Abstract
For many bacterial infections, noninherited mechanisms of resistance are responsible for extending the term of treatment and in some cases precluding its success. Among the most important of these noninherited mechanisms of resistance is the ability of bacteria to form biofilms. There is compelling evidence that bacteria within biofilms are more refractory to antibiotics than are planktonic cells. Not so clear, however, is the extent to which this resistance can be attributed to the structure of biofilms rather than the physiology and density of bacteria within them. To explore the contribution of the structure of biofilms to resistance in a quantitative way, we developed an assay that compares the antibiotic sensitivity of bacteria in biofilms to cells mechanically released from these structures. Our method, which we apply to Escherichia coli and Staphylococcus aureus each with antibiotics of five classes, controls for the density and physiological state of the treated bacteria. For most of the antibiotics tested, the bacteria in biofilms were no more resistant than the corresponding populations of planktonic cells of similar density. Our results, however, suggest that killing by gentamicin, streptomycin, and colistin is profoundly inhibited by the structure of biofilms; these drugs are substantially more effective in killing bacteria released from biofilms than cells within these structures.
INTRODUCTION
Inherited antibiotic resistance has been, remains, and, alas, will almost certainly continue to be a major reason antibiotic treatment fails. It is, however, not the only reason. For some infections with genetically susceptible bacteria, treatment fails due to mechanisms collectively known as noninherited resistance (35). Perhaps the most clinically significant form of noninherited resistance is biofilms. Bacteria embedded in these polysaccharide matrices are more refractory to antibiotics than they are when they are planktonic cells in liquid culture (2, 22, 36, 40), and a variety of mechanisms have been proposed to account for why this is the case (2, 9, 11, 16, 31, 34, 39, 52). But how, in a quantitative and clinically relevant way, does one compare the sensitivity of these embedded bacteria to corresponding populations of planktonic cells and determine the contribution of the structure of the biofilm to the sensitivity of bacteria to antibiotics?
One approach to addressing these pharmacodynamics (PD) questions has been to compare the MICs of antibiotics for bacteria in biofilms (bMIC) to the corresponding MICs of those bacteria and drugs estimated by standard CLSI (13) liquid culture protocols (5, 12, 27, 39, 45, 49–51). A limitation of this method is an inability to control for the contribution of the physiological state and density of the bacteria in the biofilms in the way they are controlled for in the CLSI MIC protocol. The susceptibility of bacteria to antibiotics varies with their physiological state; for example, bacteria at stationary phase are far more refractory to antibiotics than are exponentially growing populations (10, 19). Moreover, the MICs of antibiotics increase with densities and, for some antibiotics, profoundly so (43, 53, 57).
Another way to address these questions is to compare the antibiotic sensitivity of wild-type bacteria with mutants or strains of the same species with different capacities to form biofilms (5, 31, 39, 44, 47). This approach has been particularly useful for studies of the mechanisms underlying the decreased susceptibility to antibiotics inherent in biofilms. However, since genetically different strains are being compared, this method cannot exclude the possibility of pleiotropic effects of the genes affecting biofilm formation and structure. Thus, this approach is limited in its ability to quantify the extent to which biofilms confer increased resistance.
In this study, we use a simple, quantitative assay to determine the efficacy of different classes of bactericidal antibiotics to kill Staphylococcus aureus and Escherichia coli as planktonic cells and within biofilms. As our measure of efficacy, we consider the extent of kill over a defined period. Our method is, in some ways, similar to that used by Gilbert and colleagues (17, 18, 21) in that we compare the susceptibilities of bacteria in biofilms to those of bacteria released into the medium. Our results suggest that for most of the antibiotics examined, the primary driver of the relative antibiotic refractoriness of bacteria in biofilms is the density and/or physiological state of the cells rather than the physical structure of the habitat. For some drugs, however, the physical structure of the biofilm contributes to this phenotypic resistance. The procedures used in this study can be employed to explore the effect of structured habitats on the relative sensitivities of different species and strains of bacteria to a variety of potential antimicrobials.
MATERIALS AND METHODS
Culture and sampling media.
All bacterial strains were maintained on Luria-Bertani (LB) medium. For E. coli biofilms, bacteria were cultured in LB broth. S. aureus biofilms were grown in tryptic soy broth (TSB). For estimation of cell densities, samples were diluted in sterile saline and plated on LB plates. All medium components were purchased from Difco (Sparks, MD).
Antibiotics.
