negative regulator of the SOS response, LexA, was examined. Overexpression of lexA had no effect on the Lon-related phenotypes, but sulA overexpression recapitulated certain lon mutant phenotypes, including altered motility and cell division, indicating that Lon regulates these phenotypes through SulA. However, sulA overexpression did not affect ciprofloxacin susceptibility or biofilm formation, indicating that these properties were independently determined. Lon protease was also demonstrated to strongly influence RecA protein accumulation in the presence of ciprofloxacin. A model of DNA repair involving the Lon protease is proposed.
INTRODUCTION
Pseudomonas aeruginosa is a versatile Gram-negative bacterium that is found in many natural environments (such as soils and marshes) but also causes infections of animals and plants (33). P. aeruginosa is a major opportunistic human pathogen, being the third most common cause of nosocomial infections. It can cause pneumonia, urinary tract infections, and bacteremia, as well as morbidity and mortality in cystic fibrosis patients due to chronic infections that eventually lead to lung damage and respiratory failure. Pseudomonas infections are difficult to eradicate due to high intrinsic resistance of the bacterium, together with its ability to develop resistance to common antibiotics through adaptation and mutation. Multidrug (especially including imipenem)-resistant strains of P. aeruginosa have joined the ranks of “superbugs.” Ciprofloxacin, a fluoroquinolone, is one of the antibiotics currently used for treatment of P. aeruginosa infections (12). Fluoroquinolones target the bacterial enzymes DNA gyrase and topoisomerase IV (10), which are essential for correct DNA replication and transcription. However, the overuse of fluoroquinolones has led to the rise and spread of quinolone resistance in P. aeruginosa strains (39).
Screening of the PAO1 mini-Tn5–luxCDABE (22) and PA14 Harvard (23) mutant libraries, for altered susceptibility to ciprofloxacin revealed that dozens of genes are involved in intrinsic and mutational resistance to this antibiotic (2, 4). One gene of interest was lon, which encodes the Lon protease. Lon is an ATP-dependent cytoplasmic serine protease which associates into hexameric rings in Gram-negative bacteria. It is a homo-oligomer composed of an N-terminal domain, an ATP binding domain, a substrate sensor and discriminatory domain, and a proteolytically active C-terminal domain (5). Lon belongs to the group of the self-compartmentalized or chambered proteases. The Lon protease from P. aeruginosa has 84% similarity to the Lon protease of Escherichia coli (www.pseudomonas.com). Studies on the E. coli Lon protease have shown that Lon degrades the antitermination protein (N protein) of phage λ and is involved in the lysogenic switch of lambda (20, 26). Furthermore, Lon is involved in unfolding misfolded proteins during heat shock as well as in their degradation. Other known Lon targets in E. coli include SulA, which regulates cell division (28), and RcsA, a transcriptional activator for capsule synthesis (35). E. coli lon mutants demonstrate filamentation, sensitivity to UV light and DNA damage, as well as fluoroquinolone susceptibility (40).
The role of Lon in P. aeruginosa has not yet been studied in depth, but it appears to have the appropriate characteristics to be a central player in the complex adaptations of this organism. To date, it is known that lon mutants of P. aeruginosa show ciprofloxacin supersusceptibility, filamentation (2), and deficiencies in swarming motility, twitching motility, and biofilm formation (25). Furthermore, the ATP-dependent Lon protease is a negative regulator of quorum sensing, and a lon mutant exhibits increased hemolytic activity (34). The lon gene is induced by treatment with subinhibitory aminoglycosides (25) but not in wild-type strains in the presence of subinhibitory concentrations of ciprofloxacin.
In this study, we investigated the mechanisms by which Lon participates in the regulation of ciprofloxacin resistance in P. aeruginosa. Our results indicate that the SOS response triggered by DNA-damaging agents is suppressed in a lon mutant compared to in the wild type. This explained the increased sensitivity to fluoroquinolones and UV light in the absence of the Lon protease, and here we propose a model for that regulation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are shown in Table 1. Growth was routinely performed in Luria-Bertani broth (LB) unless indicated otherwise. For assessment of growth, overnight cultures of strains were diluted 1:100 into fresh LB and monitored by using a Tecan Spectrafluor Plus for 24 h by measuring the absorbance at 620 nm every 20 min.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description/characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| H103 (wild type) | Wild-type P. aeruginosa PAO1 strain H103 | Lab collection |
| lon (PA1803) mutant | PAO1 mini-Tn5–luxCDABE::lon; 74_D9, Tetr | 22 |
| recA (PA3617) mutant | PAO1 mini-Tn5–luxCDABE::recA; 69_C12, Tetr | 22 |
| lexA (PA3007) mutant | PA3007::ISphoA, Tetr, derived from UW-WT | 16 |
| sulA (PA3008) mutant | PA3008::ISlacZ, Tetr, derived from UW-WT | 16 |
| lonc | PAO1 lon::mini-Tn5–luxCDABE (pBBR1MCS4:lon+), Cbr | This study |
| sulA-overexpressing strain | PAO1(pBBR1MCS4::sulA+), Cbr | This study |
| lexA-overexpressing strain | PAO1(pBBR1MCS4::lexA+), Cbr | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 λ−thi-1 gyrA96 relA | Invitrogen |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kanr | Invitrogen |
| pBBR1MCS4 | Broad-host-range cloning vector; Ampr | 21 |
| pBBR1MCS4:lon+ | Cloning vector harboring lon amplicon; Ampr | This study |
| pBBR1MCS4:sulA+ | Cloning vector harboring sulA amplicon; Ampr | This study |
| pBBR1MCS4:lexA+ | Cloning vector harboring lexA amplicon; Ampr | This study |
Antibiotic resistance phenotypes: Ampr, ampicillin resistance for E. coli; Cbr, carbenicillin resistance for P. aeruginosa; Kanr, kanamycin resistance; Tetr, tetracycline resistance.
