Abstract
The role of antibiotics in treatment of enterohemorrhagic Escherichia coli (EHEC) infections is controversial because of concerns about triggering hemolytic-uremic syndrome (HUS) by increasing Shiga toxin (Stx) production. During the recent large EHEC O104:H4 outbreak, antibiotic therapy was indicated for some patients. We tested a diverse panel of antibiotics to which the outbreak strain is susceptible to interrogate the effects of subinhibitory antibiotic concentrations on induction of stx2-harboring bacteriophages, stx2 transcription, and Stx2 production in this emerging pathogen. Ciprofloxacin significantly increased stx2-harboring phage induction and Stx2 production in outbreak isolates (P values of <0.001 to <0.05), while fosfomycin, gentamicin, and kanamycin insignificantly influenced them (P > 0.1) and chloramphenicol, meropenem, azithromycin, rifaximin, and tigecycline significantly decreased them (P ≤ 0.05). Ciprofloxacin and chloramphenicol significantly upregulated and downregulated stx2 transcription, respectively (P < 0.01); the other antibiotics had insignificant effects (P > 0.1). Meropenem, azithromycin, and rifaximin, which were used for necessary therapeutic or prophylactic interventions during the EHEC O104:H4 outbreak, as well as tigecycline, neither induced stx2-harboring phages nor increased stx2 transcription or Stx2 production in the outbreak strain. These antibiotics might represent therapeutic options for patients with EHEC O104:H4 infection if antibiotic treatment is inevitable. We await further analysis of the epidemic to determine if usage of these agents was associated with an altered risk of developing HUS.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) causes diarrhea, bloody diarrhea, and hemolytic-uremic syndrome (HUS), mostly in children (21, 37). The most common EHEC serotype causing human diseases worldwide is O157:H7 (21, 37). In May to July 2011, Germany was afflicted by a massive outbreak caused by a rare EHEC serotype, O104:H4 (6, 13, 25). This outbreak involved >3,800 cases, including 855 patients with HUS and 53 deaths (33), and was the largest and most devastating outbreak of HUS in recorded history. The most likely outbreak vehicle was contaminated sprouts (6). Unusual epidemiological, clinical, and microbiological features of this outbreak were the predominance of adults and women (13, 33), an unprecedented proportion of HUS cases (∼22%) (13, 33), severe neurological symptoms (epileptic seizures, pareses, delirium, and coma) in a subset of HUS patients (18), and the hybrid nature of the outbreak strain, which had combined virulence characteristics of EHEC and enteroaggregative E. coli (EAEC) (2, 26, 32). The outbreak strain displays an extended-spectrum beta-lactamase phenotype (2) from its CTX-M-15 genotype (32), i.e., it is resistant to all penicillins and cephalosporins and susceptible to carbapenems (ertapenem, imipenem, and meropenem) (2). It is also resistant to trimethoprim-sulfamethoxazole and susceptible to fluoroquinolones (ciprofloxacin) and aminoglycosides (gentamicin and tobramycin) (2).
Antibiotic therapy is generally not recommended for EHEC infections (17, 36, 37, 38) because of no benefit (30, 37), or even harm, in particular an increased risk of HUS development in patients treated with antibiotics during the initial period of diarrhea (1, 10, 36, 37, 38). One plausible mechanism by which antibiotics increase the risk of HUS development is that they increase the production and/or release of Shiga toxin (Stx), the major virulence factor of EHEC involved in the pathogenesis of HUS (21, 37). This occurs via induction of prophages harboring Stx-encoding genes (stx) (27, 40), which enhances stx transcription, Stx production, and toxin release from the bacterial cells via phage-mediated lysis (22, 27, 40). However, the effects of different antibiotic classes on Stx production in vitro differ and are also dependent on the antibiotic concentration and the nature of the EHEC strain (e.g., O group or Stx type) (16, 22, 24, 28, 29, 39). In studies performed with EHEC O157:H7, fluoroquinolones, trimethoprim-sulfamethoxazole, and ampicillin significantly increased Stx2 production (16, 20, 22, 23, 24, 29, 39, 40), whereas macrolides (24, 29, 39), carbapenems (22, 23), aminoglycosides (22, 24), rifampin (31), rifaximin (28), and fosfomycin (22, 24, 40) either had no effect on Stx2 production or suppressed it. These in vitro data are in accordance with experiments with animal models (31, 39, 40).
During the German EHEC O104:H4 outbreak, therapeutic or prophylactic administration of antibiotics was necessary for a subset of patients. According to the recommendations of the German Society for Infectious Diseases (15), carbapenems were used for treatment of invasive complications caused by the outbreak strain or by superinfecting microorganisms, azithromycin was used for eradication of nasopharyngeal meningococcal colonization before eculizumab therapy, and rifaximin, an oral nonabsorbable drug, was used for intestinal eradication of EHEC O104 in persistently colonized individuals and for other situations without indications for systemic antimicrobial therapy. Because the effects of antibiotics on Stx production in the EHEC O104:H4 outbreak strain are unknown, we investigated a diverse panel of antibiotics for their effects on induction of stx2-harboring phages and, correspondingly, their ability to affect stx2 transcription and Stx2 production in this emerging highly virulent pathogen.
MATERIALS AND METHODS
Bacterial strains.