Gentamicin, vancomycin, oxacillin, ciprofloxacin, ampicillin, colistin, streptomycin, and tetracycline were obtained from Sigma-Aldrich Co. (St. Louis, MO). Stock solutions were prepared as a 10% solution in sterile water. Daptomycin (Cubist Pharmaceuticals, Lexington, MA) was obtained from the Emory University Hospital Pharmacy and suspended as a 10% solution in sterile water, with the weight adjusted to account for the carrier compounds. For daptomycin challenge, the medium was supplemented with 50 mg/liter CaCl2.
Bacterial strains.
E. coli strain MG1655 csrA was a gift from Tony Romeo. The strain has a kanamycin resistance cassette inserted into the csrA gene, which leads to increased biofilm formation primarily via increased production of extracellular polysaccharide (58). S. aureus strain 35556 (29), a methicillin-sensitive strain commonly used to study S. aureus biofilms (15, 26, 33, 60), was a gift from Bill Shafer.
MICs.
MICs were determined using a modification of the CLSI broth microdilution assay (13). Bacteria were grown overnight in medium used for the biofilm assay, serially diluted, and inoculated into a 2-fold antibiotic dilution series such that the final cell density was ∼105 CFU/ml. After 18 h of incubation at 37°C with agitation, the plates were assessed by eye and by optical density (OD). The MIC was determined to be the lowest concentration which had an OD630 less than or equal to 25% of the antibiotic-free value. For gentamicin, which induces clumping, the lowest concentration with no visible clumps was designated the MIC.
Biofilm growth, disruption, and estimation.
To form biofilms, 1 ml of 1:100 dilutions of overnight cultures was introduced into 24-well polystyrene microtiter plates (Fisher Scientific, Pittsburgh, PA). The plates were incubated without shaking for 24 h at the optimal temperature for biofilm formation: 25°C for E. coli and 37°C for S. aureus. Planktonic cultures were initiated and grown under identical conditions, but with vigorous rotation to reduce biofilm formation.
To prepare the biofilms, the planktonic cells were removed by decanting the medium and the wells were gently washed twice with sterile saline to remove loosely adhered cells. After air drying for 5 min, 1 ml of medium was added to each well. Cultures were disrupted by using a Tissue Tearor homogenizer (BioSpec Products, Inc., Bartlesville, OK) fitted with a sterile wooden applicator, scrubbing the sides and bottoms of the wells for 10 s. The density of the suspended cells was estimated by serial dilution and plating on LB. A crystal violet assay (46) was also used to quantitate the extent of biofilm development.
Calibration experiments.
To determine the period of disruption required for optimal cell release, biofilms were grown as described above. After the addition of fresh medium, the planktonic phase was carefully sampled so as not to disturb the biofilms (T = 0). The biofilms were then disrupted for 10 s, 20 s, or 30 s. After each disruption, the planktonic phase was sampled (T = 10, T = 20, and T = 30, respectively), and the density of the sampled cells was estimated. A complementary experiment was performed, except instead of sampling and plating the cells, the remaining attached biomass was quantified using a crystal violet assay (46).
To assess the effect of the experimental design on the stability of the biofilms, the percentage of cells within the biofilm was estimated before and after a 3-h incubation at 37°C. To estimate the cell density within biofilms, planktonic cells were carefully sampled without disturbing the biofilm. The biofilms were then disrupted for 10 s with the modified Tissue Tearor. Sampling after disruption yielded the total cell density. The biofilm density was calculated by subtracting the planktonic density from the total density. The percent biofilm was calculated as follows: (biofilm density/total density) × 100. For this experiment, biofilms were grown as described above. Overnight samples were collected without any further manipulation. T = 0 samples were taken after the biofilms had been prepared for the assay (i.e., washed, air dried, and medium replaced). T = 3 samples were taken after the biofilms had been prepared and incubated statically at 37°C for 3 h.
Time-kill experiments.
Biofilms were prepared as indicated above, replacing the medium with 1 ml of medium with or without antibiotics. The planktonic cells were prepared by diluting overnight cultures 1:20 into fresh medium with or without antibiotics. These dilutions were then aliquoted into 24-well plates, 1 ml per well. Disrupted conditions (both planktonic disrupted and biofilm disrupted) were homogenized at T = 0 and every hour thereafter. All treatment classes were incubated at 37°C for 3 h, but biofilm conditions were incubated statically and planktonic conditions were incubated with vigorous shaking. At the end of the exposure period, all of the cultures were disrupted for 10 s, and total viable cell densities were estimated. The results are presented as relative kill, which is calculated by dividing the density of cells at T = 3 by the initial cell density (T = 0).
Statistical analysis.
