Abstract
FPH-1 is a new class A carbapenemase from Francisella philomiragia. It produces high-level resistance to penicillins and the narrow-spectrum cephalosporin cephalothin and hydrolyzes these β-lactam antibiotics with catalytic efficiencies of 106 to 107 M−1 s−1. When expressed in Escherichia coli, the enzyme confers resistance to clavulanic acid, tazobactam, and sulbactam and has Ki values of 7.5, 4, and 220 μM, respectively, against these inhibitors. FPH-1 increases the MIC of the monobactam aztreonam 256-fold and the MIC of the broad-spectrum cephalosporin ceftazidime 128-fold, while the MIC of cefoxitin remains unchanged. MICs of the carbapenem antibiotics imipenem, meropenem, doripenem, and ertapenem are elevated 8-, 8-, 16-, and 64-fold, respectively, against an E. coli JM83 strain producing the FPH-1 carbapenemase. The catalytic efficiencies of the enzyme against carbapenems are in the range of 104 to 105 M−1 s−1. FPH-1 is 77% identical to the FTU-1 β-lactamase from Francisella tularensis and has low amino acid sequence identity with other class A β-lactamases. Together with FTU-1, FPH-1 constitutes a new branch of the prolific and ever-expanding class A β-lactamase tree.
INTRODUCTION
Production of β-lactamases is the major mechanism of resistance to β-lactam antibiotics in Gram-negative bacterial isolates (5, 6). β-Lactamases hydrolyze the four-membered ring of β-lactam antibiotics, rendering them incapable of inactivating their target, the bacterial penicillin-binding proteins, which are enzymes involved in the biosynthesis and restructuring of the bacterial cell wall (13). Currently, hundreds of β-lactamases have been identified in various clinical and environmental bacterial isolates, and new enzymes continue to be reported on a monthly basis. Two major classification schemes for these enzymes are based either on their functional characteristics (Bush-Jacoby nomenclature [7]) or on their amino acid sequence (Ambler nomenclature [1]). According to the Ambler nomenclature, β-lactamases are subdivided into classes A through D. β-Lactamases belonging to the A, C, and D classes comprise various serine enzymes, while class B includes numerous metallo-β-lactamases (1). Class A enzymes are abundant in both Gram-negative and Gram-positive bacteria and include, among others, the clinically important TEM-, SHV-, and CTX-type β-lactamases of Gram-negative pathogens. Multiple representatives of these enzyme types are characterized by widely varying antibiotic substrate profiles, from the narrow spectrum that covers early penicillins, to the extended and inhibitor-resistant profiles that are characterized by resistance to modern cephalosporins and clinically used β-lactam inhibitors (5, 6, 8, 12). It is thus somewhat puzzling that, despite the intensive use of carbapenems for over 3 decades, TEM-, SHV-, and CTX-type class A β-lactamases, enzymes that demonstrate remarkable evolutionary plasticity, have not yet given rise to mutants with clinically significant resistance to carbapenem antibiotics. Instead, distantly related class A β-lactamases with carbapenemase activities have been mobilized from yet-unidentified bacteria. These newly selected carbapenemases include the SFC-1, IMI/NMC-A, BIC-1, GES-, KPC-, and SME-type enzymes (4, 14, 17–25, 27). It is of interest that the first-characterized class A carbapenemases, IMI-1 and SME-1, were discovered before carbapenems were introduced into clinical use, implying that carbapenemase activity has not evolved in response to the intensive use of these antibiotics (25, 26). While some of the currently known class A carbapenemases are rare in clinical isolates, others, like KPC- and GES-type carbapenemases, are numerous and are increasingly recognized as the major threat for successful use of carbapenems, which are the antibiotics of last reserve and are used for treatment of life-threatening infections caused by multidrug-resistant bacterial pathogens. Recently we described the FTU-1 β-lactamase from Francisella tularensis, which is intrinsic in this species and possesses weak carbapenemase activity (3). In this paper we present data on the closely related FPH-1 carbapenemase from yet another Francisella species, F. philomiragia.
