Abstract
Bypass of classical penicillin-binding proteins by the l,d-transpeptidase of Enterococcus faecium (Ldtfm) leads to high-level ampicillin resistance in E. faecium mutants, whereas carbapenems remain the lone highly active β-lactams. Kinetics of Ldtfm inactivation was determined for four commercial carbapenems and a derivative obtained by introducing a minimal ethyl group at position 2. We show that the bulky side chains of commercial carbapenems have both positive and negative effects in preventing hydrolysis of the acyl enzyme and impairing drug binding.
TEXT
The last cross-linking step of peptidoglycan synthesis is catalyzed by high-molecular-weight penicillin-binding proteins (PBPs) with d,d-transpeptidase activity, which have historically been considered the lone essential targets of β-lactam antibiotics. However, we recently showed that carbapenems inactivate a second class of cross-linking enzymes (9), the l,d-transpeptidases, which bypass the classical d,d-transpeptidases by forming unusual peptidoglycan cross-links and confer high-level ampicillin resistance in mutants of Enterococcus faecium selected in vitro (10). Transpeptidases with d,d- and l,d-specificities are structurally unrelated and harbor different catalytic nucleophiles in the form of invariant Ser (14) and Cys (1, 7) residues. l,d-Transpeptidases are sporadically distributed in Gram-positive and Gram-negative bacteria and generally have nonessential roles in peptidoglycan synthesis (6, 11), except in Mycobacterium tuberculosis (4), Mycobacterium abscessus (5), and Clostridium difficile (13). Carbapenems are currently being investigated as therapeutic alternatives in the treatment of extensively drug-resistant tuberculosis (3, 8, 17), and l,d-transpeptidases are the likely targets of the drugs in M. tuberculosis (2, 4). Here, we investigated kinetics of inhibition of a model l,d-transpeptidase from E. faecium (Ldtfm) by different carbapenems in order to gain insight into the role of the antibiotic side chains in the efficiency of l,d-transpeptidase inactivation.
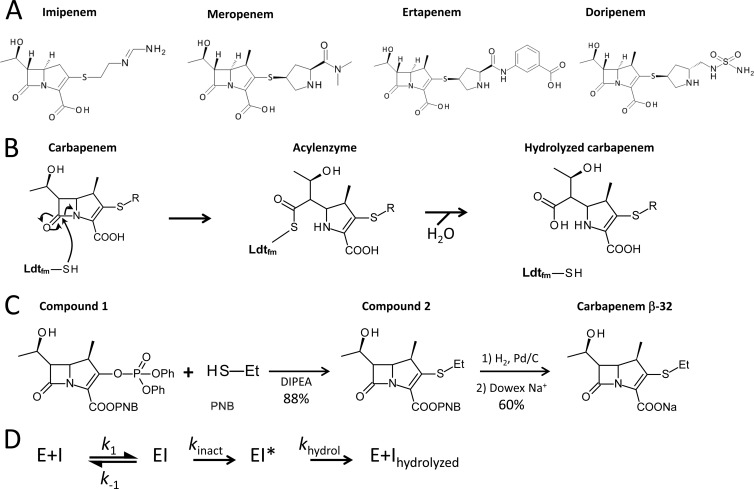
Four carbapenems—imipenem, meropenem, ertapenem, and doripenem (Fig. 1A)—are used to treat human infections in North America and Europe (12). These drugs differ by the nature of the side chain (R [Fig. 1B]) and the absence (imipenem) or presence (meropenem, doripenem, and ertapenem) of a methyl group (Fig. 1A) that has been introduced into the carbapenem ring to decrease inactivation by human renal dehydropeptidase I (12). To gain insight into the effects of structural variations in the carbapenem side chains on the efficiency of Ldtfm inactivation, we compared the four aforementioned drugs and synthesized an additional carbapenem, designated β-32, which contains a minimal ethyl side chain (Fig. 1C). Ldtfm was purified from recombinant Escherichia coli by affinity and size exclusion chromatographies (16). Kinetic constants of Ldtfm were determined by spectrofluorimetry and spectrophotometry (16) for the three steps of the reaction, which comprises noncovalent binding of the drug, acylation of the catalytic Cys residue, and hydrolysis of the corresponding acyl enzyme (Fig. 1D). Replacement of the meropenem side chain by an ethyl group (meropenem versus carbapenem β-32) led to a 15-fold increase in the second-order rate constant k1 for formation of the noncovalent complex (Fig. 2A). Thus, the bulky side chain of meropenem (Fig. 1A) impairs access of the drug to the active site. In contrast, the rate constant of the chemical step of the reaction (kinact) was similar for meropenem and β-32 (1.3 and 1.9 min−1, respectively). This observation indicates that the side chain of meropenem is nonessential for efficient acylation.