EHEC O104:H4 outbreak isolates studied for antibiotic-mediated induction of stx2-carrying phages, stx2 transcription, and Stx2 production were LB226692, first characterized and sequenced in our laboratory (2, 26), and an additional HUS isolate, LB226806 (2). Both isolates contain stx2 in combination with typical EAEC loci (aggA, aggR, set1, pic, aap, and aatA) (2). EHEC O157:H7 strain EDL933 (39) was used for comparison. Twenty additional outbreak isolates tested for susceptibility to the investigated antibiotics were recovered from patients with HUS (n = 15) or bloody diarrhea without HUS (n = 5) in different parts of Germany.
Antibiotics and antimicrobial susceptibility testing.
Ciprofloxacin, meropenem, fosfomycin (all from Sigma-Aldrich, Taufkirchen, Germany), chloramphenicol, gentamicin, and kanamycin (all from AppliChem, Darmstadt, Germany) were purchased from commercial suppliers; azithromycin, tigecycline (both from Pfizer, Groton, CT), and rifaximin (Alfa Wassermann, Bologna, Italy) were provided by their manufacturers. MICs were determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (7). For fosfomycin, for which the broth microdilution method is not approved (7), the MIC was determined using the agar dilution method (7) on Mueller-Hinton agar supplemented with 25 μg/ml of glucose-6-phosphate. The MICs (Table 1) were used to calculate subinhibitory concentrations in the range of 1/2 to 1/64 MIC.
Table 1.
MICs of nine antibiotics for EHEC strains investigated for antibiotic-mediated effects on stx2-harboring phage induction, stx2 transcription, and Stx2 production
| Antibiotic | MIC (μg/ml) |
||
|---|---|---|---|
| LB226692 (O104:H4) | LB226806 (O104:H4) | EDL933 (O157:H7) | |
| Ciprofloxacin | 0.25 | 0.25 | 0.06 |
| Meropenem | 0.016 | 0.016 | 0.016 |
| Azithromycin | 8.0 | 8.0 | 4.0 |
| Rifaximin | 128.0 | 128.0 | 64.0 |
| Tigecycline | 0.25 | 0.25 | 0.125 |
| Fosfomycin | 1.0 | 1.0 | 2.0 |
| Chloramphenicol | 8.0 | 8.0 | 8.0 |
| Gentamicin | 0.125 | 0.125 | 0.125 |
| Kanamycin | 0.5 | 0.5 | 0.25 |
Antibiotic induction.
In the initial experiment, the strains were grown (37°C, 180 rpm) in 150 ml of Luria-Bertani (LB) broth containing 5 mM CaCl2 until reaching an optical density at 600 nm (OD600) of 0.5. Each culture was divided into 10-ml aliquots, which were supplemented with 1/4 MIC of each of the nine antibiotics (Table 1) (and with 25 μg/ml of glucose-6-phosphate for fosfomycin) or with no antibiotic (control) and then grown (37°C, 180 rpm) for 15 h. After centrifugation, filtered (0.2-μm membrane filter; Corning, Lowell, MA) culture supernatants were used in phage and toxin assays. Subsequently, six antibiotics (ciprofloxacin, meropenem, azithromycin, rifaximin, tigecycline, and chloramphenicol) were also tested in dilution series of subinhibitory concentrations (1/2 to 1/16 MIC) to investigate concentration-dependent effects. Rifaximin, for which a broad range of MICs has been reported (19, 34), was also tested at 1/32 and 1/64 MIC.
Detection of stx2-carrying phages, stx2 transcription, and Stx2 production.
The presence of stx2-harboring bacteriophages in 10-fold serial dilutions of culture filtrates was determined using a plaque assay (35) followed by plaque hybridization with a digoxigenin-11-dUTP-labeled stx2 probe (DIG DNA labeling and detection kit; Roche Diagnostics, Mannheim, Germany). stx2-carrying phage titers were expressed as numbers of stx2 PFU per ml.
stx2 transcription was determined using total bacterial RNA (RNeasy miniprep kit; Qiagen, Hilden, Germany) and one-step quantitative real-time reverse transcription-PCR (3) performed with a CFX96 real-time PCR system (Bio-Rad, Munich, Germany) and a SensiMix SYBR No-ROX one-step kit (Peqlab Biotechnologie, Erlangen, Germany). Primers RT-stx2F (5′-CGACCCCTCTTGAACATA-3′) and RT-stx2R (5′-TAGACATCAAGCCCTCGTAT-3′) were used to amplify stx2, and primers GapA_forward and GapA_reverse (4) amplified the reference gene gapA (for normalization). A melting curve analysis to confirm the specificity of the amplification products was performed with continuous fluorescence reading from 65°C to 95°C. Data were analyzed using CFX Manager software, version 1.6 (Bio-Rad).
Stx2 titers were determined using a Vero cell assay (2) and were expressed as numbers of 50% cytotoxic doses (CD50; the highest filtrate dilutions that caused cytotoxicity in 50% of cells after 72 h) per ml. The specific role of Stx2 in Vero cell cytotoxicity was verified by immunodetection of Stx2 in the filtrates, using a reversed passive latex agglutination test (VTEC-RPLA; Denka Seiken, Co., Ltd., Niigata, Japan).