MATERIALS AND METHODS
Cloning of the fph1 gene.
The gene for a putative class A carbapenemase from F. philomiragia (GenBank accession number ZP_05249935.1) was optimized for expression in Escherichia coli and custom synthesized (GenScript). The predicted leader peptide of the FPH-1 β-lactamase was replaced with the signal sequence of the outer membrane protein A (OmpA) to ensure efficient transport of the mature enzyme into the periplasm, and unique NdeI and HindIII restriction sites were introduced at the 5′ and 3′ ends, respectively. The gene for FPH-1 was cloned between the NdeI and HindIII sites of the pHF016 vector (pHF016:FPH-1), and this plasmid was transformed into E. coli JM83 for antibiotic susceptibility testing (15). For protein expression and purification, the gene for the FPH-1 enzyme was cloned into the pET24a(+) expression vector between the NdeI and HindIII sites [pET24a(+):FPH-1] and transformed into E. coli BL21(DE3) cells.
Antibiotic susceptibility testing.
MICs of various β-lactam antibiotics and combinations of antibiotics with β-lactamase inhibitors were determined in Mueller-Hinton II broth by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (9). Additionally, MICs of some β-lactam antibiotics were measured in the presence of fixed concentrations of β-lactamase inhibitors. The MIC values were determined for E. coli JM83 cells harboring the pHF016:FPH-1 plasmid, and the E. coli JM83 strain carrying the pHF016 vector only was used as a control. The optical densities of the cultures were equalized and diluted to result in a final inoculum of 5 × 105 CFU/ml. The MICs were determined in triplicate in 96-well plates containing 100 μl/well of Mueller-Hinton II broth with 2-fold serial dilutions of antibiotics. The plates were incubated at 37°C for 16 to 20 h before the results were analyzed.
Protein expression and purification.
An E. coli BL21(DE3) strain harboring the pET24a(+):FPH-1 plasmid was grown in a shaker/incubator at 37°C in LB broth supplemented with 50 μg/ml kanamycin A. When the optical density at 600 nm of the bacterial culture reached 0.6, protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside, and the culture was incubated for an additional 20 h at 24°C. The bacteria were removed by centrifugation (20,000 × g, 20 min, 4°C), and the supernatant containing the enzyme was concentrated from 1 liter to 50 ml by using a 10-kDa cutoff Centricon Plus 70 concentrator (Millipore). After dialysis overnight at 4°C against 25 mM HEPES (pH 7.5) buffer, the concentrated supernatant was loaded onto a DEAE anion exchange column (Bio-Rad) equilibrated with the same buffer. Under these conditions, the majority of E. coli proteins that were present in the supernatant were bound to the DEAE resin, while the FPH-1 enzyme was recovered in the flowthrough fractions. FPH-1 was identified by reaction with the chromogenic β-lactam nitrocefin, and the enzyme's purity was evaluated by 12% SDS-PAGE. FPH-1-containing fractions were pooled together, and the protein was further purified by CM cation exchange chromatography (Bio-Rad). Bound proteins were eluted with a 0-to-500 mM gradient of NaCl in 25 mM HEPES (pH 7.5) buffer, and the FPH-1 enzyme was detected in the fractions containing 180 to 240 mM NaCl. Nitrocefin-positive fractions were analyzed by 12% SDS-PAGE, and those containing the pure enzyme were pooled, concentrated, and stored at −80°C. Protein concentration was measured spectrophotometrically using the predicted extinction coefficient (Δε280, 31,922 cm−1 M−1) (http://web.expasy.org/protparam).
Enzyme kinetics.