Fig 1.
(A) Structures of carbapenems. (B) Mechanism of inactivation of E. faecium l,d-transpeptidase (Ldtfm) by carbapenems. The sulfhydryl (SH) of the active-site cysteine of Ldtfm attacks the carbonyl of the β-lactam ring to form a covalent adduct (acyl enzyme). Hydrolysis of the resulting thioester bond leads to native enzyme and hydrolyzed carbapenem. R, carbapenem side chain. (C) Synthesis of carbapenem β-32. An addition-elimination reaction in N,N-diisopropylethylamine (DIPEA) afforded compound 2 from commercially available compound 1 with an 88% yield (15). Palladium-catalyzed (Pd/C) hydrogenation followed by ion exchange (Dowex resin) (15) afforded the sodium salt of carbapenem β-32 with a 60% yield. Et, ethyl; PNB, paranitrobenzoate. (D) Reaction scheme used in kinetic analyses. Binding of carbapenem (I) to Ldtfm (E) leads to reversible formation of a noncovalent complex (EI). Formation of the acyl enzyme (EI*) and its hydrolysis are irreversible. k1 and k−1 are second- and first-order constants for formation and dissociation of the noncovalent complex. kinact and khydrol are the rate constants for the acylation and hydrolysis reactions.
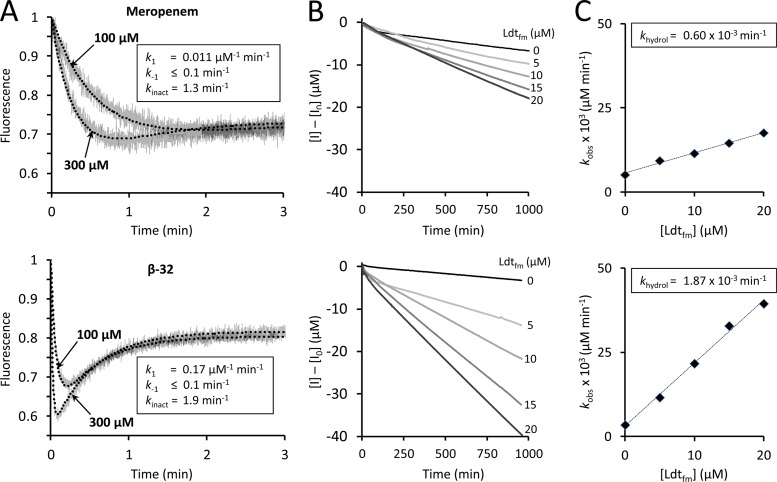
Fig 2.
Determination of kinetic constants of Ldtfm. (A) The kinetic constants k1, k−1, and kinact were determined by spectrofluorimetry, as previously described (16). Ldtfm (10 μM) was incubated with meropenem or carbapenem β-32 (100 and 300 μM) in sodium phosphate buffer (100 mM; pH 6.0) and fluorescence was recorded at 10°C on a Cary Eclipse spectrofluorimeter (Varian SA) equipped with a stopped-flow apparatus (RX-2000; Applied Photophysics). Trp residues were excited at 225 nm with a slit of 5 nm and an optical path length of 2 mm. Fluorescence emission was determined at 335 nm with a slit of 5 nm and an optical path length of 10 mm (gray lines). The detector voltage was set to 600 V. Fluorescence was normalized to the fluorescence intensity of free Ldtfm, which was the lone form of the enzyme at the onset of the reaction. Kinetic constants were determined by fitting simulations (dotted lines) to experimental data (gray lines), as previously described (16). Fitting was simultaneously performed with four drug concentrations (30, 100, 300, and 1,000 μM; results for only two concentrations are shown for clarity). (B) The kinetic constant khydrol was determined spectrophotometrically. Carbapenems (100 μM) were incubated with Ldtfm (0, 5, 10, 15, and 20 μM) in sodium phosphate buffer (100 mM; pH 6.0), and absorbance at 299 nm was recorded at 20°C on a Cary 100-Bio spectrophotometer (Varian SA). Alkaline hydrolysis of the β-lactam ring of meropenem and of β-32 led to similar decreases in the molar extinction coefficient (Δε = −7,200 and −7,400 M−1 cm−1, respectively). The velocity of the reaction (kobs) increased with Ldtfm concentration, indicating that the enzyme catalyzed carbapenem hydrolysis in addition to spontaneous drug hydrolysis. (C) Hydrolysis velocity (kobs) was plotted as a function of Ldtfm concentration, and enzyme turnover (khydrol) was deduced from the slope.