Statistical analysis was performed in JMP 9 (SAS Institute, Inc., Cary, NC) and SPSS (IBM, Armonk, NY). All drug treatments were assessed with one-way analysis of variance (ANOVA) comparing growth conditions with Tukey's HSD post hoc analysis for all pairwise comparisons. For drugs where Levene's test for homogeneity of variance was significant, Welch's ANOVA was used followed by Games-Howell post hoc analysis. Statistical outliers, as determined by a Grubb's test, were removed from the data set prior to analysis.
RESULTS
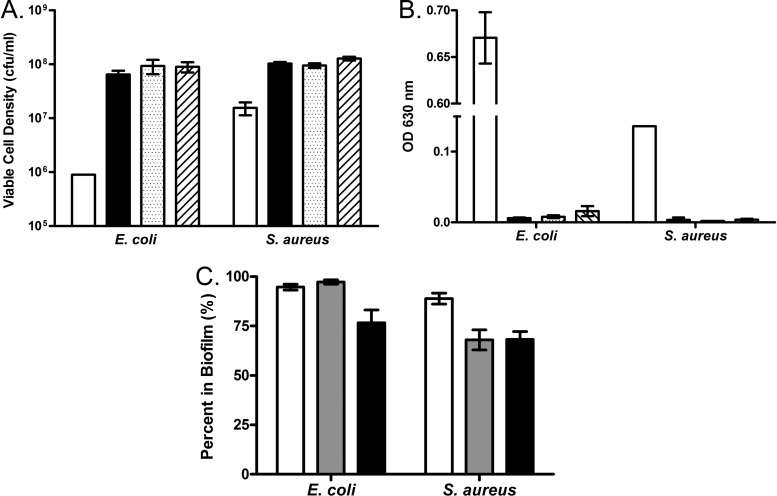
Figure 1 presents the results of calibration experiments to determine the effects of the experimental conditions on both E. coli and S. aureus biofilms. The efficacy of disruption was measured in two ways: as an increase in the density of viable cells in the planktonic phase (Fig. 1A) and as a decline in the surface-attached biomass as measured by a crystal violet assay (Fig. 1B). By both measures, a 10-s disruption was sufficient for the maximal release of biofilm cells; the slight increases seen with longer disruptions were not statistically significant.
Fig 1.
Calibration experiments for biofilm growth and disruption. (A, B) Extent of disruption over time. Biofilms were grown and prepared as described for the time-kill experiments, followed by disruption for 0, 10, 20, or 30 s. White bars, no disruption; black bars, 10-s disruption; dotted bars, 20-s disruption; hatched bars, 30-s disruption. (A) Extent of disruption as measured by an increase in viable cells in the planktonic phase; (B) extent of disruption as measured by a loss of crystal violet staining of attached biomass; (C) fraction of cells in the biofilm over the experimental time course. White bars in panel C represent the fraction of cells in the biofilm at 24 h, prior to any experimental manipulation. Gray bars represent biofilms after being washed and air dried and the addition of fresh medium. Black bars represent biofilms after being washed and air dried, the addition of fresh medium, and incubation at 37°C for 3 h. Error bars represent the standard errors.
Since our goal was to compare bacteria within biofilms to planktonic cells, it was critical to determine the extent to which our protocol (washing, addition of fresh medium, and incubation at 37°C) contributes to the release of cells from the biofilms (30). To answer this question, the fraction of cells in the biofilm was followed throughout the experimental procedure. Prior to manipulation, both E. coli and S. aureus are biased toward biofilm formation; 94.7% and 88.9% of cells, respectively, in these 24-hour cultures are in the biofilm (Fig. 1C). Washing the biofilms and adding fresh medium had no apparent effect on the distribution of biofilm and planktonic cells for E. coli (97.3% cells in biofilm) (Fig. 1C) but induced substantial shedding in S. aureus (68% remaining in the biofilm). As anticipated from reference 30, incubation at 37°C for 3 h induced shedding in the E. coli biofilms; however, the majority of the cells (76.6%) are still in the biofilm phase. S. aureus biofilms did not show the same propensity to shed during this 3-h incubation; 68.3% were present in the biofilm fraction.
The MICs for the antibiotics and bacteria used in this study were estimated with a serial dilution procedure as described in reference 13, but using the medium that promotes biofilm formation: LB for E. coli and TSB for S. aureus (Table 1). Both LB and TSB resulted in higher estimated MICs for some drugs relative to that observed in standard Mueller-Hinton II medium (data not shown).
Table 1.