The hydrolysis of the β-lactam ring was monitored spectrophotometrically, and the data were collected either with a Cary 50 spectrophotometer (Varian) or with a BioLogic SFM-300 stopped flow instrument. Activities of the FPH-1 β-lactamase against β-lactam substrates were measured at room temperature in reaction buffer containing 50 mM sodium phosphate (pH 7.0) and 100 mM NaCl. The reactions were initiated with 1 to 400 nM enzyme. The following extinction coefficients for the indicated wavelengths were used to calculate the enzyme activity: ampicillin (Δε240, −538 M−1 cm−1), oxacillin (Δε260, 440 M−1 cm−1), piperacillin (Δε235, −1,070 M−1 cm−1), ticarcillin (Δε235, −660 M−1 cm−1), cephalothin (Δε262, −7,960 M−1 cm−1), cefuroxime (Δε262, −7,800 M−1 cm−1), ceftazidime (Δε260, −10,500 M−1 cm−1), cefotaxime (Δε265, −6,260 M−1 cm−1), aztreonam (Δε318, −640 M−1 cm−1), imipenem (Δε297, −10,930 M−1 cm−1), meropenem (Δε298, −7,200 M−1 cm−1), ertapenem (Δε295, −9,970 M−1 cm−1), doripenem (Δε296, −7,540 M−1 cm−1), nitrocefin (Δε500, 15,900 M−1 cm−1). The steady-state velocities were determined from the linear phase of the reaction progress curves and plotted as a function of substrate concentration. The Km and kcat values were calculated by nonlinear regression using the Michaelis-Menten equation and Prism 5 software (GraphPad Software, Inc.): v = (Vmax[S])/(Km + [S]), where v is the initial velocity, [S] is the substrate concentration, and Vmax = kcat[E], where [E] is the enzyme concentration. When saturation could not be reached due to a high Km, the kcat/Km ratio was determined by fitting the progress curve of the reaction with the following equation: At = A∞ + (A0 − A∞)e−kt, where At is the absorbance at time t and A0 is the initial and A∞ is the final absorbance. In this equation the observed first-order rate constant k = kcat/Km[E].
The inhibitor dissociation constant (Ki) values for clavulanic acid, sulbactam, and tazobactam were measured using the competitive reporter substrate nitrocefin. The reaction buffer contained nitrocefin and inhibitor (25 and 50 μM nitrocefin for 0 to 8 μM clavulanic acid; 150 and 200 μM nitrocefin for 0 to 500 μM sulbactam; 200 and 280 μM nitrocefin for 0 to 15 μM tazobactam), and the reactions were initiated with the enzyme (0.2 nM final concentration). The FPH-1 enzyme was diluted and stored in reaction buffer containing 1 mg/ml bovine serum albumin at 4°C. Progress curves of the reactions were fitted using the following equation for time-dependent inhibition (10): At = A∞ + vsst + [(vi −vss)/kobs](1 − e−kobst), where At is the absorbance at time t, A0 is the initial absorbance, vi and vss are the initial and steady-state velocities, respectively, and the observed rate constant is kobs. The Ki values were determined using the method of Dixon (11).
RESULTS AND DISCUSSION
Cloning and amino acid sequence analysis of the FPH-1 β-lactamase.