MIC.
The standard broth microdilution method was used for measurement of the MIC of ciprofloxacin in LB (38).
Assessing killing by ciprofloxacin.
Cells of P. aeruginosa were grown to mid-log phase (optical density at 600 nm [OD600] of 0.5) in LB medium. The cultures were pelleted by centrifugation (3,000 × g for 10 min) and resuspended in 1× BM2 minimal salts [62 mM potassium phosphate buffer, pH 7.0, 7 mM (NH4)2SO4, 2 mM MgSO4, 10 μM FeSO4], and 1× MIC of ciprofloxacin was added to the samples. Samples were shaken at 37°C, and aliquots were taken at specified times up to 90 min after antibiotic addition, plated for survivors on LB agar, and grown overnight at 37°C. All experiments were repeated at least three times.
DNA microarray experiment.
Each microarray experiment was performed as previously described (27), comparing five independent cultures of the P. aeruginosa PAO1 wild type and the lon mutant. The cultures were grown to mid-logarithmic phase (OD600 of 0.5) with subinhibitory (one-half the MIC) concentrations of ciprofloxacin (0.05 μg/ml for the wild type and 0.0125 μg/ml for the mutant). Briefly, RNA was isolated, reverse transcribed into cDNA, and then labeled with Cy3 or Cy5 dye. Labeled sample pairs were applied to a spotted microarray provided by the J. Craig Venter Institute, hybridized overnight, and scanned on a ScanArray Express scanner and analyzed using Imagene software and ArrayPipe version 1.7. Only statistically significant changes in gene expression were considered here. Changes in gene expression for several genes of interest were confirmed using real-time quantitative PCR (RT-qPCR).
Real-time quantitative PCR.
The ABI Prism 7000 sequence detection system together with SYBR green I dye (Applied Biosystems) was used to perform RT-qPCR to validate microarray data. For primer design, the ABI Primer Express program (v2.0) and the gene sequence derived from the Pseudomonas database (www.pseudomonas.com) were used. cDNA was prepared as described above.
UV irradiation sensitivity assay.
To test UV sensitivity, 200 μl of an overnight culture was diluted 1:10 in LB and grown for 4 h at 37°C with shaking. After 4 h, the culture was diluted again 1:10 and transferred to a petri dish. UV irradiation was performed with a Chemigenius2 Bio imaging system, which contains 10 UV lamps, from Syngene. The total power was 16 W, the radiant energy was 12 W, and the sample was placed at a distance of 0.5 inches from the UV lamp. The sample was exposed to the UV light for 10 s, after which it was serially diluted and plated on LB agar. The number of CFU was counted the next day. The assay was repeated in triplicate.
Overexpression studies.
The low-copy-number vector pBBR1MCS4 contains a lac promoter just upstream of the multiple-cloning site (MCS) (21). For overexpression studies, the respective genes lexA (PA3007) and the sulA homolog (PA3008) were PCR amplified and cloned into TopoBlunt. This was followed by restriction digestion with the enzymes HindIII and XbaI. These fragments were then ligated into pBBR1MCS4 and transformed into E. coli DH5α competent cells. After plasmid isolation and transformation into wild-type P. aeruginosa PAO1, the constructs were checked by PCR, sequencing, and assessment of appropriate size after plasmid isolation. Experiments performed include RNA isolation, growth curve, MIC, swarming, swimming, twitching, and biofilm formation experiments.
Motility assays.
Swarming motility was measured as described previously (29) by inoculating 1 μl of a mid-logarithmic-phase bacterial culture onto a swarm plate containing modified BM2 minimal medium, containing 0.5% Casamino Acids instead of 7 mM ammonium sulfate and 0.5% agar. Swarming behavior was compared to that of the wild type after 20 h of incubation at 37°C. Swimming motility was evaluated on BM2 plates containing 0.3% agar; 1 μl of a culture was inoculated on a swimming plate, and the resultant diameters of the swim zones were measured after 20 h of incubation at 37°C. The ability of Pseudomonas strains to twitch on a solid surface or interface was evaluated by stabbing 1 μl of an overnight culture through a 1% agar plate to the interface between the plastic and the bottom of the agar. The twitching zone was measured after 24 h of incubation at 37°C.
Biofilm assay.
An abiotic solid surface assay was used to measure mature biofilm formation and has been previously described (11). Overnight cultures were diluted 1:100 in LB and inoculated into 96-well polystyrene microtiter plates and incubated at 37°C for 20 h without shaking. Crystal violet was used to stain the biofilms, and the absorbance was measured at 595 nm with a microtiter plate reader (Bio-Tek Instruments).
Mutagenesis experiments.
The formation of resistant colonies induced by ciprofloxacin was studied. Briefly, bacterial overnight cultures were diluted to an OD600 of 0.1 in 0.7% NaCl before they were spread onto an LB plate. A hole was punched in the middle of the agar plate, and it was filled with a total of 5 μg of ciprofloxacin. Plates were incubated at 37°C for 48 to 72 h, and the resistant colonies formed in the clearance zone were counted. The assay was repeated in 3 independent experiments.
Protein gel and Western blot.
Whole bacterial cells were denatured by boiling at 100°C for 5 min in 2× protein solubilization buffer (0.5 M Tris [pH 6.8], glycerol, 10% SDS, 0.5 M EDTA, distilled water [dH2O]). Denatured samples were then separated on a 12% SDS-PAGE gel. Gels were stained overnight in Coomassie blue. Western blotting was carried out by transferring the protein bands from an unstained SDS-polyacrylamide gel to a polyvinylidene fluoride (PVDF) membrane (Amersham) by using a tank blot apparatus, blocked for 1 h in TBS (20 mM Tris, pH 7.4, 150 mM NaCl) containing 5% skim milk powder and 1% bovine serum albumin (BSA), and incubated with a rabbit polyclonal antibody against E. coli RecA protein (1:1,000; Abcam) (this antibody recognizes RecA in multiple species due to the high homology of the forms of this protein between species) overnight at 4°C in TBS plus 5% skim milk powder. Membranes were washed 8 times for 10 min in TBST (TBS plus 0.1% Tween 20) and then incubated with anti-rabbit IgG horseradish peroxidase (HRP)-linked (1:3,000; Cell Signaling Technology) antibody (secondary antibody) for 30 min in TBS plus 5% skim milk powder. Washing the membranes 5 times for 10 min in TBST allowed for the removal of unbound secondary antibody. The ECL kit (Amersham) was used to develop protein bands.