MICs for strains and media used in this studya
| Drug | MIC |
|
|---|---|---|
| S. aureus 35556 | E. coli MG1655 csrA | |
| Gentamicin | 1.25 | |
| Daptomycinb | 2.50 | |
| Vancomycin | 0.63 | |
| Oxacillin | 0.16 | |
| Ciprofloxacin | 0.31 | 0.08 |
| Ampicillin | 5.00 | |
| Colistin | 0.31 | |
| Streptomycin | 10.00 | |
| Tetracycline | 1.25 | |
Seed cultures and assay cultures were grown in biofilm media: LB for E. coli and TSB for S. aureus. All antibiotic concentrations are in grams/liter.
Supplemented with 50 mg/liter CaCl2 and compared to a control culture containing 50 mg/liter CaCl2.
To assess the contribution of physical structure to the antibiotic resistance of biofilms, the intact and disrupted biofilm cultures were exposed to 40× MIC of drugs from different classes. The disrupted subset was subjected to mechanical disruption at T = 0 and every hour throughout the course of the experiment to minimize reseeding of the biofilm. To determine whether the cells released from the disrupted biofilms are physiologically similar to cells in an aerated planktonic culture, an equivalent density (∼108 CFU/ml) of cells grown in well-agitated liquid culture were also challenged with antibiotics. To control for the effects of cell damage due to disruption, planktonic cultures were also subjected to periodic disruption. In sum, there are four classes with the following designations: (i) biofilm, cells within biofilms; (ii) disrupted biofilm, cells mechanically released from biofilms; (iii) planktonic, cells from cultures grown under planktonic conditions; and (iv) planktonic disrupted, planktonic cultures with periodic disruption.
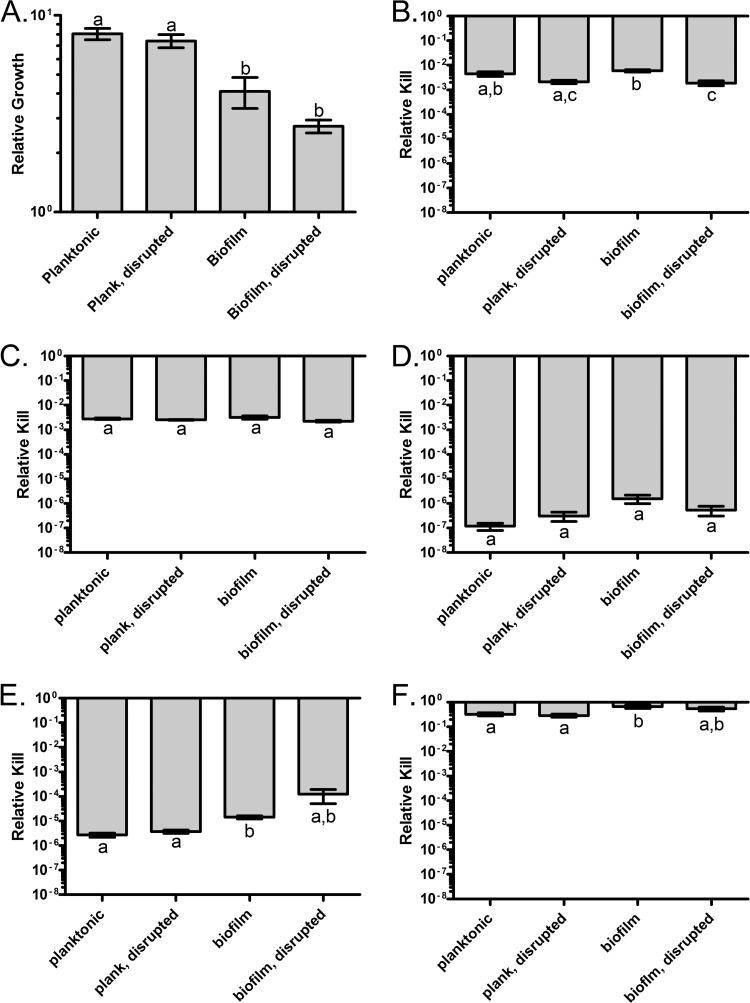
The results of this experiment with E. coli are presented in Fig. 2. In the absence of drugs, there is a net increase in the densities of the bacteria in all cultures, though planktonic conditions showed a greater increase in density than biofilm conditions (Fig. 2A). Mechanical disruption did not affect the number of viable cells in either class. For ciprofloxacin (Fig. 2C) and colistin (Fig. 2D), there were no significant differences in the fraction of cells surviving exposure for any of the four treatment classes. There were, however, substantial differences in the extent to which these two antibiotics kill, an ∼2.5-log decrease for ciprofloxacin and a 7-log reduction in the density of viable cells exposed to colistin. For both streptomycin (Fig. 2E) and tetracycline (Fig. 2F), the cells in the biofilm were more refractory to treatment than the corresponding planktonic and disrupted planktonic controls. Due to the high variance in survival, the cells released from the biofilm were statistically indistinguishable from the other treatment classes. However, the absolute extent of kill of these two antibiotics differed considerably, with streptomycin being far more bactericidal than tetracycline. The last pattern of susceptibility is that seen with ampicillin (Fig. 2B). For this drug, mechanical disruption increased the susceptibility to killing, but there was no evidence that cells in biofilms were less susceptible to this beta-lactam antibiotic than planktonic cells.