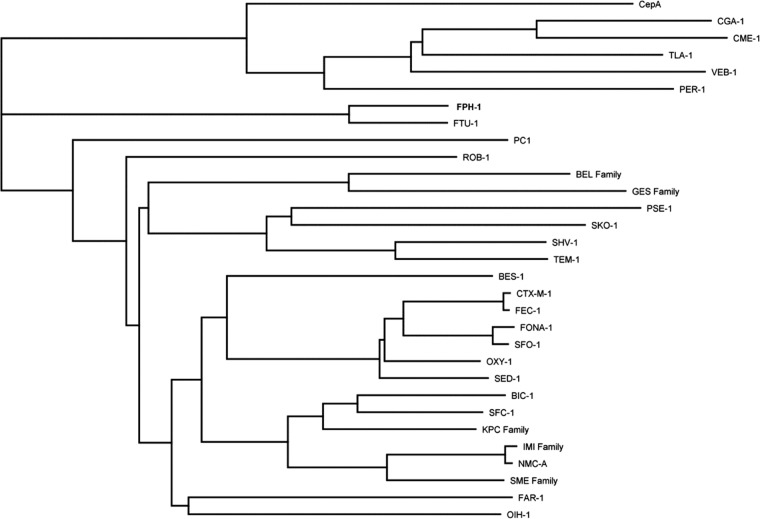
We recently reported the antibiotic resistance profile and steady-state kinetics of the class A β-lactamase FTU-1 from F. tularensis (3). All 14 genomes of the F. tularensis strains currently deposited in the BioProject Database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/bioproject) encode FTU-1 or its mutant derivatives. Available genome sequences are represented by all four known F. tularensis subspecies (F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, F. tularensis subsp. novicida, and F. tularensis subsp. mediasiatica), an indication that FTU-1 is intrinsic in F. tularensis. This database also includes three genomic sequences from another species of Francisella, F. philomiragia. Our analysis of these sequences revealed that all three encode a putative class A β-lactamase that is closely related to FTU-1. In this study, we cloned one of these enzymes, FPH-1, from the environmental isolate F. philomiragia subsp. philomiragia ATCC 25017. This is the only other Francisella species (apart from F. tularensis) whose genome has been sequenced. Similar to the FTU-1 β-lactamase, the gene for FPH-1 was optimized for E. coli codon usage and the sequence corresponding to the leader peptide of FPH-1 was replaced by the leader sequence of the OmpA protein. Amino acid sequence alignment of FPH-1 and FTU-1 showed that the mature enzymes are 77% identical (Fig. 1). Amino acid sequences of the mature FPH-1 and FTU-1 enzymes were almost perfectly aligned, except for one amino acid insertion close to the C terminus of FPH-1. FPH-1 possesses all the conserved features characteristic of class A β-lactamases, namely, S-X-X-K (with the catalytic Ser70), S-D-N (this motif is S-D-S in FTU-1 but S-D-N in the majority of known class A β-lactamases), and K-T-G motifs (Fig. 1). E166, the residue essential for the deacylation of the acyl-enzyme intermediate, and R220, the residue involved in anchoring the carboxylate of the substrate, are also conserved in FPH-1, as well as cysteines 69 and 238, which are a characteristic conserved feature for all class A carbapenemases. FPH-1 shares only 24 to 33% amino acid sequence identities with other class A carbapenemases (Table 1), while the amino acid sequence identities with known class A β-lactamases are even lower. A dendrogram of representative class A β-lactamases (Fig. 2) indicates that FPH-1 and the closely related FTU-1 enzymes represent a new branch (with a 75.6% probability) on the prolific and fast-growing class A β-lactamase tree.
Fig 1.
Amino acid sequence alignment of the FTU-1 and FPH-1 β-lactamases. In bold are the conserved motifs, S70-X-X-K, S130-D-N/S, and K234-T/S-G, and E166 and R220. Underlined are the conserved cysteine 69 and 238 residues. The residues highlighted in gray indicate the predicted leader sequences of the FTU-1 and FPH-1 β-lactamases.
Table 1.
Identities of class A carbapenemases
| Enzyme | % identity with: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FPH-1 | FTU-1 | GES-1 | SME-1 | IMI-1 | NMC-A | KPC-2 | BIC-1 | SFC-1 | |
| FPH-1 | 77.6 | 24.4 | 30.7 | 33.0 | 33.0 | 30.4 | 28.1 | 31.1 | |
| FTU-1 | 77.6 | 24.1 | 31.5 | 31.8 | 31.8 | 30.7 | 29.2 | 29.7 | |
| GES-1 | 24.4 | 24.1 | 37.7 | 34.6 | 35.0 | 37.4 | 36.9 | 36.8 | |
| SME-1 | 30.7 | 31.5 | 37.7 | 73.6 | 73.6 | 58.3 | 59.9 | 62.5 | |
| IMI-1 | 33.0 | 31.8 | 34.6 | 73.6 | 97.7 | 53.0 | 57.4 | 63.0 | |
| NMC-A | 33.0 | 31.8 | 35.0 | 73.6 | 97.7 | 57.4 | 57.7 | 63.4 | |
| KPC-2 | 30.4 | 30.7 | 37.4 | 58.3 | 57.0 | 57.4 | 65.8 | 66.7 | |
| BIC-1 | 28.1 | 29.2 | 36.9 | 59.9 | 57.4 | 57.7 | 65.8 | 71.1 | |
| SFC-1 | 31.1 | 29.7 | 36.8 | 62.5 | 63.0 | 63.4 | 66.7 | 71.1 | |
Fig 2.