Microarray accession number.
The ArrayExpress accession number corresponding to this work is E-MTAB-1000.
RESULTS
Supersusceptibility phenotype.
In agreement with previous data (2), MIC measurements showed that the lon mini-Tn5–luxCDABE mutant displayed a 4-fold increased susceptibility to ciprofloxacin compared to the wild type and the complemented strain. This supersusceptibility phenotype was also evident when susceptibility was analyzed by means of killing curves. After 10 min of killing in the presence of 1× MIC (0.1 μg/ml) ciprofloxacin, the lon mutant, the complemented strain, and the wild type showed survival rates of 10%, 70%, and 60%, respectively (Fig. 1). The same trend was observed when killing was assessed at 2× MIC. The increased susceptibility of the mutant compared to that of the wild type was even more accentuated at later time points in that after 45 min, only 0.1% of the lon mutant cells survived exposure to ciprofloxacin, whereas the wild type and the complemented strain showed survival rates of around 5 to 10%.
Fig 1.
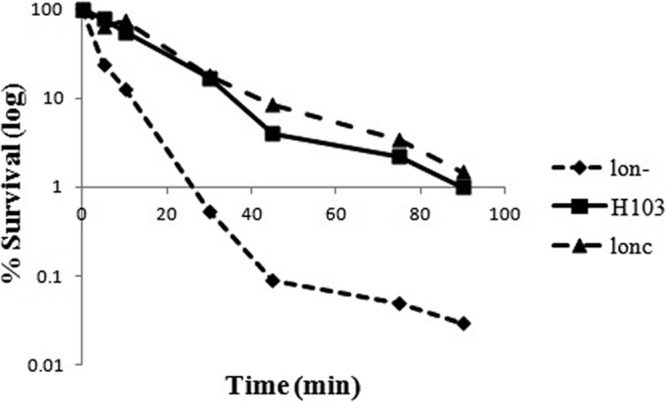
Ciprofloxacin-mediated killing of P. aeruginosa at 1× MIC. Cultures of P. aeruginosa wild-type strain H103, the lon mutant, and the complemented mutant (lonc) were grown to mid-log phase, washed, and then exposed to 0.1 μg/ml ciprofloxacin (1× MIC). After plating and overnight incubation, residual CFU were determined over an incubation period of 90 min. Results from 1 out of three representative experiments are shown.
Microarray analysis of the P. aeruginosa lon mutant under subinhibitory concentrations of ciprofloxacin.
To understand the molecular basis for the ciprofloxacin supersusceptibility phenotype displayed by lon mutants, gene expression was investigated during growth in subinhibitory concentrations of ciprofloxacin in order to trigger any adaptive mechanisms. Microarray analysis revealed that in the presence of ciprofloxacin, there was differential expression of 230 genes between the mutant and the wild type (∼4% of all P. aeruginosa genes) using a cutoff of >2-fold change and a P value of <0.05 (see Table 2 for selected genes and Table S1 in the supplemental material for a complete list of dysregulated genes). Genes involved in the SOS response, including recA, lexA, sulA, recN, prtR, and prtN, and the heat shock genes (dnaK, hslV, hslU, and groES) are known to be upregulated by ciprofloxacin in the wild type (3). Here we demonstrated that, except groES, all of these genes were less expressed in the lon mutant under the conditions utilized here (Tables 2 and 3). Furthermore, 13 out of the 15 LexA-regulated genes (phl, dnaE2, imuB, sulA2, PA0922, PA1044, dinG, PA2288, lexA, sulA, yebG, PA3414, recX, recA, and recN) identified by Cirz et al. (6) were less expressed in the lon mutant under the conditions used here (Table 2; see Table S1 in the supplemental material). This further emphasized that the SOS damage response is majorly impacted in a lon mutant.
Table 2.
Selected P. aeruginosa genes differentially expressed in the lon mutant in the mid-log-phase microarray under subinhibitory (one-half MIC) concentrations of ciprofloxacin
| PA no. | Gene name | Gene function/product | Mid-log phase |
|
|---|---|---|---|---|
| Fold changea | P value | |||
| PA0069 | Conserved hypothetical protein | −5.5 | 0.00 | |
| PA0610 | prtN | Transcriptional regulator PrtN | −12.7 | 0.00 |
| PA0611 | prtR | Transcriptional regulator PrtR | −3.0 | 0.00 |
| PA0625 | Hypothetical protein in phage pyocin operon | −5.6 | 0.00 | |
| PA0649 | trpG | Anthranilate synthase component II | −3.2 | 0.00 |
| PA0650 | trpD | Anthranilate phosphoribosyltransferase | −3.5 | 0.01 |
| PA0651 | trpC | Indole-3-glycerol-phosphate synthase | −2.4 | 0.00 |
| PA0669 | Probable DNA polymerase alpha chain | −3.1 | 0.00 | |
| PA0670 | Hypothetical protein | −5.3 | 0.00 | |
| PA0671 | sulA2 | Hypothetical protein | −5.6 | 0.00 |
| PA0865 | hpd | 4-Hydroxyphenylpyruvate dioxygenase | 2.8 | 0.00 |
| PA0872 | phhA | Phenylalanine-4-hydroxylase | 2.8 | 0.00 |
| PA0922 | Hypothetical protein | −2.4 | 0.00 | |
| PA2008 | fahA | Fumarylacetoacetase | 3.0 | 0.00 |
| PA2009 | hmgA | Homogentisate 1,2-dioxygenase | 8.1 | 0.00 |
| PA2288 | Hypothetical protein | −2.1 | 0.00 | |
| PA3007 | lexA | Repressor protein LexA | −2.2 | 0.01 |
| PA3413 | Hypothetical protein | −2.4 | 0.00 | |
| PA3414 | Hypothetical protein | −2.7 | 0.00 | |
| PA3616 | recX | Conserved hypothetical protein | −2.2 | 0.00 |
| PA4761 | dnaK | DnaK protein | −2.1 | 0.01 |
| PA5053 | hslV | Heat shock protein | −5.0 | 0.00 |
| PA5054 | hslU | Heat shock protein | −2.3 | 0.00 |
A negative fold change represents reduced upregulation, whereas a positive change represents increased upregulation.
