Abstract
A total of 316 toxigenic Clostridium difficile clinical isolates of known PCR ribotypes from patients in North America were screened for resistance to clindamycin, metronidazole, moxifloxacin, and rifampin. Clindamycin resistance was observed among 16 different ribotypes, with ribotypes 017, 053, and 078 showing the highest proportions of resistance. All isolates were susceptible to metronidazole. Moxifloxacin resistance was present in >90% of PCR-ribotype 027 and 053 isolates but was less common among other ribotypes. Only 7.9% of the C. difficile isolates were resistant to rifampin. Multidrug resistance (clindamycin, moxifloxacin, and rifampin) was present in 27.5% of PCR-ribotype 027 strains but was rare in other ribotypes. These results suggest that antimicrobial resistance in North American isolates of C. difficile varies by strain type and parallels rates of resistance reported from Europe and the Far East.
INTRODUCTION
Clostridium difficile is a major health care-associated pathogen that is responsible for a wide spectrum of disease, ranging from mild diarrhea to life-threatening complications, such as pseudomembranous colitis and toxic megacolon (19). The severity and outcome of C. difficile infection (CDI) is influenced by a multiplicity of factors, including patient demographics, such as age and immune status, length of hospitalization, and, most of all, receipt of antimicrobial therapy (23, 25). Antimicrobial therapy, often given for treatment of other infectious diseases, can render the patient susceptible to CDI if the patient is exposed to a toxigenic strain of the organism. When CDI was first reported in the 1970s, prior use of clindamycin was established as a significant risk factor. However, by 1980, cephalosporins replaced clindamycin as the major risk factor (4). In the next 20 years, expanded- and extended-spectrum cephalosporins became associated with a high risk of CDI (6). More recently, fluoroquinolones have been linked to CDI and to severe epidemics, particularly those caused by PCR-ribotype 027 (27). Between 5% and 30% of patients receiving antimicrobial agents may develop health care-associated diarrhea, with C. difficile causing up to 30% of those cases (22). Antimicrobial stewardship programs can assist in curtailing these selective pressures (17, 18). The emergence of C. difficile strains that are resistant to multiple antimicrobial agents can complicate prevention programs and potentially, in the case of metronidazole, treatment (13). Thus, knowing the prevalence of antimicrobial-resistant strains in an institution can be helpful for optimizing antimicrobial stewardship programs.
(These data were presented in part at the 111th General Meeting of the American Society for Microbiology, 21 to 24 May 2011, New Orleans, LA.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 316 toxigenic clinical isolates of C. difficile were received from 7 hospitals in the United States (including hospitals located in California, Illinois, Indiana, North Carolina, and Washington) and Canada (Quebec) during 2008 and 2009. These isolates have been described elsewhere (32). Organisms were isolated using broth-enrichment toxigenic culture and typed by PCR-ribotyping (PCR-R), pulsed-field gel electrophoresis (PFGE), and restriction endonuclease analysis of whole-cell DNA (REA). The isolates were preserved at −80°C in 15% glycerol-Brucella broth and were subcultured twice on prereduced anaerobically sterilized (PRAS) Brucella blood agar (Anaerobe Systems, Morgan Hill, CA) prior to antimicrobial susceptibility testing.
Antimicrobial susceptibility testing.
Isolates were tested for susceptibility to clindamycin, metronidazole, moxifloxacin, and rifampin using Etest strips (bioMérieux, Marcy-l'Étoile, France), as described in the Etest technical guide. The MIC results were rounded up to the next doubling dilution and interpreted using Clinical and Laboratory Standards Institute (CLSI) breakpoints for susceptibility testing of anaerobic bacteria (8). Individual colonies from 24- to 48-h Brucella blood agar plates were suspended in Brucella broth to the turbidity of 1.0 McFarland standard, and the inoculum was applied to 150-mm Brucella blood agar plates (Anaerobe Systems, Morgan Hill, CA). Plates were incubated anaerobically at 35°C in a Bactron anaerobic chamber (Sheldon Manufacturing Inc., Cornelius, OR). MICs were read and recorded at 24 h, and clindamycin results were confirmed after 48 h of incubation to ensure detection of inducible resistance (as per the Etest package insert). Bacteroides fragilis ATCC 25285, Bacteroides thetaiotaomicron ATCC 29741, and Eubacterium lentum ATCC 43055 were used for quality control (9). All control results were within acceptable ranges.