Dendrogram of mature class A β-lactamases. The tree was built using neighbor-joining analysis with the Jukes-Cantor model. The nodal robustness was evaluated by bootstrap analysis (1,000 replicates). GenBank accession numbers (in parentheses) for the evaluated carbapenemases were as follows: FPH-1 (ZP_05249935.1), FTU-1 (YP_513599.1), GES-1 (YP_003799986.1), BIC-1 (GQ260093.1), KPC-2 (AY034847.1), SFC-1 (AY354402.1), IMI-1 (U50278.1), NMC-A (Z21956.1), and SME-1 (Z28968.1). Other class A β-lactamases included BES-1 (AF234999.1), CepA (L13472.1), CGA-1 (AAL55262.1), CME-1 (CAB55427.1), CTX-M-1 (X92506.1), FAR-1 (AAB81957.1), FEC-1 (BAC53608.1), FONA-1 (CAB61635.1), OIH-1 (NP_693715.1), OXY-1 (P22391.1), PC1 (NP_878023.1), PER-1 (Z21957.1), PSE-1 (Q03170.1), ROB-1 (X52872.1), SED-1 (AAK63223.1), SFO-1 (AB003148.1), SHV-1 (AAD18054.1), SKO-1 (AAL85333.1), VEB-1 (AF010416.1), and TEM-1 (AAG47772.1).
MICs of β-lactam antibiotics and β-lactam–inhibitor combinations.
Production of the FPH-1 β-lactamase by E. coli JM83 resulted in the increase of MICs of all β-lactam antibiotics tested with the single exception of cefoxitin (Table 2). FPH-1 is evidently a more robust enzyme than the closely related FTU-1 β-lactamase. The FPH-1 β-lactamase produces very high levels of resistance to penicillins (ampicillin, oxacillin, amoxicillin, piperacillin, and ticarcillin; MICs of 16,000, 32,000, >2,000, 4,000, and 32,000 μg/ml, respectively), similar to those produced by the robust TEM-1 enzyme (16). Unlike the TEM-1 penicillinase, FPH-1 also elevates the MIC of ceftazidime (a broad-spectrum cephalosporin) 128-fold and the MIC of aztreonam (a monobactam) 256-fold, to 32 and 16 μg/ml, respectively. The ability to produce resistance to ceftazidime and aztreonam, along with the high level of resistance to the narrow-spectrum cephalosporin cephalothin, also distinguish FPH-1 from the closely related enzyme FTU-1 of F. tularensis. The latter enzyme does not elevate the MICs of either aztreonam or cephalothin and only slightly (4-fold) decreases the susceptibility of E. coli JM83 to ceftazidime (3). While production of FTU-1 does not change the MICs of carbapenem antibiotics (except for the 2-fold increase in the MIC of imipenem), FPH-1-producing E. coli JM83 is 8-fold less susceptible to imipenem and meropenem, 16-fold less susceptible to doripenem, and 64-fold less susceptible to ertapenem than the parental β-lactamase-negative strain. The elevations in the MICs of carbapenem antibiotics produced by FPH-1 are very similar to those produced by the clinically important GES-5 β-lactamase, expressed under identical conditions (in the same E. coli strain from the same plasmid and under the same promoter) (15). While the MIC values of various penicillins conferred by FPH-1 slightly decrease in the presence of β-lactamase inhibitors, they remain above the levels that define strains as inhibitor resistant according to CLSI guidelines (9).