Table 3.
Expression of selected genes involved in SOS response and DNA damage repair under various conditions
| PA no. | Gene name | Gene function/product | Fold change in gene expressiona |
|||
|---|---|---|---|---|---|---|
| WT sub-cipro vs WT | lon mutant vs WT | lon mutant sub-cipro vs lon mutant | lon mutant sub-cipro vs WT sub-cipro | |||
| PA0610 | prtN | Transcriptional regulator PrtN | 11.08 ± 2.85 | 2.40 ± 0.14 | 0.77 ± 0.29 | 0.15 ± 0.03 |
| PA0611 | prtR | Transcriptional regulator PrtR | 4.06 ± 0.72 | 1.65 ± 0.36 | 0.84 ± 0.03 | 0.33 ± 0.00 |
| PA0625 | Hypothetical protein | 28.40 ± 8.13 | 2.95 ± 0.16 | 1.30 ± 0.30 | 0.16 ± 0.05 | |
| PA3007 | lexA | Repressor protein LexA | 9.01 ± 0.81 | 0.99 ± 0.22 | 3.27 ± 0.76 | 0.35 ± 0.05 |
| PA3008 | sulA | Hypothetical protein | 9.35 ± 1.77 | 1.38 ± 0.34 | 3.42 ± 0.46 | 0.50 ± 0.09 |
| PA3617 | recA | RecA protein | 6.07 ± 1.68 | 1.29 ± 0.06 | 2.35 ± 0.65 | 0.50 ± 0.00 |
| PA4763 | recN | DNA repair protein RecN | 14.85 ± 2.35 | 0.83 ± 0.25 | 5.44 ± 1.85 | 0.29 ± 0.06 |
Each fold change is represented as the average of 3 biological repeats with the standard deviation (the first listed element relative to the second listed element). WT, wild type; sub-cipro, subinhibitory concentration of ciprofloxacin.
The microarray also revealed that in the presence of subinhibitory concentrations of ciprofloxacin, Lon was a repressor of the phenylalanine degradation pathway and was likely affecting anthranilate synthesis in the presence of ciprofloxacin based on the dysregulation in the lon mutant of the following genes: phhA, hpc, hpd, hmgA, fahA, trpG, trpD, and trpC.
Role of Lon in the SOS response in the presence of ciprofloxacin.
To further investigate if some of the dysregulated genes mentioned above were responsible for the supersusceptible phenotype of the lon mutant, we determined whether selected genes were dysregulated to a lesser extent in the lon mutant than in the wild type in the presence of subinhibitory concentrations of ciprofloxacin.
The expression levels of selected genes were determined by RT-qPCR, comparing the lon mutant strains with and without subinhibitory concentrations of ciprofloxacin as well as the wild-type strains with and without subinhibitory ciprofloxacin. This demonstrated that the induction of genes involved in the SOS response and DNA repair was indeed significantly impaired in the lon mutant, as this mutant showed only a minor response or no response in the presence of ciprofloxacin, compared to the situation in the wild type. For example, recA, a major SOS response gene, was upregulated 6.1-fold in the wild type in the presence of ciprofloxacin; however, the mutant showed just a 2.4-fold increase in the transcription of this gene. The differential induction of the expression of genes involved in the SOS response and DNA repair was also shown (Table 3) for lexA, sulA, and recA as well as prtN, prtR, and the entire RecA/PrtR/PrtN-regulated phage pyocin region, e.g., PA0625, which is involved in ciprofloxacin susceptibility (3).
This is consistent with the explanation that subinhibitory ciprofloxacin caused DNA damage in the wild type, eliciting an SOS response in an attempt to repair this damage, but that the lon mutant was substantially deficient in the induction of this response. We thus suggest that this limited inducibility of the SOS DNA repair responses in the lon mutant explains its observed phenotype of supersusceptibility to the DNA-damaging agent ciprofloxacin, consistent with previous observations that mutants in the recA, recN, and recG genes showed increased supersusceptibility to ciprofloxacin and/or UV light (2, 18). Intriguingly, Brazas et al. (3) previously reported that RecA controlled the expression of a large phage pyocin operon in response to ciprofloxacin and that this influenced susceptibility to ciprofloxacin. Here, we were able to demonstrate that genes in this operon, e.g., PA0625 (and the direct regulators prtRN), were substantially downregulated (Table 2), although this would actually counteract ciprofloxacin susceptibility in the lon mutant.
Effect of Lon on ciprofloxacin-mediated stimulation of mutagenesis.
Ciprofloxacin is known to be a good inducer of mutagenesis, as it interferes with DNA replication (6). The importance of the Lon protease in this mutagenic activity was evaluated. In a plate assay, ciprofloxacin (total, 5 μg) was used to induce mutagenesis, and the ability of cells to grow in the clearing zone was evaluated for the wild type, the lon mutant, and the complemented strain. The results demonstrated that Lon was necessary for the induction of mutagenesis, since 2 ± 1 resistant colonies were observed in the case of the lon mutant, whereas 77 ± 8 and 61 ± 11 colonies were observed for the wild type and complemented strain, respectively (Fig. 2). These results were consistent with an effect of Lon on DNA repair through the SOS system.