The effects of each antibiotic on stx2-harboring phage induction, stx2 transcription, and Stx2 production are expressed as fold increases of the stx2-harboring phage titer, stx2 transcription level, and Stx2 titer, respectively, in the antibiotic-treated culture compared to a control, untreated culture (reflecting baseline, spontaneous stx2-harboring phage induction, stx2 transcription, and Stx2 production, each defined as 1). The baseline stx2-carrying phage and Stx2 titers in the control cultures were as follows (range [mean ± standard deviation]): for strain LB226692, stx2-harboring phage titers of 2.9 × 104 to 8.4 × 104 (5.63 × 104 ± 2.24 × 104) and Stx2 titers of 640 to 1,280 (960 ± 369); for strain LB226806, stx2-harboring phage titers of 4.5 × 104 to 1 × 105 (7.15 × 104 ± 2.25 × 104) and Stx2 titers of 320 to 1,280 (880 ± 480); and for strain EDL933, stx2-harboring phage titers of 5.2 × 103 to 7.8 × 104 (6.55 × 104 ± 1.1 × 104) and Stx2 titers of 1,280 to 5,120 (3,520 ± 1,920). For stx2 transcription, the stx2/gapA mRNA ratio of the control culture of each strain was considered 1.
Statistical analysis.
The experiments were performed three (stx2 transcription) or four (stx2-harboring phage induction and Stx2 production) times, and results are expressed as means ± standard deviations for the repeated experiments. The differences in stx2-harboring phage induction, stx2 transcription, and Stx2 production caused by the different antibiotics were analyzed using unpaired Student's t test. P values of ≤0.05 were considered significant.
RESULTS
Effects of subinhibitory antibiotic concentrations on induction of stx2-harboring bacteriophages and Stx2 production.
The antibiotics were initially investigated for their effects on stx2-harboring phage induction and Stx2 production at 1/4 MIC. Ciprofloxacin, an inducer of stx2-harboring phages in EHEC O157:H7 (40), significantly increased stx2-harboring phage titers and Stx2 titers in both EHEC O104:H4 outbreak isolates and EHEC O157:H7 strain EDL933 (P < 0.001) (Table 2). This demonstrated that each of the outbreak isolates contains an inducible stx2-harboring bacteriophage. Meropenem, azithromycin, fosfomycin, gentamicin, and kanamycin at 1/4 MIC had only slight, and statistically insignificant, effects on the baseline stx2-harboring phage titers and Stx2 titers in all strains (Table 2). In contrast, rifaximin, tigecycline, and chloramphenicol significantly decreased the phage and/or Stx2 titer for both outbreak isolates (P ≤ 0.05) (Table 2). For EHEC O157:H7 strain EDL933, decreases in the phage and toxin titers mediated by the last three antibiotics did not reach statistical significance (Table 2).
Table 2.
Effects of antibiotics at 1/4 MIC on induction of stx2-harboring bacteriophages and Stx2 productiona
| Antibioticb | LB226692 (O104:H4) |
LB226806 (O104:H4) |
EDL933 (O157:H7) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| stx2-harboring phage titer | P value | Stx2 titer | P value | stx2-harboring phage titer | P value | Stx2 titer | P value | stx2-harboring phage titer | P value | Stx2 titer | P value | |
| CIP | 6.4 × 104 ± 7.5 × 103c | <0.001 | 128.0 ± 0c | <0.001 | 1.4 × 105 ± 9.3 × 103c | <0.001 | 128.0 ± 0c | <0.001 | 6 × 103 ± 8.8 × 102c | <0.001 | 128.0 ± 0c | <0.001 |
| MEM | 0.98 ± 0.2 | 0.68 | 0.67 ± 0.18 | 0.28 | 0.95 ± 0.10 | 0.68 | 0.51 ± 0.09 | 0.13 | 1.0 ± 0.1 | 0.79 | 0.50 ± 0.06 | 0.12 |
| AZM | 0.72 ± 0.1 | 0.43 | 0.50 ± 0 | 0.12 | 0.51 ± 0.11 | 0.11 | 0.54 ± 0.13 | 0.14 | 0.63 ± 0.09 | 0.22 | 0.59 ± 0.04 | 0.19 |
| RIF | 0.12 ± 0.01d | 0.011 | 0.13 ± 0d | 0.012 | 0.39 ± 0.04 | 0.062 | 0.25 ± 0.06d | 0.025 | 0.61 ± 0.11 | 0.21 | 0.93 ± 0.28 | 0.65 |
| TGC | 0.21 ± 0.02d | 0.05 | 0.58 ± 0.1 | 0.17 | 0.16 ± 0.04d | 0.014 | 0.29 ± 0.11d | 0.031 | 0.85 ± 0.12 | 0.52 | 0.50 ± 0.14 | 0.12 |
| FOF | 0.97 ± 0.22 | 0.71 | 1.2 ± 0.42 | 0.95 | 0.96 ± 0.26 | 0.72 | 0.75 ± 0.23 | 0.37 | 1.8 ± 0.48 | 0.45 | 0.87 ± 0.28 | 0.54 |
| CHL | 0.25 ± 0.06d | 0.024 | 0.13 ± 0.02d | 0.012 | 0.13 ± 0.03d | 0.012 | 0.04 ± 0.02d | 0.0068 | 0.61 ± 0.09 | 0.21 | 0.55 ± 0.06 | 0.15 |
| GEN | 0.57 ± 0.12 | 0.17 | 0.83 ± 0.18 | 0.53 | 1.3 ± 0.23 | 0.89 | 0.67 ± 0.18 | 0.29 | 2.4 ± 0.12 | 0.24 | 1.0 ± 0.18 | 0.96 |
| KAN | 1.56 ± 0.42 | 0.37 | 0.83 ± 0.18 | 0.51 | 0.48 ± 0.09 | 0.21 | 0.58 ± 0.18 | 0.33 | 1.0 ± 0.11 | 0.99 | 0.83 ± 0.18 | 0.51 |
Fold increases of stx2-harboring phage titers (indicating stx2-harboring phage induction) and Stx2 titers (indicating Stx2 production) in antibiotic-treated cultures compared to those in control, untreated cultures (defined as 1) are presented (for the actual stx2-harboring phage and Stx2 titers in the control cultures, see Materials and Methods). Data are means ± standard deviations for four independent experiments. P values (by unpaired Student's t test) of ≤0.05 are statistically significant (data shown in bold).