RESULTS
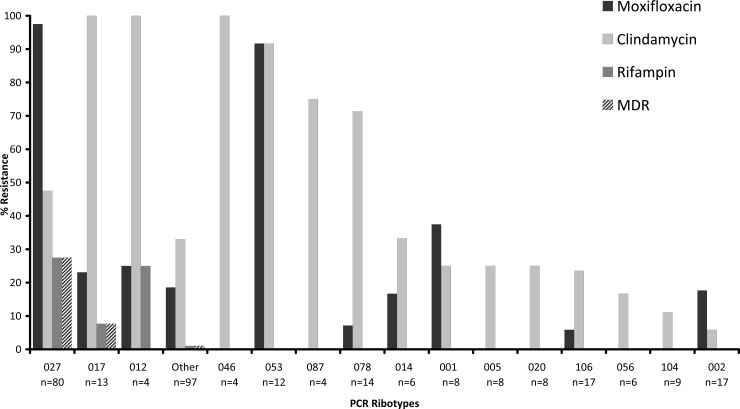
The MIC50 and MIC90 results for clindamycin, metronidazole, moxifloxacin, and rifampin when tested against 316 C. difficile strains are presented in Table 1. Clindamycin resistance was present in 41.5% of isolates, while moxifloxacin resistance was present in 38.0% of isolates; only 7.9% of isolates were resistant to rifampin. All isolates were susceptible to metronidazole. There were no inner colonies suggestive of heteroresistance within the zones of inhibition around the metronidazole Etest strips. Table 2 shows the MIC data for the seven most common PCR ribotypes observed among the study isolates. Moxifloxacin resistance was present in >90% of PCR-ribotype 027 and 053 isolates but was less common among other ribotypes (Fig. 1). Clindamycin resistance was observed among several ribotypes, with ribotypes 017, 053, and 078 showing the highest proportions of resistance (100%, 91.7%, and 71.4%, respectively). Rifampin resistance (MIC ≥ 32 μg/ml) was observed in 27.5% of ribotype 027 isolates and sporadically among ribotypes 017 and 012 isolates (Fig. 1). Overall, 27.5% of ribotype 027 isolates were resistant to clindamycin, moxifloxacin, and rifampin. This multidrug resistance pattern was also observed among several isolates of ribotype 017.
Table 1.
MIC range, MIC50, and MIC90 for antimicrobial agents tested against 316 C. difficile isolates
| Drug | MIC (μg/ml) |
No. resistant | % resistant | ||
|---|---|---|---|---|---|
| Range | 50% | 90% | |||
| Clindamycin | 0.25–>256 | 4 | >256 | 131 | 41.50 |
| Metronidazole | 0.125–4 | 0.25 | 0.5 | 0 | 0 |
| Rifampin | ≤0.002–>32 | ≤0.002 | ≤0.002 | 25 | 7.90 |
| Moxifloxacin | 0.5–>32 | 2 | >32 | 120 | 38.00 |
Table 2.