Fig 2.
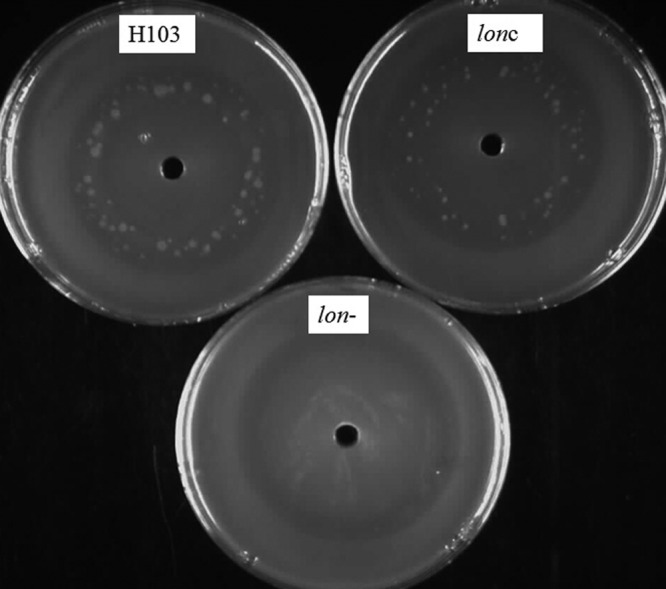
Effect of Lon on ciprofloxacin-mediated stimulation of mutagenesis. Cultures of P. aeruginosa wild-type strain H103, the lon mutant, and the complemented mutant (lonc) were diluted to an OD600 of 0.1 and spread onto an LB plate. A hole was punched in the middle of the agar plate, and it was filled with a total of 5 μg of ciprofloxacin. Plates were incubated at 37°C for 48 to 72 h, and the resistant colonies formed in the clearing zone were counted. Results from one of three representative experiments are shown.
The lon mutant demonstrated increased sensitivity to UV irradiation.
Like ciprofloxacin, UV light is a DNA-damaging agent, and thus, we predicted that the lon mutant would be more sensitive to UV killing than the wild type, as previously shown for lon mutants of E. coli and P. fluorescens Pf-5 (13, 14, 37). For the P. aeruginosa lon mutant, a 20-fold increase in susceptibility to UV irradiation was demonstrated after 10 s of UV light exposure. This susceptibility phenotype could be reversed by complementation. A mini-Tn5–luxCDABE recA mutant (22) was included as a control of the experimental conditions, confirming the known susceptibility of recA mutants to UV damage (Fig. 3) (18).
Fig 3.
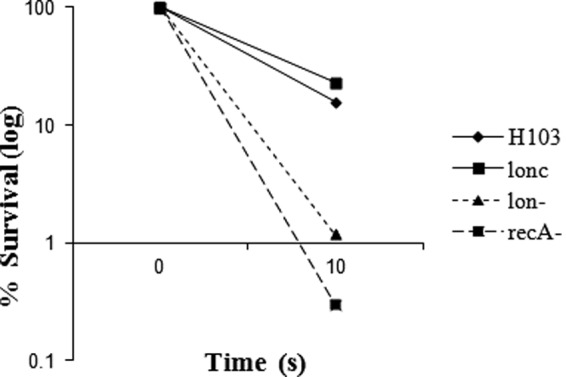
UV-mediated killing of P. aeruginosa. Cultures of P. aeruginosa wild-type strain H103, the lon mutant, the complemented mutant (lonc), and the recA mutant were grown to mid-log phase, diluted, and then exposed to UV light for 10 s and residual CFU determined. Results from one of three representative experiments are shown.
Dysregulation of lon during lethal UV light and ciprofloxacin exposure.
Treatment of the PAO1 wild-type strain of P. aeruginosa with either a lethal dose of UV light (30 s exposure) or 4× MIC (0.4 μg/ml) of ciprofloxacin resulted in a moderate downregulation of lon expression (0.25 ± 0.16-fold and 0.42 ± 0.20-fold, respectively); conversely, a gene involved in the SOS response (lexA) was highly upregulated ([8 ± 2.57]-fold) and served as an experimental control.
Phenotypic effects of sulA and lexA overexpression.