CIP, ciprofloxacin; MEM, meropenem; AZM, azithromycin; RIF, rifaximin; TGC, tigecycline; FOF, fosfomycin; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin.
Significantly increased (P < 0.001) compared to control.
Significantly decreased (P ≤ 0.05) compared to control.
The six antibiotics that most profoundly changed stx2-harboring phage induction and/or Stx2 production in the outbreak isolates at 1/4 MIC and/or were proposed as therapeutic options during the EHEC O104:H4 outbreak (15) were tested for their effects on phage titers (Fig. 1) and toxin titers (Fig. 2) at a broader range of subinhibitory concentrations. Ciprofloxacin significantly increased both stx2-harboring phage and Stx2 titers (P value of <0.001 or <0.05) at all subinhibitory concentrations (1/2 to 1/16 MIC), in a dose-dependent manner (Fig. 1 and 2). Chloramphenicol caused a concentration-dependent decrease in stx2-harboring phage and Stx2 titers in the range of 1/2 to 1/8 MIC; the most pronounced and statistically significant suppressive effects (P < 0.05) of chloramphenicol were regularly observed at 1/2 MIC (Fig. 1 and 2). The remaining four antibiotics (meropenem, azithromycin, rifaximin, and tigecycline) showed no clear concentration-dependent effects on stx2-harboring phage and Stx2 titers. For EHEC O157:H7 strain EDL933, none of these antibiotics at any concentration significantly changed the baseline phage titers (P = 0.27 to 0.92) (Fig. 1) and toxin titers (P = 0.11 to 0.89) (Fig. 2). In contrast, with the EHEC O104:H4 outbreak isolates, meropenem, rifaximin, and tigecycline (both isolates), as well as azithromycin (LB226806), significantly decreased stx2-harboring phage titers (P ≤ 0.05), regularly at 1/2 MIC and sometimes also at lower concentrations (Fig. 1). Phage titer decreases were usually accompanied by Stx2 titer decreases, though the latter attained statistical significance less often (Fig. 2). None of these four antibiotics at any concentration significantly increased the phage titer (Fig. 1) or toxin titer (Fig. 2) of the outbreak isolates.
Fig 1.
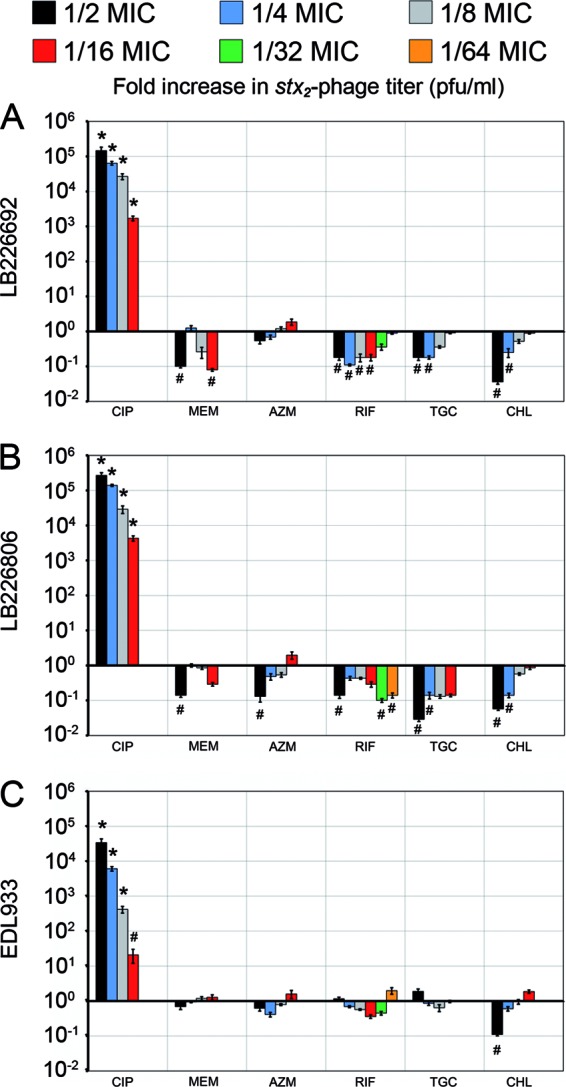
Effects of various subinhibitory concentrations of ciprofloxacin (CIP), meropenem (MEM), azithromycin (AZN), rifaximin (RIF), tigecycline (TGC), and chloramphenicol (CHL) on stx2-harboring phage induction in EHEC O104:H4 outbreak isolates LB226692 (A) and LB226806 (B) and EHEC O157:H7 strain EDL933 (C). Data show fold increases in stx2-harboring phage titers (indicating stx2-harboring phage induction) compared to those of antibiotic-free controls (defined as 1) and are means ± standard deviations for four independent experiments. All antibiotics were tested at 1/2 to 1/16 MIC, and rifaximin was also tested at 1/32 and 1/64 MIC. *, P < 0.001; #, P ≤ 0.05 (unpaired Student's t test).