Fig 2.
Survival of E. coli MG1655 csrA biofilm and planktonic cells in the presence of high concentrations of antibiotics. Cells were exposed to 40× MIC of the indicated drugs, and viable cell densities were estimated at 3 h. (A) Control, no drug; (B) ampicillin; (C) ciprofloxacin; (D) colistin; (E) streptomycin; (F) tetracycline. Bars represent the standard errors. Letters represent statistically homogeneous subsets with an α value of 0.05.
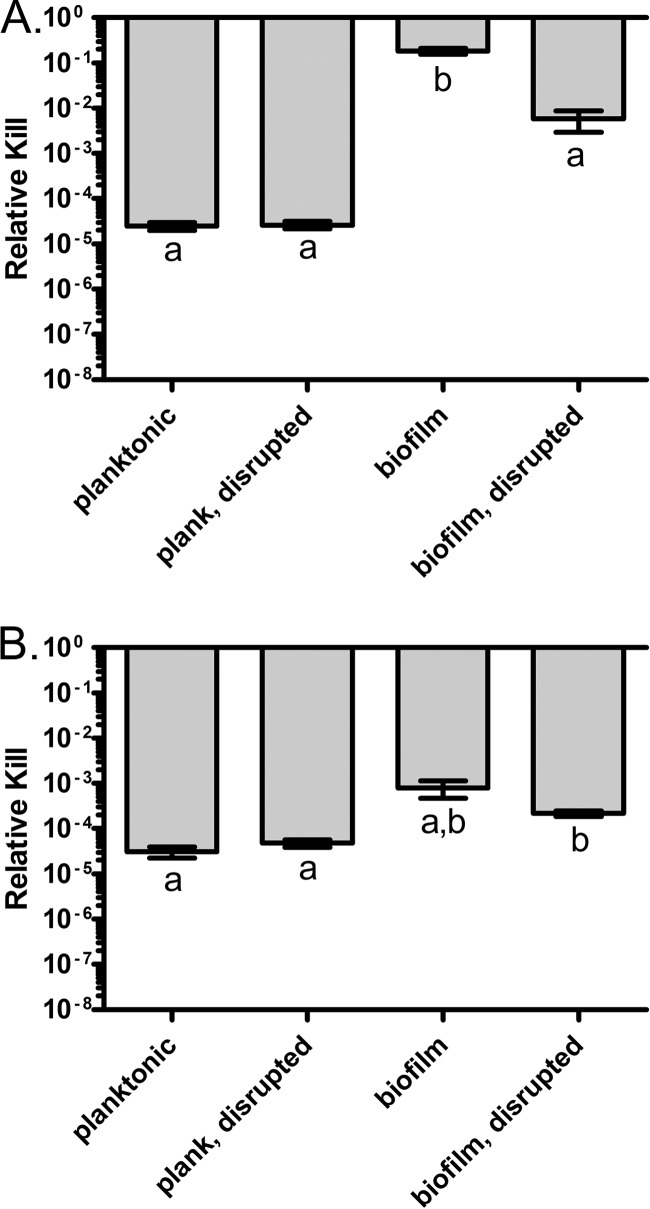
Due to the extent of kill by streptomycin and colistin in the above 40× MIC challenge experiments, differences in susceptibility among the four treatment classes may not be detectable. To determine if this was the case, we performed similar time-kill experiments with these drugs at 10× MIC (Fig. 3). At this concentration, colistin was far more effective in killing planktonic cells and cells released from the biofilm than those in intact biofilms (Fig. 3A). While it is clear that streptomycin at 10× MIC was slightly more effective in killing planktonic cells than cells released from biofilms, it is less clear whether there is any difference in killing between planktonic cells and cells within biofilms (Fig. 3B). By a Games-Howell post hoc analysis (23), this difference in sensitivity is not statistically significant, but with the less conservative Tukey's honestly significant difference post hoc analysis (28), intact biofilms are statistically less sensitive to streptomycin than are the other three classes.
Fig 3.