Table 2.
Antimicrobial susceptibility profile of E. coli JM83 expressing the FPH-1 β-lactamase
| Antimicrobial(s) | MIC(s) (μg/ml) |
|
|---|---|---|
| FPH-1 | Controla | |
| Ampicillin | 16,000 | 2 |
| Ampicillin-clavulanic acidb | 4,000 | 1 |
| Ampicillin-sulbactamb | 8,000 | 2 |
| Ampicillin-tazobactamb | 8,000 | 2 |
| Ampicillin-sulbactam (2:1)c | 500/250 | 2/1 |
| Amoxicillind | >2,000 | 4 |
| Amoxicillin-clavulanic acid (2:1)c | 32/16 | 4/2 |
| Oxacillin | 32,000 | 128 |
| Piperacillin | 4,000 | 2 |
| Piperacillin-clavulanic acidb | 1,000 | 2 |
| Piperacillin-sulbactamb | 2,000 | 2 |
| Piperacillin-tazobactamb | 2,000 | 2 |
| Ticarcillin | 32,000 | 2 |
| Ticarcillin-clavulanic acide | 4,000 | 2 |
| Cephalothin | 1,000 | 4 |
| Cefuroxime | 16 | 2 |
| Ceftazidime | 32 | 0.25 |
| Cefotaxime | 0.25 | 0.03 |
| Aztreonam | 16 | 0.06 |
| Cefoxitin | 4 | 4 |
| Imipenem | 2 | 0.25 |
| Meropenem | 0.25 | 0.03 |
| Doripenem | 0.5 | 0.03 |
| Ertapenem | 0.25 | 0.004 |
Value for the control strain E. coli JM83 with the pHF016 vector.
Clavulanic acid, sulbactam, and tazobactam were used at a constant concentration of 4 μg/ml.
The β-lactam and β-lactamase inhibitor (sulbactam or clavulanic acid) ratio was held at 2:1 according to the CLSI standard recommendation.
The maximum solubility of amoxicillin in Mueller-Hinton II broth is 2,000 μg/ml.
Clavulanic acid was used at a constant concentration of 2 μg/ml according to the CLSI standard recommendation.
Kinetic analysis of the FPH-1 β-lactamase.
The FPH-1 β-lactamase was purified to apparent homogeneity (greater than 95% as evaluated by SDS-PAGE) for kinetics studies. In agreement with the MIC values, FPH-1 exhibited robust catalytic efficiency against penicillin antibiotics (Table 3). Despite the low relative affinity of the enzyme for ampicillin, oxacillin, piperacillin, and ticarcillin (in the range of 77 to 220 μM), high turnover numbers (kcat values of 3,700, 1,000, 530, and 620 s−1 for ampicillin, oxacillin, piperacillin, and ticarcillin, respectively) are responsible for the high catalytic efficiencies of the enzymes with these antibiotics (the kcat/Km ratios for ampicillin, oxacillin, piperacillin, and ticarcillin are 1.6 × 107, 1.3 × 107, 6.3 × 106, and 7.7 × 106 M−1 s−1, respectively). The catalytic efficiency of FPH-1 with cephalothin is 3 orders of magnitude higher than that of FTU-1, in good correlation with the high MIC value of this narrow-spectrum cephalosporin against FPH-1-producing E. coli JM83. The extended-spectrum cephalosporin cefuroxime is hydrolyzed by FPH-1 more than 200-fold less efficiently than cephalothin, due to a 5-fold increase in the Km value and an approximate 47-fold decrease in the kcat value against this antibiotic. The kcat/Km value of FPH-1 for aztreonam is 3.2 × 104 M−1 s−1, and the production of the enzyme results