Fig 2.
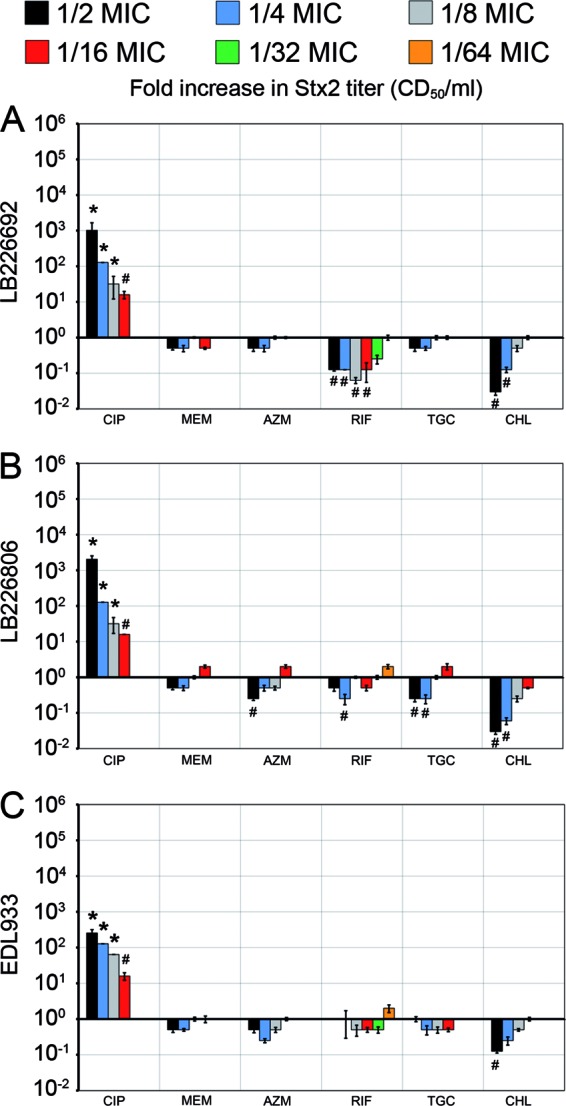
Effects of various subinhibitory concentrations of ciprofloxacin (CIP), meropenem (MEM), azithromycin (AZN), rifaximin (RIF), tigecycline (TGC), and chloramphenicol (CHL) on Stx2 production in EHEC O104:H4 outbreak isolates LB226692 (A) and LB226806 (B) and EHEC O157:H7 strain EDL933 (C). Data show fold increases in Stx2 titers (indicating Stx2 production) compared to those of antibiotic-free controls (defined as 1) and are means ± standard deviations for four independent experiments. All antibiotics were tested at 1/2 to 1/16 MIC, and rifaximin was also tested at 1/32 and 1/64 MIC. *, P < 0.001; #, P ≤ 0.05 (unpaired Student's t test).
Effects of subinhibitory antibiotic concentrations on stx2 transcription.
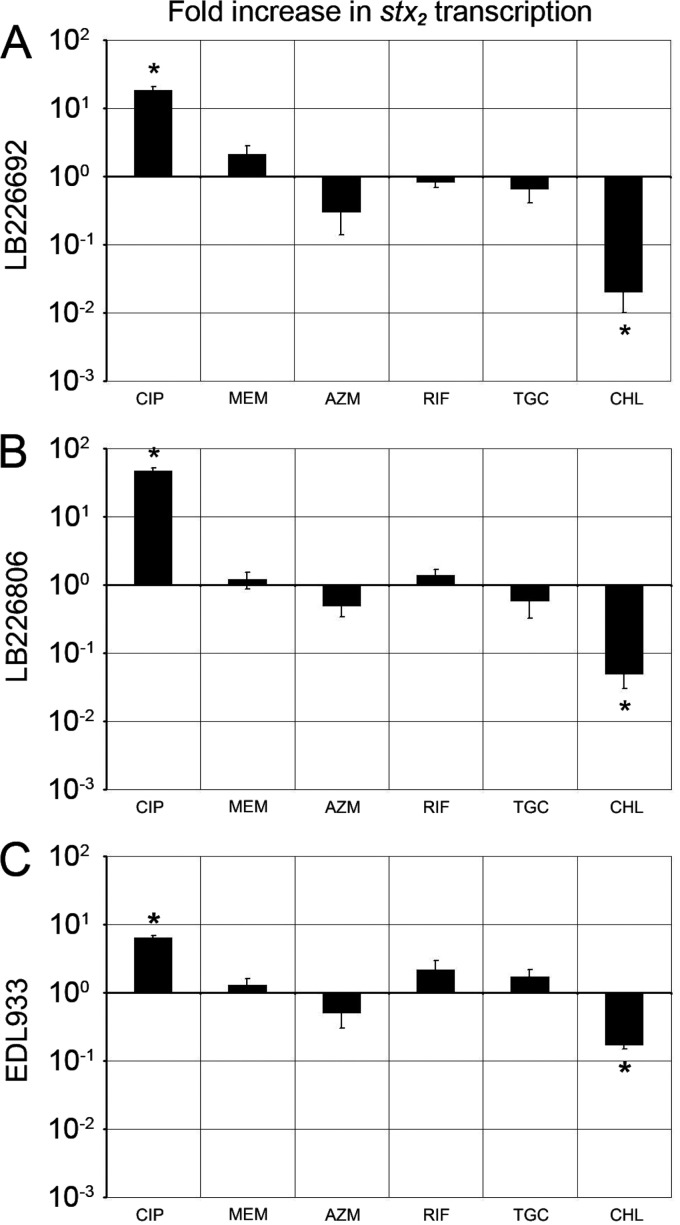
The above six antibiotics at 1/2 MIC, which most considerably influenced stx2-harboring phage induction (Fig. 1) and Stx2 production (Fig. 2), were tested for their effects on stx2 transcription. Ciprofloxacin significantly increased (P < 0.01) and chloramphenicol significantly decreased (P < 0.01) stx2 transcription in the EHEC O104:H4 outbreak isolates and strain EDL933 (Fig. 3). Meropenem, azithromycin, rifaximin, and tigecycline had only slight effects (P = 0.39 to 0.72), and none of them significantly upregulated stx2 transcription in any strain (Fig. 3).