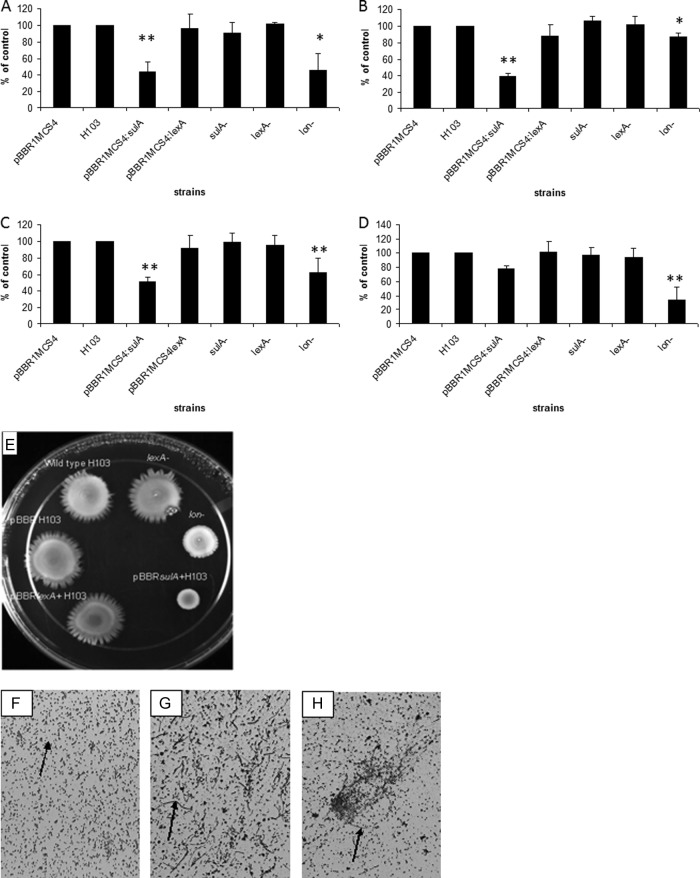
LexA is the primary repressor of the SOS DNA damage response and a target of RecA coprotease (24). SulA acts downstream of LexA and RecA in the SOS response to repress cell division while damage is being repaired (15). For E. coli, it has been shown to be a direct target for the protease action of Lon (28). If Lon were acting directly on either the LexA or SulA protein in P. aeruginosa, then removal of Lon by mutation would have the effect of increasing its function, which we attempted to mimic by overexpressing the cloned genes. Overexpression of sulA (PA3008) and lexA (PA3007) was achieved by constitutive expression of the cloned genes behind the lacZ promoter (constitutive in Pseudomonas) of the low-copy-number vector pBBR1MCS4. The expression level was checked by RT-qPCR and revealed a 31-fold upregulation for the sulA-overexpressing clone compared to the vector alone and 101-fold upregulation in the case of the lexA-overexpressing clone. Overexpression of sulA or lexA did not result in any major growth defect as measured using the Tecan Spectrafluor Plus. Cells overexpressing LexA showed a normal swarming behavior like the wild type and a lexA mutant. In contrast, overexpression of sulA led to a swarming deficiency similar to that exhibited by a lon mutant, whereas a sulA mutant did not have a swarming defect. The same observations were made for the other motilities affected by Lon, namely, swimming (mediated by flagella in an aqueous environment) and twitching (on solid surfaces and interfaces mediated by type IV pili). No major changes in ciprofloxacin MIC or biofilm formation were noted for the constructs overexpressing either sulA or lexA compared to the control, even though biofilm formation is another form of social behavior mediated by flagella and type IV pili. As expected, the strain overexpressing sulA formed, like a lon mutant, long filaments, since SulA is a cell division inhibitor (Fig. 4). This is consistent with the explanation that Lon degrades SulA also in P. aeruginosa and that lon mutants likely accumulate SulA. Accumulation of SulA in cells would lead to the formation of long filaments, and such cells might demonstrate motility defects. However, SulA accumulation clearly did not affect other Lon phenotypes.
Fig 4.
Effect of sulA and lexA overexpression on various types of motility. Cultures of P. aeruginosa overexpressing either lexA or the sulA homolog (PA3008) were grown to mid-log phase and then inoculated onto swarming, swimming, or twitching plates. Swarming motility (A and E), swimming (B), twitching (C), and biofilm formation (D) were assayed compared to the wild type. The filamentation phenotype was analyzed by light microscopy. The wild type (F), the sulA-overexpressing strain (G), and the lon mutant (H) are shown. The bars and error bars represent the averages and standard deviations of 3 independent experiments. A statistically significant difference was observed for the sulA-overexpressing strain at the indicated cases with a P value of ≤0.003 (Student's t test, indicated by **; for lon- the P value of <0.05 is indicated by *).
To investigate if Lon amplified the SOS response through SulA, transcriptional regulation was investigated for the strain overexpressing sulA. The sulA-overexpressing strain showed a transcriptional profile for the genes recA and lexA, in the presence of subinhibitory concentrations of ciprofloxacin, that was similar to the transcriptional profile of the control. Under nondamaging conditions, sulA was, as expected, highly upregulated in the strain overexpressing sulA. However, in the presence of DNA damage, sulA was less expressed in the overexpressing strain, indicating the autoregulation of sulA itself (Table 4).
Table 4.
Effects on SOS transcriptional regulation of the sulA-overexpressing strain compared to the vector control after subinhibitory ciprofloxacin induction
| Gene | Fold change in gene expressiona |
|
|---|---|---|
| Vector control plus ciprofloxacinb vs vector control (no antibiotic) | sulA-overexpressing strain plus ciprofloxacin vs sulA-overexpressing strain | |
| recA | 4.0 ± 1.5 | 5.0 ± 1.9 |
| lexA | 5.9 ± 0.8 | 11.0 ± 4.1 |
| sulA | 4.3 ± 1.3 | 1.4 ± 0.9 |
Fold changes represent the increase in the first listed element relative to the second listed element.
Induction was with a subinhibitory concentration of ciprofloxacin.
Effect of ciprofloxacin on protein expression in the lon mutant.
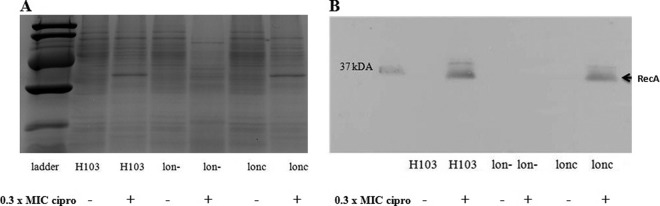
Since no apparent difference in transcriptional regulation was observed for the sulA-overexpressing strain, we hypothesized that the Lon protease influenced protein accumulation in the presence of ciprofloxacin. Whole-cell lysates were used to determine if Lon impacted the major protein of the SOS response (RecA) under DNA-damaging conditions, such as subinhibitory concentrations of ciprofloxacin. SDS-polyacrylamide gel electrophoresis and Western blot analysis demonstrated that expression of RecA (37 kDa) was induced under subinhibitory concentrations of ciprofloxacin in the wild type and the complemented lon strain; however, it failed to be expressed in the lon mutant, thus highlighting that RecA production was dependent on the Lon protease (Fig. 5). Under noninducing conditions, no RecA band was visible in any of the strains tested, as RecA is accumulated only upon DNA damage.