MIC range, MIC50, and MIC90 among isolates of the seven most frequent PCR ribotypes
| PCR ribotype (no. of isolates) | Measureb | Resulta |
|||
|---|---|---|---|---|---|
| Clindamycin | Metronidazole | Moxifloxacin | Rifampin | ||
| 002 (17) | MIC50 | 4 | 0.25 | 2 | ≤0.002 |
| MIC90 | 8 | 0.5 | >32 | ≤0.002 | |
| No. R | 1 | 0 | 3 | 0 | |
| % R | 5.9 | 0 | 17.7 | 0 | |
| 017 (13) | MIC50 | >256 | 0.5 | 2 | ≤0.002 |
| MIC90 | >256 | 0.5 | >32 | 0.004 | |
| No. R | 13 | 0 | 3 | 1 | |
| % R | 100 | 0 | 23.1 | 7.7 | |
| 027 (80) | MIC50 | 4 | 0.5 | >32 | ≤0.002 |
| MIC90 | >256 | 1 | >32 | >32 | |
| No. R | 38 | 0 | 78 | 22 | |
| % R | 47.5 | 0 | 97.5 | 27.5 | |
| 053 (12) | MIC50 | >256 | 0.5 | >32 | ≤0.002 |
| MIC90 | >256 | 0.5 | >32 | ≤0.002 | |
| No. R | 11 | 0 | 11 | 0 | |
| % R | 91.7 | 0 | 91.7 | 0 | |
| 078 (14) | MIC50 | 8 | 0.25 | 1 | ≤0.002 |
| MIC90 | >256 | 0.5 | 2 | 0.003 | |
| No. R | 10 | 0 | 1 | 0 | |
| % R | 71.4 | 0 | 7.1 | 0 | |
| 104 (9) | MIC50 | 6 | 0.5 | 2 | ≤0.002 |
| MIC90 | 6 | 0.50 | 2 | ≤0.002 | |
| No. R | 1 | 0 | 0 | 0 | |
| % R | 11.1 | 0 | 0 | 0 | |
| 106 (17) | MIC50 | 4 | 0.50 | 2 | 0.003 |
| MIC90 | 8 | 0.50 | 2 | 0.003 | |
| No. R | 4 | 0 | 1 | 0 | |
| % R | 23.5 | 0 | 5.8 | 0 | |
MIC results are in μg/ml.
No. R and % R are the number and percentage of resistant isolates, respectively.
Fig 1.
Proportions of antimicrobial resistance among 307 isolates of C. difficile according to their PCR ribotypes. The graph excludes a total of nine isolates of PCR ribotypes 003 and 075 for which no resistance was observed.
A comparison of the proportions of resistance observed in this study and those reported in 15 other studies of C. difficile resistance is shown in Table 3. Moxifloxacin resistance ranged from 2 to 87%. A report of 82% resistance was from a single site, where 69% of isolates were pulsed-field gel electrophoresis types NAP1 or NAP2 (7). Clindamycin resistance ranged from 15 to 97% (the latter in a study of 1,613 isolates from Scotland). Rifampin resistance was reported infrequently, and only a single metronidazole-resistant isolate was reported in these other studies.
Table 3.