Fig 3.
Effects of ciprofloxacin (CIP), meropenem (MEM), azithromycin (AZN), rifaximin (RIF), tigecycline (TGC), and chloramphenicol (CHL) at 1/2 MIC on stx2 transcription in EHEC O104:H4 outbreak isolates LB226692 (A) and LB226806 (B) and EHEC O157:H7 strain EDL933 (C). Data show fold increases in stx2 transcription compared to that in antibiotic-free cultures and are means ± standard deviations for three independent experiments. *, P < 0.01 (unpaired Student's t test).
Susceptibilities of outbreak isolates to the investigated antibiotics.
Each of 20 additionally tested EHEC O104:H4 outbreak isolates was susceptible to ciprofloxacin, meropenem, fosfomycin, chloramphenicol, gentamicin, and kanamycin according to CLSI guidelines (8) and to tigecycline according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) (11) guidelines (no CLSI breakpoints are available for this antibiotic), as were isolates LB226692 and LB226806. The MICs for azithromycin and rifaximin, for which there are no interpretative categories for E. coli, were in the ranges reported previously for other EHEC strains (19, 24).
DISCUSSION
The necessity of using antibiotic therapy for a subset of patients during the recent German E. coli O104:H4 outbreak prompted us to investigate a panel of antibiotics to which the outbreak strain is susceptible for their effects on induction of stx2-harboring phages, stx2 transcription, and Stx2 production in this pathogen. Except for ciprofloxacin (32), no other antibiotics have previously been investigated thusly. We were particularly interested in those antibiotics that had been proposed for indicated use during the O104:H4 outbreak (15) based on their inability to induce stx2-harboring phages and Stx2 production in EHEC O157:H7 (23, 24, 28, 39), i.e., azithromycin, rifaximin, and meropenem (though another carbapenem, imipenem, not meropenem itself, was investigated with EHEC O157:H7 in this respect) (23). We also included tigecycline because of its broad activity against Gram-positive and Gram-negative bacteria (14) and because, to our knowledge, its effects on induction of stx-harboring phages and Stx production have not yet been studied. We used various subinhibitory antibiotic concentrations, the methodology commonly applied to investigate effects of antibiotics on Stx production and/or stx-harboring phage induction in other EHEC strains (16, 23, 24, 28, 39), which allows determination of the concentration dependence of the effects studied. In contrast, therapeutic concentrations rapidly inhibit or kill bacteria in vitro, thus hampering unbiased measurement of antibiotic effects.
Among the nine antibiotics tested, only ciprofloxacin, an agent previously shown to induce stx-harboring phages in EHEC O157:H7 via triggering of the SOS response (22, 40), significantly increased stx2-harboring phage induction, stx2 transcription, and Stx2 production in EHEC O104:H4 outbreak isolates. These effects of ciprofloxacin are in accordance with the structural similarities between the stx2-harboring phage of EHEC O104:H4 and the stx2-harboring phage 933W of EHEC O157:H7 strain EDL933 reported by Rasko et al. (32). These authors also showed a significant (∼80-fold) ciprofloxacin-mediated increase in stx2 transcription in the EHEC O104:H4 outbreak strain, but no attempts were made to demonstrate increases in the stx2-harboring phage titer and the amount of biologically active Stx2 as shown in our study. Because of its unfavorable effects on phage induction and Stx production, which may increase the risk of development of HUS, ciprofloxacin should not be used in patients with diarrhea if an EHEC etiology cannot be excluded. The administration of ciprofloxacin to some patients with diarrhea at the beginning of the EHEC O104:H4 outbreak (our unpublished data), before the EHEC etiology was determined, was due mainly to the predominant involvement of adults, which did not lead to a suspicion of an EHEC-mediated outbreak because EHEC infections typically affect children (21, 37). However, no data are yet available from the German outbreak about the impact of ciprofloxacin treatment on the clinical outcome of infection, in particular the progression of infection to HUS. Similarly, in a study from Denmark, where four of five patients with HUS caused by the German EHEC O104:H4 outbreak strain received ciprofloxacin upon first presenting with diarrhea to their physicians (9), no analysis determined any association between this antibiotic treatment and development of HUS.