In order to compare the hydrolysis rates (khydrol) of the acyl enzymes, meropenem and β-32 were incubated with increasing concentrations of Ldtfm, and rupture of the β-lactam ring was monitored by spectrophotometry over 1,000 min (Fig. 2B). Under the conditions used, Ldtfm was totally converted to acyl enzyme in less than 3 min, and the subsequent decrease in absorbance revealed slow enzyme turnover (Fig. 2B). The hydrolysis rate (khydrol) was determined by subtracting the rate of spontaneous hydrolysis observed in the absence of enzyme. Replacing the meropenem side chain by an ethyl side chain led to a 3-fold increase in khydrol indicating that the meropenem side chain slows acyl enzyme hydrolysis. Together, these results show that the bulky side chain of meropenem has a negative effect on drug binding but a positive effect on acyl enzyme stability.
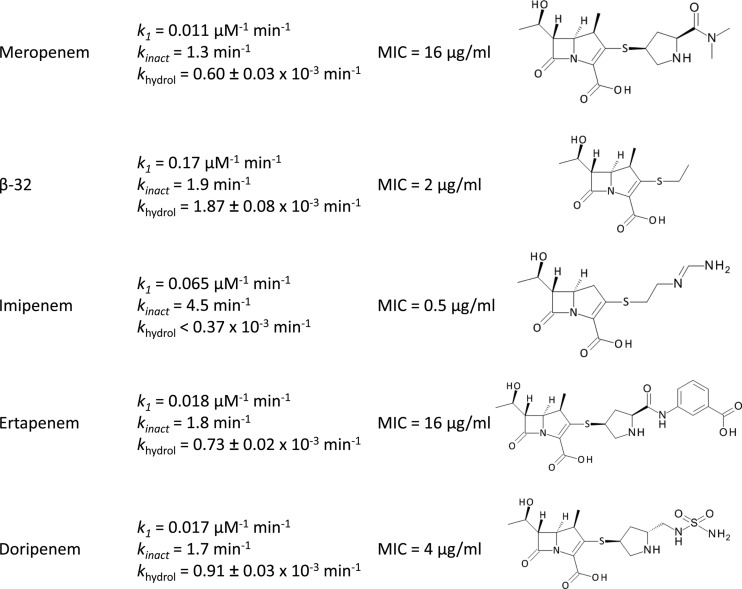
Since kinetic analyses of Ldtfm inactivation were previously performed only with imipenem (16), we determined kinetic constants for Ldtfm inactivation of commercially available carbapenems (Fig. 3). Imipenem was the most efficient Ldtfm inhibitor, with respect to both drug binding (k1 = 0.065 μM−1 min−1) and acylation (kinact = 4.5 min−1). The other carbapenems, meropenem, ertapenem, and doripenem, bound to Ldtfm less rapidly (k1 = 0.011 to 0.018 μM−1 min−1). The rates of acylation were also lower (kinact = 1.3 to 1.8 min−1). Thus, kinetics of Ldtfm inactivation by meropenem, ertapenem, and doripenem were very similar. This observation confirms that the carbapenem side chains do not contribute to efficient drug binding or acylation, in agreement with the aforementioned comparison of meropenem and β-32. The absence of a methyl group on the carbapenem ring of imipenem may account for the ca. 3-fold-higher Ldtfm acylation rate observed for this drug (kinact = 4.5 min−1) in comparison to rates for other carbapenems (kinact = 1.3 to 1.9 min−1). The rate constant for binding of imipenem (k1 = 0.065 μM−1 min−1) was intermediate between that of β-32 (k1 = 0.17 μM−1 min−1) and those of meropenem, ertapenem, and doripenem (k1 = 0.011 to 0.018 μM−1 min−1). The rate constant k1 therefore appears to be inversely correlated with the size of the side chain (Fig. 1A).