Published reports of antimicrobial resistance rates in C. difficilea
| Yr isolated, location | No. | % resistant to: |
AST method(s) used | Most common ribotypes | % that were PCR ribotype: |
Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mox | Clinda | Met | Rif | 027 | 078 | 017 | |||||
| 2008–2009, U.S. and Canada | 316 | 38 | 41 | 0 | 8 | Etest | 027, 106, 002 | 25 | 4 | 4 | This study |
| 2001–2009, Taiwan | 113 | 16 | 46 | 0 | Agar dilution | NA | 0 | 0 | 21 | ||
| 2002–2004, Germany | 317 | 40 | 65 | 0 | Etest | NA | 30 | ||||
| 2004, Quebec, Canada | 258 | 82 | 15 | 0 | 0 | Agar dilution | NAb | 69 | 7 | ||
| 2004–2006, Poland (hospital 1) | 153 | 39 | 54 | 0 | Etest | NA | 28 | ||||
| 2004–2006, Poland (hospital 2) | 177 | 38 | 48 | 0 | Etest | NA | 28 | ||||
| 2005, 14 EU countries | 349 | 38 | 46 | 0 | Etest | 001, 014, 027 | 6 | ∼2 | 5 | 3 | |
| 2005, Scotland | 116 | 87 | 63 | 0 | Agar dilution | 001, 106 | 0 | 0 | 0 | 24 | |
| 2006–2007, Austria | 142 | 38 | 57 | 1c | Etest | AI-5,d 014, 053 | 1 | 0 | 16 | ||
| 2007–2009, Scotland | 1613 | 64 | 97 | 0 | Agar dilution | 106, 001, 027 | 13 | 3 | 0 | 33 | |
| 2008, Sweden | 585 | 20 | 16 | 0 | Disk diffusion and Etest | 012, SE37,d 017 | <1 | ∼4 | 1 | ||
| 2009, Sweden | 364 | 16 | 16 | 0 | Disk diffusion and Etest | SE21,d 001, 020 | 0 | ∼5 | 1 | ||
| 2009, Ireland | 133 | 57 | 22 | Etest | 027, 001, 106 | 19 | 11 | 31 | |||
| 2009, New Zealand | 101 | 2 | 61 | 0 | Agar dilution | 014, 002, 005 | 0 | 1 | 29 | ||
| 2009, Shanghai, China | 75 | 45 | 85 | 0 | 17 | Agar dilution | 017, 012, A | 0 | 0 | 19 | 14 |
| 2009, Stockholm, Sweden | 80 | 15 | 65 | 0 | 4 | Agar dilution | 005, 014, 023 | 0 | 0 | 14 | |
Mox, moxifloxacin; Clinda, clindamycin; Met, metronidazole; Rif, rifampin; AST, antimicrobial susceptibility testing method; NA, not available.
These are primarily isolates of pulsed-field types NAP1 and NAP2.
Isolate reverted to metronidazole susceptibility after storage.
AI-5, SE21, and SE37 are locally defined strain types.
DISCUSSION
Multidrug resistance (i.e., resistance to clindamycin, moxifloxacin, and rifampin) was present in 22 of 80 (27.5%) C. difficile PCR-ribotype 027 isolates from the United States and Canada but was unusual among other ribotypes (Fig. 1). All rifampin-resistant strains were also resistant to clindamycin and moxifloxacin. This is in contrast to the report of Curry et al. (12), in which 81.5% of ribotype 027 isolates from a hospital in Pittsburgh were resistant to rifampin. The range of antibiograms observed in our study indicates that not all ribotype 027 isolates, which were obtained from seven laboratories across the United States and Canada, are clonal. This is consistent with the data presented by Killgore et al. (20), which indicated that subtypes could be defined within ribotype 027 isolates by other typing techniques. Unfortunately, rifampin resistance is rarely reported in other resistance surveys of C. difficile isolates, limiting comparisons to other data sets (Table 3). Although clindamycin and moxifloxacin resistance were both relatively common among our isolates (41.5% and 38.0% of isolates, respectively), clindamycin resistance was observed in all but two of the 16 ribotypes in our study (where there were at least 4 isolates of that ribotype tested), while moxifloxacin resistance was limited to 10 strain types. Yet, only in ribotype 053 isolates did the two resistances appear tightly linked (i.e., both were present in 11/12 isolates). The ribotype 053 strains came from four different laboratories located in California, Indiana, Illinois, and North Carolina and thus did not appear to be associated with a clonal outbreak of CDI. Overall, though, no resistance pattern was consistent enough to be a useful strain marker.
A review of antimicrobial resistance among C. difficile isolates by Huang et al. in 2009 (15) did not contain data specifically on ribotype 053 strains but did confirm high rates of clindamycin resistance in a variety of ribotypes, including ribotype 017 isolates from Europe (there were no data on ribotypes 012 or 046 reported). Among other surveys of C. difficile resistance not covered by Huang et al. (i.e., those cited in Table 3), only 7 of 15 reports showed higher rates of moxifloxacin resistance than was observed in our study, but 11 of 15 showed higher rates of clindamycin resistance. PCR ribotype 001 was the most common ribotype reported in these surveys, followed by ribotypes 014, 027, and 106. Modest levels of resistance were seen among our isolates of these ribotypes as well.