The crucial finding in our study is that none of the antibiotics proposed for urgent therapeutic or prophylactic use during the EHEC O104:H4 outbreak (15), specifically meropenem, azithromycin, and rifaximin, induced stx2-harboring phages or increased stx2 transcription or Stx2 production in the outbreak isolates in an in vitro system. Indeed, these antibiotics, regularly at 1/2 MIC but also at lower concentrations, significantly decreased one or more of these processes. The same holds true for tigecycline. However, despite these promising in vitro results, it is premature to advocate, based solely on these data, the use of these antibiotics for treatment of patients with EHEC O104:H4 infection. Further studies using animal models are necessary to determine whether or not the favorable effects on stx2-harboring phages and Stx2 production observed in vitro can be confirmed in vivo. Moreover, other effects of these antibiotics, e.g., the extent to which they reduce the normal intestinal flora or their influence on intestinal adherence of the pathogen, might modulate the course of EHEC infection. Indeed, reduced adherence to epithelial cells after their pretreatment with rifaximin has been reported for EAEC (5). Finally, thorough analyses of clinical outcomes of patients who were administered these antibiotics during the outbreak are necessary for ultimate evaluation of their potential usefulness for treatment of humans infected with EHEC O104:H4 in cases where antibiotic therapy is inevitable. A recent study demonstrated that treatment with azithromycin significantly decreased frequency of long-term carriage of the EHEC O104:H4 outbreak strain (27a).
Rifaximin, which is licensed in many countries to treat traveler's diarrhea (34), might be considered an option, in contrast to ciprofloxacin, for treatment of diarrhea of unclear etiology because it does not induce stx-harboring phages and Stx production (28; this study) and is thus supposed to have no adverse effect on systemic progression of infection in cases of unrecognized EHEC infection. Although chloramphenicol is not used extensively anymore in the developed world (12), the significant suppressive effects of this antibiotic on stx2-harboring phage induction, stx2 transcription, and Stx2 production in all tested strains in our study are important findings which should be kept in mind.
In conclusion, we demonstrated that an antibiotic-mediated effect on stx2-harboring phage induction is the critical process through which Stx2 production is modulated in EHEC O104:H4. Several antibiotics decrease or do not influence baseline levels of stx2-harboring phage induction, stx2 transcription, and Stx2 production in the EHEC O104:H4 outbreak strain and might therefore be potentially therapeutically useful in indicated cases. These data are the first step in the investigation of the effects of antibiotics on the outbreak strain regarding their potential benefit or harm to patients. We await further analysis of the outbreak to determine if usage of these agents was associated with a reduced risk of developing HUS.
ACKNOWLEDGMENTS
This study was supported by grants from the Interdisciplinary Center of Clinical Research (IZKF) Münster (Me2/021/12), from the Medical Faculty of the University of Münster (BD9817044), and from the EU FP7 ANTIGONE program (278976).
We thank Phillip I. Tarr, Washington University School of Medicine, St. Louis, MO, for fruitful discussions on the manuscript and N. Brandt, M. Junge, R. Fischer, and A. Lagemann, Institute of Hygiene, and B. Grünastel, M. Tigges, S. Weber, and M. Ressel, Institute of Medical Microbiology, for technical assistance. We are grateful to Pfizer (Groton, CT) for the gift of azithromycin and tigecycline and to Alfa Wassermann (Bologna, Italy) for the gift of rifaximin.
Footnotes
Published ahead of print 5 March 2012
REFERENCES
- 1. Bell BP, et al. 1997. Predictors of hemolytic uremic syndrome in children during a large outbreak of Escherichia coli O157:H7 infections. Pediatrics 100:E12. [DOI] [PubMed] [Google Scholar]
- 2. Bielaszewska M, et al. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11:671–676 [DOI] [PubMed] [Google Scholar]
- 3. Bielaszewska M, et al. 2011. Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol. Microbiol. 79:1024–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blumer C, et al. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287–3298 [DOI] [PubMed] [Google Scholar]
- 5. Brown EL, Xue Q, Jiang ZD, Xu Y, Dupont HL. 2010. Pretreatment of epithelial cells with rifaximin alters bacterial attachment and internalization profiles. Antimicrob. Agents Chemother. 54:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buchholz U, et al. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365:1763–1770 [DOI] [PubMed] [Google Scholar]
- 7. Clinical Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M07-A8. Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21. Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Colic E, Dieperink H, Titlestad K, Tepel M. 2011. Management of an acute outbreak of diarrhoea-associated haemolytic uraemic syndrome with early plasma exchange in adults from southern Denmark: an observational study. Lancet 378:1089–1093 [DOI] [PubMed] [Google Scholar]
- 10. Dundas S, et al. 2001. The central Scotland Escherichia coli O157:H7 outbreak: risk factors for the hemolytic uremic syndrome and death among hospitalized patients. Clin. Infect. Dis. 33:923–931 [DOI] [PubMed] [Google Scholar]