in a 256-fold increase in the MIC for this monobactam antibiotic, to a level of 16 μg/ml. It is of interest that the catalytic efficiency of FPH-1 against cefotaxime is almost an order of magnitude higher than that for ceftazidime, while FPH-1 produces a much higher increase in the MIC for ceftazidime than for cefotaxime (128- versus 8-fold increase). These seemingly controversial results can be explained by the poor penetration rate of ceftazidime through the bacterial outer membrane in comparison to the penetration rates of other broad-spectrum cephalosporins (18a). As we demonstrated recently, even the deacylation-deficient β-lactamase, when produced in large quantities, is capable of producing resistance to ceftazidime by a covalent trapping mechanism (2). Of importance, FPH-1 was capable of hydrolyzing all four carbapenem antibiotics tested, the time courses for which were linear. Despite the sluggish turnover numbers (kcat of 0.05 to 0.15 s−1) for imipenem, meropenem, ertapenem, and doripenem, FPH-1 exhibited catalytic efficiencies against these carbapenem antibiotics in the range of 3 × 104 to 3.5 × 105 M−1 s−1. Thus, FPH-1 is a much more robust carbapenemase than the closely related FTU-1, whose catalytic efficiency against the corresponding carbapenems is 2 to 3 orders of magnitude lower (3). The catalytic efficiency of FPH-1 against carbapenem antibiotics and the corresponding MIC values very closely match those reported for the GES-5 carbapenemase (15). The dissociation constants for the β-lactamase inhibitors tazobactam, clavulanic acid, and sulbactam with FPH-1 are 4, 7.5, and 220 μM, respectively. Thus, all three inhibitors have significantly higher affinity for FPH-1 than the closely related FTU-1 β-lactamase, whose corresponding affinities to tazobactam, clavulanic acid, and sulbactam are 300, 1,000, and 2,300 μM (3).
Table 3.
Kinetic substrate profilea of the FPH-1 β-lactamase
| Antimicrobial | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Ampicillin | 3,700 ± 200 | 220 ± 30 | (1.6 ± 0.3) × 107 |
| Oxacillin | 1,000 ± 100 | 77 ± 6 | (1.3 ± 0.1) × 107 |
| Piperacillin | 530 ± 20 | 83 ± 8 | (6.3 ± 0.7) × 106 |
| Ticarcillin | 620 ± 20 | 80 ± 6 | (7.7 ± 0.6) × 106 |
| Cephalothin | 280 ± 10 | 59 ± 6 | (4.7 ± 0.5) × 106 |
| Cefuroxime | 6.0 ± 0.3 | 290 ± 20 | (2.1 ± 0.2) × 104 |
| Ceftazidime | NAb | NA | (2.3 ± 0.2) × 103 |
| Cefotaxime | NA | NA | (2.0 ± 0.2) × 104 |
| Aztreonam | NA | NA | (3.2 ± 0.2) × 104 |
| Imipenem | 0.15 ± 0.01 | 0.42 ± 0.04 | (3.5 ± 0.4) × 105 |
| Meropenem | 0.093 ± 0.003 | 3.1 ± 0.3 | (2.9 ± 0.3) × 104 |
| Ertapenem | 0.11 ± 0.01 | 1.4 ± 0.1 | (7.8 ± 0.6) × 104 |
| Doripenem | 0.052 ± 0.002 | 0.8 ± 0.1 | (6 ± 1) × 104 |
| Nitrocefin | 1,000 ± 100 | 54 ± 8 | (1.9 ± 0.3) × 107 |
Values are means ± standard deviations.
NA, not applicable.
Conclusion.