Fig 5.
Induction of the P. aeruginosa RecA protein by subinhibitory concentrations of ciprofloxacin (cipro) in the wild type, the complemented lon strain, and the lon mutant. A Coomassie brilliant blue-stained SDS-PAGE gel (A) and an immunoblot of an identical gel are shown (B). Bacterial strains were grown overnight in the presence or absence of subinhibitory concentrations of ciprofloxacin, and whole-cell protein was loaded onto SDS-PAGE gels for analysis. For Western blotting, samples separated by SDS-PAGE were transferred to a PVDF membrane and incubated with a polyclonal antibody against RecA and a secondary antibody conjugated to HRP. Protein bands were developed with the ECL kit. Results from one of three repeats with similar data are shown.
DISCUSSION
This study demonstrates for the first time that the ATP-dependent Lon protease (PA1803) plays a major role in regulating the SOS response and DNA repair in P. aeruginosa. Studies on the ATP-dependent Lon protease in different bacteria have shown its involvement in such diverse processes as cell division (2, 30), flagellar biosynthesis (31), capsule synthesis (35), UV tolerance (37), motility (25), and antibiotic resistance (2). However, the phenotypes of the lon mutants vary between different bacterial species. In P. aeruginosa PAO1, lon mutants exhibit a motility defect, show extreme filamentation and greater hemolytic activity, and of particular interest, are supersusceptible to ciprofloxacin (2, 25, 34). Explaining this supersusceptibility was of great interest to us.
Previous studies showed that subinhibitory and inhibitory concentrations of ciprofloxacin lead to dramatic changes in global transcription. Most striking were the upregulation of a susceptibility determinant, the R2/F2 pyocins (PA0613 to PA0648), and the genes involved in DNA repair and SOS response (3).
Overall, our microarray and RT-qPCR results showed that the Lon protease of P. aeruginosa was important for full induction of the SOS response upon exposure to DNA-damaging agents, including ciprofloxacin. Brazas et al. (2) previously demonstrated that preexposure to ciprofloxacin led to adaptive resistance in the P. aeruginosa wild-type PAO1 but failed to induce adaptive resistance in the lon mutant. It was concluded that the extreme cell elongation phenotype of the mutant contributed to this phenotype. In the present study, we were able to demonstrate another explanation, namely, the weaker induction of the SOS response and DNA repair observed in the lon mutant. The cell elongation phenotype triggered through the SOS response is controlled by SulA (17), and overexpression of sulA did not recapitulate ciprofloxacin susceptibility, indicating that elongation likely did not explain altered ciprofloxacin susceptibility. The negative and the positive regulators of the SOS response, LexA and RecA, respectively, are common in all bacterial species and are highly upregulated in P. aeruginosa in response to environmental stress. While these genes and several other genes involved in the SOS response are upregulated in the wild type after ciprofloxacin treatment to overcome the DNA damage, the present study demonstrated that if lon was mutated, this SOS response was considerably weaker. Consistent with this deficiency in this DNA repair system, it was demonstrated that Lon protease was important in ciprofloxacin-mediated mutagenesis. Some SOS response genes were still upregulated by ∼3-fold in the lon mutant, but this induction was substantially lower than in the wild type. We propose a model (Fig. 6) whereby Lon protease normally inhibits the action of RecA repressors (RecX, RecR, and RdgC, etc.) (8, 9, 32, 36), leading to the autoamplification of RecA. In the situation where Lon is inactive, the repressor stays intact and no RecA amplification would occur, leading to supersusceptibility to DNA-damaging agents, such as fluoroquinolones and UV light, as damage cannot be repaired. Consistent with this, mutants in genes involved in the SOS response and DNA repair (recA, recN, and recG) exhibit an increased susceptibility to ciprofloxacin and UV light (2, 18).
Fig 6.
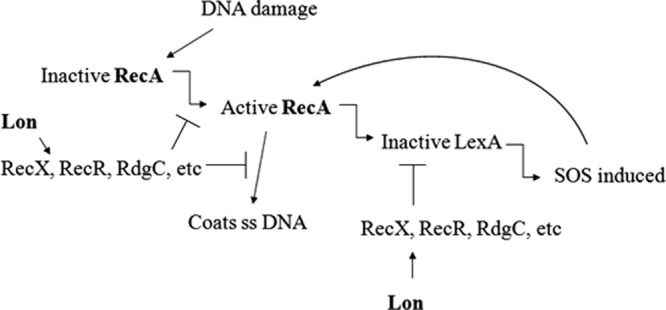
Proposed model for the involvement of the Lon protease in the DNA damage response. Under DNA-damaging conditions, the Lon protease is proposed to cleave and thus antagonize one or more of RecX, RecR, and RdgC, etc., which are known repressors of RecA in E. coli (8, 9, 32, 36). This inhibition leads to autoamplification of RecA, and the SOS response is induced. However, if the Lon protease is inactive, the repressors are proposed to inhibit RecA function and autoamplification.
Under normal conditions, the LexA repressor binds to the SOS box located in the promoter region of genes involved in the SOS response and represses the expression of genes involved in DNA repair. The SOS response includes 43 genes in E. coli (7), 33 genes in Bacillus subtilis (1), and at least 15 genes in P. aeruginosa (6). Upon DNA damage, the resulting single-stranded DNA (ssDNA) is recognized by RecA, and RecA in turn forms filaments around the ssDNA; at the same time, RecA induces the autocleavage of the repressor LexA, enabling the damage repair genes to be transcribed and the repair of DNA damage. In a lon-deficient mutant, the damage likely would not be repaired as effectively as in the wild type, due to the reduced induction of genes involved in the SOS response. Interestingly, upon exposure to 30 s of UV light and 4× MIC of ciprofloxacin (representing lethal conditions), the lon gene itself is slightly downregulated. Thus, it can be hypothesized that the Lon protease plays a role in controlling the expression level of the SOS response genes, possibly to suppress potential lethality associated with overexpression of recA and other SOS response genes and especially to limit the induction of cell death mediated by the phage pyocin operon (3). We were able to demonstrate that Lon does not act through sulA at the transcriptional level but likely works through SulA to regulate cell division. Therefore, we hypothesize that Lon acts through cleavage of one or more of the control elements of the SOS response. Since protein accumulation of RecA did not occur in the lon mutant upon ciprofloxacin exposure, RecA would not be able to cleave the repressor LexA to activate the SOS system. Furthermore, reduced RecA expression in the lon mutant would reduce binding to ssDNA, and thus, DNA damage could not be repaired. Lon is therefore important for modulating RecA function. It is also possible that the Lon protease might be important for cleaving other elements of the SOS response.