Survival of E. coli MG1655 csrA biofilm and planktonic cells in the presence of low concentrations of colistin and streptomycin. Cells were exposed to 10× MIC of colistin or streptomycin, and viable cell densities were estimated at 3 h. (A) Colistin; (B) streptomycin. Bars represent the standard error. Letters represent statistically homogeneous subsets with an α value of 0.05.
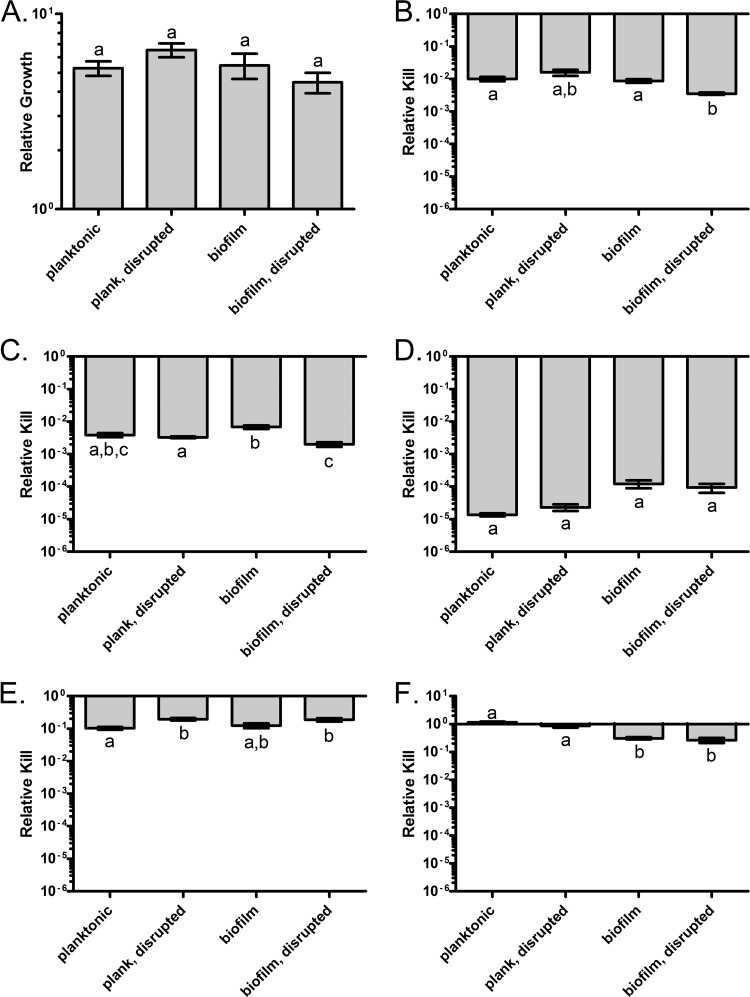
In Fig. 4, the results of time-kill experiments with S. aureus challenged with 40× MIC of various antibiotics are presented. In the absence of drug, the cells in all four conditions grew equivalently, with no indication of damage due to disruption (Fig. 4A). Each antibiotic had a unique pattern of susceptibility with respect to the four treatment groups. For ciprofloxacin, intact biofilms were as sensitive to killing as were planktonic cells (both intact and disrupted), but cells released from the biofilm were more susceptible to the drug, though the difference was not great (Fig. 4B). In contrast, daptomycin was slightly less effective against intact biofilms than planktonic cells and, as with ciprofloxacin, disruption of the biofilm structure increased the extent of killing (Fig. 4C). Gentamicin was the most effective antibiotic against S. aureus under all conditions (Fig. 4D). However, no differences were seen between the four culture conditions. As seen with ampicillin and E. coli (Fig. 2B), disruption influenced the susceptibility of the cells to oxacillin, where intact planktonic and biofilm cells were equally sensitive to killing, while disrupted cells were slightly less sensitive (Fig. 4E). For vancomycin, disruption had no effect on drug susceptibility; intact and disrupted planktonic cells formed one homogeneous subset that did not overlap intact and disrupted biofilms (Fig. 4F). Unexpectedly, biofilms, either intact or disrupted, were more sensitive to drug killing than were planktonic cells.
Fig 4.
Survival of S. aureus 35556 biofilm and planktonic cells in the presence of high antibiotic concentrations. Cells were exposed to 40× MIC of the indicated drugs, and viable cell densities were estimated at 3 h. (A) Control, no drug; (B) ciprofloxacin; (C) daptomycin; (D) gentamicin; (E) oxacillin; (F) vancomycin. Bars represent the standard errors. Letters represent statistically homogeneous subsets with an α value of 0.05.