Resistance to the antimicrobial agents most commonly used to treat C. difficile infections, i.e., vancomycin and metronidazole (5, 10), is reported rarely in the literature (15). However, both metronidazole heteroresistance (26) and reduced susceptibility to metronidazole have been reported (2). We did not observe either of these phenomena in our study, but this may have been due to the limited number of medical centers that contributed isolates to this study.
In summary, while resistance to clindamycin and moxifloxacin is widespread in C. difficile isolates from North America, multidrug resistance (i.e., resistance to clindamycin, moxifloxacin, and rifampin) was limited primarily to ribotype 027 isolates. Although these antimicrobial agents are not used for therapy (with the possible exception of rifaximin, which is in an antimicrobial agent class related to rifampin), they may still play a key role in enabling patients taking these antimicrobial agents to become colonized and infected with C. difficile if exposed to these resistant strains (11).
ACKNOWLEDGMENTS
We thank Ellen Jo Baron and Diane Citron for helpful comments.
F.C.T., I.A.T., and D.H.P. are employees and shareholders of Cepheid. Funding for this study was provided by Cepheid.
Footnotes
Published ahead of print 12 March 2012
REFERENCES
- 1. Akerlund T, et al. 2011. Geographical clustering of cases of infection with moxifloxacin-resistant Clostridium difficile PCR-ribotypes 012, 017 and 046 in Sweden, 2008 and 2009. Euro Surveill. 16:pii=19813 [DOI] [PubMed] [Google Scholar]
- 2. Baines SD, et al. 2008. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J. Antimicrob. Chemother. 62:1046–1052 [DOI] [PubMed] [Google Scholar]
- 3. Barbut F, et al. 2007. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin. Microbiol. Infect. 13:1048–1057 [DOI] [PubMed] [Google Scholar]
- 4. Bartlett JG, Taylor NS, Chang T, Dzink J. 1980. Clinical and laboratory observations in Clostridium difficile colitis. Am. J. Clin. Nutr. 33:2521–2526 [DOI] [PubMed] [Google Scholar]
- 5. Bauer MP, Kuijper EJ, van Dissel JT. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI). Clin. Microbiol. Infect. 15:1067–1079 [DOI] [PubMed] [Google Scholar]
- 6. Bignardi GE. 1998. Risk factors for Clostridium difficile infection. J. Hosp. Infect. 40:1–15 [DOI] [PubMed] [Google Scholar]
- 7. Bourgault AM, Lamothe F, Loo VG, Poirier L. 2006. In vitro susceptibility of Clostridium difficile clinical isolates from a multi-institutional outbreak in Southern Quebec, Canada. Antimicrob. Agents Chemother. 50:3473–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing of anaerobic bacteria; informational supplement. CLSI document M11-S1. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Cohen SH, et al. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455 [DOI] [PubMed] [Google Scholar]
- 11. Coia JE. 2009. What is the role of antimicrobial resistance in the new epidemic of Clostridium difficile? Int. J. Antimicrob. Agents 33(Suppl 1):S9–S12 [DOI] [PubMed] [Google Scholar]
- 12. Curry SR, et al. 2009. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin. Infect. Dis. 48:425–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deneve C, Janoir C, Poilane I, Fantinato C, Collignon A. 2009. New trends in Clostridium difficile virulence and pathogenesis. Int. J. Antimicrob. Agents 33(Suppl 1):S24–S28 [DOI] [PubMed] [Google Scholar]
- 14. Huang H, Fang H, Weintraub A, Nord CE. 2009. Distinct ribotypes and rates of antimicrobial drug resistance in Clostridium difficile from Shanghai and Stockholm. Clin. Microbiol. Infect. 15:1170–1173 [DOI] [PubMed] [Google Scholar]