The ATP-dependent Lon protease investigated in this study controls the DNA stress response and fluoroquinolone susceptibility and is upregulated by aminoglycosides. P. aeruginosa has additional ATP-dependent proteases that exhibit similarity to PA1803. One of these proteases is the recently identified AsrA (aminoglycoside-induced stress response) ATP-dependent protease (PA0779), which is 60% similar to the Lon protease PA1803 (19). AsrA is highly upregulated by bacteriostatic and lethal concentrations of tobramycin and controls the heat shock stress response to tobramycin in Pseudomonas. It is thus becoming clear that ATP-dependent proteases are not only involved in general stress responses but have very specific roles and regulate and control important stress responses in Pseudomonas. The significant role of the Lon protease in virulence-related processes and antibiotic resistance makes it an attractive antimicrobial target to combat P. aeruginosa infections.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institute of Health Research and Cystic Fibrosis Canada. E.B.M.B. was a recipient of a scholarship from CFC. R.E.W.H. holds a Canada Research Chair.
We thank Lucía Fernández for insightful discussions and editorial help and Reza Falsafi for technical assistance with the Western blot. We also thank the J. Craig Venter Institute for providing the microarray slides.
Footnotes
Published ahead of print 26 March 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Au N, et al. 2005. Genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 187:7655–7666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brazas MD, Breidenstein EBM, Overhage J, Hancock REW. 2007. Role of lon, an ATP-dependent protease homolog, in resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 51:4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brazas MD, Hancock REW. 2005. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butler SM, Festa RA, Pearce MJ, Darwin KH. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 60:553–562 [DOI] [PubMed] [Google Scholar]
- 6. Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 188:7101–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. 2006. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J. Biol. Chem. 281:4708–4717 [DOI] [PubMed] [Google Scholar]
- 9. Drees JC, Lusetti SL, Cox MM. 2004. Inhibition of RecA protein by the Escherichia coli RecX protein: modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 279:52991–52997 [DOI] [PubMed] [Google Scholar]
- 10. Drlica K, Zhao X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 [DOI] [PubMed] [Google Scholar]
- 12. Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 13. Gottesman S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465–506 [DOI] [PubMed] [Google Scholar]
- 14. Gottesman S, Maurizi MR. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huisman O, D'Ari R, Gottesman S. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc. Natl. Acad. Sci. U. S. A. 81:4490–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobs MA, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162–168 [DOI] [PubMed] [Google Scholar]
- 18. Kidambi SP, Booth MG, Kokjohn TA, Miller RV. 1996. recA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology 142:1033–1040 [DOI] [PubMed] [Google Scholar]
- 19. Kindrachuk KN, Fernandez L, Bains M, Hancock REW. 2011. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1874–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobiler O, Oppenheim AB, Herman C. 2004. Recruitment of host ATP-dependent proteases by bacteriophage lambda. J. Struct. Biol. 146:72–78 [DOI] [PubMed] [Google Scholar]
- 21. Kovach ME, et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 22. Lewenza S, et al. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Little JW, Mount DW, Yanisch-Perron CR. 1981. Purified LexA protein is a repressor of the recA and lexA genes. Proc. Natl. Acad. Sci. U. S. A. 78:4199–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marr AK, Overhage J, Bains M, Hancock REW. 2007. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153:474–482 [DOI] [PubMed] [Google Scholar]
- 26. Maurizi MR. 1987. Degradation in vitro of bacteriophage lambda N protein by Lon protease from Escherichia coli. J. Biol. Chem. 262:2696–2703 [PubMed] [Google Scholar]
- 27. McPhee JB, et al. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mizusawa S, Gottesman S. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc. Natl. Acad. Sci. U. S. A. 80:358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Overhage J, Lewenza S, Marr AK, Hancock REW. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 189:2164–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoemaker JM, Gayda RC, Markovitz A. 1984. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J. Bacteriol. 158:551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stewart BJ, Enos-Berlage JL, McCarter LL. 1997. The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 179:107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stohl EA, et al. 2003. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 278:2278–2285 [DOI] [PubMed] [Google Scholar]
- 33. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 34. Takaya A, Tabuchi F, Tsuchiya H, Isogai E, Yamamoto T. 2008. Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease. J. Bacteriol. 190:4181–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torres-Cabassa AS, Gottesman S. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umezu K, Kolodner RD. 1994. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 269:30005–30013 [PubMed] [Google Scholar]
- 37. Whistler CA, Stockwell VO, Loper JE. 2000. Lon protease influences antibiotic production and UV tolerance of Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 66:2718–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 39. Wu YL, Scott EM, Po AL, Tariq VN. 1999. Development of resistance and cross-resistance in Pseudomonas aeruginosa exposed to subinhibitory antibiotic concentrations. APMIS 107:585–592 [PubMed] [Google Scholar]
- 40. Yamaguchi Y, Tomoyasu T, Takaya A, Morioka M, Yamamoto T. 2003. Effects of disruption of heat shock genes on susceptibility of Escherichia coli to fluoroquinolones. BMC Microbiol. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.