To determine whether the failure to detect differences in susceptibility to gentamicin among classes is an artifact of the high rate of kill of this drug at 40× MIC, this experiment was repeated with gentamicin at 10× MIC. The results are presented in Fig. 5. With the lower overall killing, the resistance inherent to biofilms is readily apparent, with no loss of viable cells after a 3-h exposure. In contrast, the cells released from the biofilm were as sensitive to killing as the cells in either planktonic class.
Fig 5.
Survival of S. aureus biofilms and planktonic cells in the presence of low concentrations of gentamicin. Cells were exposed to 10× MIC of gentamicin, and viable cell densities were estimated at 3 h. Bars represent the standard errors. Letters represent statistically homogeneous subsets with an α value of 0.05.
DISCUSSION
We examined the relative contributions of the physiological state of cells within biofilms and the physical structure of their habitat to the sensitivity of bacteria to antibiotics. For this, we compared the relative extent of antibiotic-mediated killing of E. coli and S. aureus within biofilms to that of cells released from these structures. As additional controls, we estimated the extent of kill of a similar density of planktonic bacteria that were or were not subjected to the disruption procedure used to suspend the bacteria within the biofilms. Five classes of antibiotics were used for each of these two species.
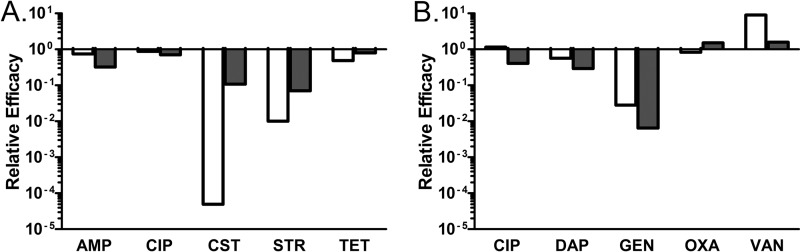
For the majority of the drugs tested, the relative susceptibility to antibiotics of bacteria within biofilms was comparable to that of the corresponding planktonic population of similar density. There were exceptions, however, which are summarized in Fig. 6. Colistin, streptomycin, and gentamicin were substantially more effective in killing planktonic cells than those embedded within biofilms. Ampicillin, tetracycline, and ciprofloxacin (for S. aureus) also manifest drug-related differences in the efficacy against bacteria in the different growth conditions. Although statistically significant, these differences were too small to be biologically or clinically important.
Fig 6.
Relative efficacy of antibiotics under different culture conditions. Relative efficacy is the ratio of the average relative kill of the given antibiotic in the given condition to the average relative kill of the antibiotic in intact biofilm conditions. Values less than 1 indicate that the drug is less effective against bacteria in biofilms than bacteria of the comparison state. The colistin, streptomycin, and gentamicin values were calculated from the experiments with 10× MIC; all other values were calculated from the experiments with 40× MIC. White bars are these ratios for planktonic cells, and the dark gray bars are these ratios for disrupted biofilm. (A) E. coli; (B) S. aureus.
For colistin, streptomycin, and gentamicin, the structure of the biofilm was important, putatively providing protection for the cells within. Surprisingly, residence within a biofilm was not always protective in this way; vancomycin was more effective in killing S. aureus in biofilms than it was in killing planktonic cells; however, the difference is not great and unlikely to be biologically relevant.
The results of our experiments also suggest that E. coli cells released from biofilms and suspended in liquid are physiologically different from planktonic cells that have never been within a biofilm. As measured by the extent of kill, colistin and streptomycin are less effective in killing cells released from biofilms than the corresponding population of planktonic cells. The opposite is true for ampicillin, though to a lesser degree. This difference between cells released from biofilms and planktonic cells of the same density was not observed for any of the antibiotics used in the S. aureus experiments. However, we do not interpret this result to mean that mechanically released S. aureus biofilm cells are physiologically identical to planktonic cells. Rather, with the antibiotics tested here, we were not able to detect such a difference.
The procedure used here was designed to assess the contribution of physical structure to the phenotypic resistance of biofilms without genetic or chemical methods that could be confounded by pleiotropic effects. While we cannot exclude the possibility that the mechanical disruption of the biofilm damaged the cells and changed their sensitivities to antibiotics, it is unlikely that such damage accounts for our results. Most of the drugs showed no change in sensitivity upon disruption of either planktonic or biofilm conditions, as would be expected if cell damage contributed to the observed differences in susceptibility to antibiotics. Only oxacillin differentially killed undisturbed and disrupted planktonic cells, but this disruption resulted in less rather than greater killing (Fig. 4E). Moreover, among the cultures not exposed to drugs, the disrupted cells grew at the same rate as the intact cells, indicating no gross loss of viability due to disruption.