- 11. European Committee on Antimicrobial Susceptibility Testing 2011. Breakpoint tables for interpretation of MICs and zone diameters, version 1.3, 5 January 2011. http://www.eucast.org/clinical_breakpoints/ Accessed 15 October 2011
- 12. Falagas ME, Kopterides P. 2007. Old antibiotics for infections in critically ill patients. Curr. Opin. Crit. Care 13:592–597 [DOI] [PubMed] [Google Scholar]
- 13. Frank C, et al. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany—preliminary report. N. Engl. J. Med. 365:1771–1780 [DOI] [PubMed] [Google Scholar]
- 14. Garrison MW, Mutters R, Dowzicky MJ. 2009. In vitro activity of tigecycline and comparator agents against a global collection of Gram-negative and Gram-positive organisms: tigecycline evaluation and surveillance trial 2004 to 2007. Diagn. Microbiol. Infect. Dis. 65:288–299 [DOI] [PubMed] [Google Scholar]
- 15. German Society for Infectious Diseases 4 June 2011. EHEC infection and antibiotic therapy. http://www.dgi-net.de/images/stories/DGI-position_paper_EHECantibiotics_English_version_plus_references_20110604.pdf Accessed 15 October 2011
- 16. Grif K, Dierich MP, Karch H, Allerberger F. 1998. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 17:761–766 [DOI] [PubMed] [Google Scholar]
- 17. Holtz LR, Neill MA, Tarr PI. 2009. Acute bloody diarrhea: a medical emergency for patients of all ages. Gastroenterology 136:1887–1898 [DOI] [PubMed] [Google Scholar]
- 18. Jansen A, Kielstein JT. 2011. The new face of enterohaemorrhagic Escherichia coli infections. Euro Surveill. 16:19898. [PubMed] [Google Scholar]
- 19. Jiang ZD, DuPont HL. 2005. Rifaximin: in vitro and in vivo antibacterial activity—a review. Chemotherapy 51(Suppl 1):67–72 [DOI] [PubMed] [Google Scholar]
- 20. Karch H, Strockbine NA, O'Brien AD. 1986. Growth of Escherichia coli in the presence of trimethoprim-sulfamethoxazole facilitates detection of Shiga-like toxin producing strains by colony blot assay. FEMS Microbiol. Lett. 35:141–145 [Google Scholar]
- 21. Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405–418 [DOI] [PubMed] [Google Scholar]
- 22. Kimmitt PT, Harwood CR, Barer MR. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JH, Stein BD. 2009. Antimicrobials effective for inhibition of enterohaemorrhagic Escherichia coli strains O26, O111, and O157 and their effects on Shiga toxin releases. J. Microbiol. Biotechnol. 19:1238–1243 [PubMed] [Google Scholar]
- 24. McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob. Agents Chemother. 54:3790–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mellmann A, et al. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mellmann A, et al. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751 doi:10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mühldorfer I, et al. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect. Immun. 64:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a. Nitschke M, et al. 2012. Association between azithromycin therapy and duration of bacterial shedding among patients with Shiga toxin-producing enteroaggregative Escherichia coli O104:H4. JAMA 303:1046–1052 [DOI] [PubMed] [Google Scholar]
- 28. Ochoa TJ, Chen J, Walker CM, Gonzales E, Cleary TG. 2007. Rifaximin does not induce toxin production or phage-mediated lysis of Shiga toxin-producing Escherichia coli. Antimicrob. Agents Chemother. 51:2837–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pedersen MG, Hansen C, Riise E, Persson S, Olsen KE. 2008. Subtype-specific suppression of Shiga toxin 2 released from Escherichia coli upon exposure to protein synthesis inhibitors. J. Clin. Microbiol. 46:2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Proulx F, Turgeon JP, Delage G, Lafleur L, Chicoine L. 1992. Randomized, controlled trial of antibiotic therapy for Escherichia coli O157:H7 enteritis. J. Pediatr. 121:299–303 [DOI] [PubMed] [Google Scholar]
- 31. Rahal EA, Kazzi N, Kanbar A, Abdelnoor AM, Matar GM. 2011. Role of rifampicin in limiting Escherichia coli O157:H7 Shiga-like toxin expression and enhancement of survival of infected BALB/c mice. Int. J. Antimicrob. Agents 37:135–139 [DOI] [PubMed] [Google Scholar]
- 32. Rasko DA, et al. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robert Koch Institute (RKI) 2011. Final presentation and evaluation of the epidemiological findings in the EHEC O104:H4 outbreak, Germany 2011. RKI, Berlin, Germany: http://www.rki.de Accessed 20 November 2011 [Google Scholar]
- 34. Ruiz J, et al. 2007. In vitro antimicrobial activity of rifaximin against enteropathogens causing traveler's diarrhea. Diagn. Microbiol. Infect. Dis. 59:473–475 [DOI] [PubMed] [Google Scholar]
- 35. Schmidt H, Bielaszewska M, Karch H. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage Φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith KE, et al. 2012. Antibiotic treatment of Escherichia coli O157 infection and the risk of hemolytic uremic syndrome, Minnesota. Pediatr. Infect. Dis. J. 31:37–41 [DOI] [PubMed] [Google Scholar]
- 37. Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086 [DOI] [PubMed] [Google Scholar]
- 38. Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Q, et al. 2009. Gnotobiotic piglet infection model for evaluating the safe use of antibiotics against Escherichia coli O157:H7 infection. J. Infect. Dis. 199:486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, et al. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670 [DOI] [PubMed] [Google Scholar]