Fig 3.
Inactivation of Ldtfm by various carbapenems. Kinetic constants were determined by spectrofluorimetry (k1 and kinact) and spectrophotometry (khydrol). MICs of carbapenems for E. faecium M512 were determined at 37°C in brain heart infusion broth (Difco) containing 200 μg/ml of ampicillin to inactivate the d,d-transpeptidases (9).
Comparison of acyl enzyme stability (Fig. 3) indicated that the rate constant for hydrolysis was 2- to 3-fold higher for carbapenem β-32 (1.87 × 10−3 min−1) than for ertapenem, doripenem, and meropenem (0.60 to 0.91 × 10−3 min−1). These results confirmed that the bulky side chain of carbapenems stabilizes the acyl enzyme. Hydrolysis of imipenem by Ldtfm was undetectable (<0.37 × 10−3 min−1).
In order to compare the antibacterial activities of carbapenems, MICs were determined for E. faecium M512, which is resistant to ampicillin due to activation of the l,d-transpeptidation pathway. M512, formerly designated D344M512 (10), was obtained by serial selection on media containing increasing concentrations of ampicillin and derives from hypersusceptible E. faecium D344S, which does not harbor the gene encoding low-affinity PBP5. MICs of carbapenems were determined in the presence of ampicillin (200 μg/ml), a drug concentration which was chosen to fully inactivate the d,d-transpeptidation pathway (9). Under such conditions, M512 relies exclusively on Ldtfm for peptidoglycan cross-linking, thereby providing an estimate of in vivo enzyme inhibition (9). The lowest MIC (0.5 μg/ml) was observed for imipenem that combines relatively high k1 and kinact. For β-32, the large increase in k1 was not associated with the expected decrease of the MIC, indicating, as expected, that acyl enzyme stability and efficient drug binding are both important for antibacterial activity of carbapenems. Together, these data show that the bulky side chains of carbapenems have both positive and negative effects in preventing hydrolysis of the acyl enzyme and impairing drug binding.
ACKNOWLEDGMENTS
The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 261378, and the National Institute of Allergy and Infectious Diseases (grant RO1 AI046626).
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Biarrotte-Sorin S, et al. 2006. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 359:533–538 [DOI] [PubMed] [Google Scholar]
- 2. Gupta R, et al. 2010. The Mycobacterium tuberculosis protein Ldt(Mt2) is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16:466–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, III, Blanchard JS. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lavollay M, et al. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 190:4360–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lavollay M, et al. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J. Bacteriol. 193:778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. 2008. Identification of the l,d-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190:4782–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mainardi JL, et al. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280:38146–38152 [DOI] [PubMed] [Google Scholar]
- 8. Mainardi JL, Hugonnet JE, Gutmann L, Arthur M. 2011. Fighting resistant tuberculosis with old compounds: the carbapenem paradigm. Clin. Microbiol. Infect. 17:1755–1756 [DOI] [PubMed] [Google Scholar]
- 9. Mainardi JL, et al. 2007. Unexpected inhibition of peptidoglycan LD-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J. Biol. Chem. 282:30414–30422 [DOI] [PubMed] [Google Scholar]
- 10. Mainardi JL, et al. 2000. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J. Biol. Chem. 275:16490–16496 [DOI] [PubMed] [Google Scholar]
- 11. Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32:386–408 [DOI] [PubMed] [Google Scholar]
- 12. Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 55:4943–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peltier J, et al. 2011. Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3-3 cross-links. J. Biol. Chem. 286:29053–29062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 15. Tewari N, et al. 2007. An improved procedure for preparation of carbapenem antibiotic: meropenem. Org. Process Res. Dev. 11:773–775 [Google Scholar]
- 16. Triboulet S, et al. 2011. Inactivation kinetics of a new target of beta-lactam antibiotics. J. Biol. Chem. 286:22777–22784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Veziris N, Truffot C, Mainardi JL, Jarlier V. 2011. Activity of carbapenems combined with clavulanate against murine tuberculosis. Antimicrob. Agents Chemother. 55:2597–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]