In this study we have described the substrate profile and steady-state kinetic characteristics of a novel class A β-lactamase from F. philomiragia, FPH-1. This enzyme shares 77% amino acid sequence identity with the FTU-1 β-lactamase from F. tularensis, an enzyme that is intrinsic in this species. Like FTU-1, FPH-like proteins have been found in all F. philomiragia isolates sequenced to date, a strong indication that FTU-/FPH-type β-lactamases are native to the genus Francisella. These enzymes form a new and distinct branch of the class A β-lactamase tree. Despite their close amino acid sequence homologies and their belonging to the same bacterial genus, these enzymes have vastly different antibiotic resistance profiles. The FTU-1 β-lactamase confers a narrow spectrum of resistance to β-lactams, limited mainly to penicillins. In stark contrast, FPH-1 confers a much broader spectrum and higher levels of resistance that include, in addition to penicillins, extended-spectrum and broad-spectrum cephalosporins and the monobactam aztreonam. Unlike FTU-1, which elevates the MIC of imipenem 2-fold exclusively, FPH-1 elevates the MICs of imipenem, meropenem, ertapenem, and doripenem 8- to 64-fold. The abilities of the closely related FTU-1 and FPH-1 enzymes to confer distinct antibiotic resistance profiles represent yet another example of the evolutionary plasticity of class A β-lactamases.
ACKNOWLEDGMENT
This work was supported by grant 1R01AI089726 from the National Institutes of Health.
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Ambler RP, et al. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antunes NT, Frase H, Toth M, Mobashery S, Vakulenko SB. 2011. Resistance to the third-generation cephalosporin ceftazidime by a deacylation-deficient mutant of the TEM β-lactamase by the uncommon covalent-trapping mechanism. Biochemistry 50:6387–6395 [DOI] [PubMed] [Google Scholar]
- 3. Antunes NT, Frase H, Toth M, Vakulenko SB. 2012. The class A β-lactamase FTU-1 is native to Francisella tularensis. Antimicrob. Agents Chemother. 56:666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bae IK, et al. 2007. Genetic and biochemical characterization of GES-5, an extended-spectrum class A β-lactamase from Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 58:465–468 [DOI] [PubMed] [Google Scholar]
- 5. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant enterobacteria. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 6. Bush K. 2010. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit. Care 14:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaibi EB, Sirot D, Paul G, Labia R. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic, and biochemical characteristics. J. Antimicrob. Chemother. 43:447–458 [DOI] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement, approved standard, 7th ed, CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Copeland RA. 2000. Enzymes: a practical introduction to structure, mechanism, and data analysis, 2nd ed John Wiley & Sons, New York, NY [Google Scholar]
- 11. Dixon M. 1953. The determination of enzyme inhibitor constants. Biochem. J. 55:170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Falagas ME, Karageorgopoulos DE. 2009. Extended-spectrum β-lactamase-producing organisms. J. Hosp. Infect. 73:345–354 [DOI] [PubMed] [Google Scholar]
- 13. Fisher JF, Meroueh SO, Mobashery S. 2005. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105:395–424 [DOI] [PubMed] [Google Scholar]
- 14. Fonseca F, et al. 2007. Biochemical characterization of SFC-1, a class A carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 51:4512–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frase H, Shi Q, Testero SA, Mobashery S, Vakulenko SB. 2009. Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of β-lactamases. J. Biol. Chem. 284:29509–29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giakkoupi P, et al. 2001. Substitution of Thr for Ala-237 in TEM-17, TEM-12 and TEM-26: alterations in β-lactam resistance conferred on Escherichia coli. FEMS Microbiol. Lett. 201:37–40 [DOI] [PubMed] [Google Scholar]
- 17. Girlich D, Poirel L, Nordmann P. 2010. Novel Ambler class A carbapenem-hydrolyzing β-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 54:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Naas T, Vandel L, Sougakoff W, Livermore DM, Nordmann P. 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38:1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a. Nikaido H, Normark S. 1987. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol. Microbiol. 1:29–36 [DOI] [PubMed] [Google Scholar]
- 19. Nordmann P, Poirel L. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321–331 [DOI] [PubMed] [Google Scholar]
- 20. Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poirel L, et al. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Queenan AM, et al. 2006. SME-3, a novel member of the Serratia marcescens SME family of carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 50:3485–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Queenan AM, et al. 2000. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44:3035–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rasmussen BA, et al. 1996. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang YJ, Wu PJ, Livermore DM. 1990. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 34:755–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yigit H, et al. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]