- 15. Huang H, Weintraub A, Fang H, Nord CE. 2009. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 34:516–522 [DOI] [PubMed] [Google Scholar]
- 16. Indra A, et al. 2008. Characterization of clinical Clostridium difficile isolates by PCR ribotyping and detection of toxin genes in Austria, 2006–2007. J. Med. Microbiol. 57:702–708 [DOI] [PubMed] [Google Scholar]
- 17. Jarvis WR, Schlosser J, Jarvis AA, Chinn RY. 2009. National point prevalence of Clostridium difficile in US health care facility inpatients, 2008. Am. J. Infect. Control 37:263–270 [DOI] [PubMed] [Google Scholar]
- 18. Kallen AJ, et al. 2009. Complete restriction of fluoroquinolone use to control an outbreak of Clostridium difficile infection at a community hospital. Infect. Control Hosp. Epidemiol. 30:264–272 [DOI] [PubMed] [Google Scholar]
- 19. Kelly CP, LaMont JT. 2008. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 359:1932–1940 [DOI] [PubMed] [Google Scholar]
- 20. Killgore G, et al. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin YC, et al. 2011. Antimicrobial susceptibilities and molecular epidemiology of clinical isolates of Clostridium difficile in Taiwan. Antimicrob. Agents Chemother. 55:1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McFarland LV. 2007. Diarrhoea associated with antibiotic use. BMJ 335:54–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mullane KM, et al. 2011. Efficacy of fidaxomicin versus vancomycin as therapy for Clostridium difficile infection in individuals taking concomitant antibiotics for other concurrent infections. Clin. Infect. Dis. 53:440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mutlu E, Wroe AJ, Sanchez-Hurtado K, Brazier JS, Poxton IR. 2007. Molecular characterization and antimicrobial susceptibility patterns of Clostridium difficile strains isolated from hospitals in south-east Scotland. J. Med. Microbiol. 56:921–929 [DOI] [PubMed] [Google Scholar]
- 25. Owens RC. 2007. Clostridium difficile-associated disease: changing epidemiology and implications for management. Drugs 67:487–502 [DOI] [PubMed] [Google Scholar]
- 26. Pelaez T, et al. 2008. Metronidazole resistance in Clostridium difficile is heterogeneous. J. Clin. Microbiol. 46:3028–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pepin J, et al. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41:1254–1260 [DOI] [PubMed] [Google Scholar]
- 28. Pituch H, et al. 2011. Characterization and antimicrobial susceptibility of Clostridium difficile strains isolated from adult patients with diarrhoea hospitalized in two university hospitals in Poland, 2004–2006. J. Med. Microbiol. 60:1200–1205 [DOI] [PubMed] [Google Scholar]
- 29. Roberts S, et al. 2009. Molecular epidemiology and susceptibility profiles of Clostridium difficile in New Zealand. N. Z. Med. J. 124:45–51 [PubMed] [Google Scholar]
- 30. Schmidt C, Loffler B, Ackermann G. 2007. Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn. Microbiol. Infect. Dis. 59:1–5 [DOI] [PubMed] [Google Scholar]
- 31. Solomon K, et al. 2011. PCR ribotype prevalence and molecular basis of macrolide-lincosamide-streptogramin B (MLSB) and fluoroquinolone resistance in Irish clinical Clostridium difficile isolates. J. Antimicrob. Chemother. 66:1976–1982 [DOI] [PubMed] [Google Scholar]
- 32. Tenover FC, et al. 2010. Impact of strain types on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48:3719–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wiuff C, et al. 2011. The epidemiology of Clostridium difficile in Scotland. J. Infect. 62:271–279 [DOI] [PubMed] [Google Scholar]