It is well known that the efficacy of many antibiotics is correlated with the growth rate of the bacteria (10, 14, 56). The conditions utilized in this study do not allow for substantial growth during the challenge period; however, some growth does occur under all conditions (Fig. 2A and 4A). For E. coli, the bacteria in the biofilm or released from the biofilm grew to a lower density than planktonic cells that had never been within a biofilm. This raises the possibility that differences in rate of growth or capacity to saturate the resources (broadly, their physiological state) could contribute to differences in the extent of kill between planktonic and biofilm conditions. This is clearly not the case for S. aureus, where in the absence of antibiotics, under all four conditions considered, the bacteria grew to the same extent.
How do the results of the experiments performed here compare to those of other quantitative studies of the pharmacodynamics of antibiotics and bacteria in biofilms? In a series of studies in the 1990s, Gilbert and colleagues explored the contribution of the structure of biofilms to the antibiotic sensitivity of bacteria (17, 18, 21, 24). For their investigations, biofilms were formed and maintained by continuous perfusion of membranes (24). As in the present study, they compared the extent of kill over a defined period for the bacteria within the biofilms to that of cells mechanically or naturally released from the biofilms and to planktonic cells. They made these comparisons for four pairs of bacteria and antibiotics. As observed here, the structure of the biofilm did not contribute to the sensitivity of E. coli or staphylococci to ciprofloxacin (18, 21). In their study with tobramycin and Staphylococcus epidermidis, the bacteria within the biofilm were more resistant to the drugs than were those released from the biofilm (17), which is consistent with our results for gentamicin and S. aureus (Fig. 5).
The most surprising result of the current study was that the resistance of bacteria in biofilms to the majority of antibiotics studied can be attributed to the density and physiological state of the culture rather than their residence within biofilms. Neither the extent nor the dynamics (data not shown) of killing differed considerably between planktonic and biofilm cells for seven of the 10 drug-bacteria combinations. This is in line with the observation of Qu and colleagues (47) that multilayer biofilms of S. epidermidis were no more resistant to antibiotic challenge than were monolayer biofilms. Although the authors concluded that the increased resistance could be attributed to adherence of cells to surfaces, their experimental design does not enable them to distinguish the effects of adhesion from that which can be attributed to culture density. Our results show that high-density planktonic growth stimulates the same level of resistance as adherent biofilms, and disruption of biofilms (and loss of adherence) does not necessarily reduce resistance.
Our observation that bacteria released from biofilms are more susceptible than planktonic cells to colistin and aminoglycosides is particularly interesting in light of recent information on biofilm physiology. Unlike the other drugs tested, these antibiotics either target the cell membrane (colistin) (32) or are dependent on membrane function for uptake (streptomycin, gentamicin) (54). Together with the lack of structure-specific resistance to the other drugs, these results suggest that the membranes of biofilm cells may be different from those of planktonic cells and that the structure (i.e., the matrix) of the biofilm acts as a shield for these more sensitive cells. This is in line with several recent studies that have shown that the cell membrane physiology changes significantly during biofilm growth (1, 12, 45, 49, 55). Thus, a picture is beginning to emerge wherein membrane charge and physiology are different in biofilm cells relative to their planktonic counterparts, and this difference can contribute to the efficacy of antibiotic therapy. One interpretation of our results with the aminoglycosides and colistin is that the structure of the biofilm may confound, to some extent, the intrinsic sensitivity of the bacteria within biofilms to these drugs.
At present, the primary and often unique measure of the pharmacodynamics of antibiotics and their target bacteria is the MIC (3, 7, 25), which is also used as a measure of susceptibility (resistance) (13, 20, 42). While this in vitro measure of the potential clinical efficacy of antibiotics has the virtue of being readily estimated in a standardized way, it is restricted to relatively low densities of planktonic cells growing exponentially under conditions that are optimal for action of the drug (13). These conditions are limited and unlikely to be met in populations of bacteria in infected patients. Among the additional factors affecting the efficacy of antibiotics (4, 6–8, 10, 25, 37, 38, 41, 48, 53, 56, 57, 59) is the physical structure of the target population of bacteria and in particular their residence in the polysaccharide matrices of biofilms. The method employed in this study provides a simple and broadly applicable way to evaluate the PD of antibiotics and bacteria in biofilms. The application of this measure in combination with other in vitro measures of the PD of antibiotics will provide a broader foundation for the rational design of antibiotic therapy than that currently employed.
ACKNOWLEDGMENTS
This work was supported by a grant from the U.S. National Institutes of Health GM091875 (B.R.L.) and a grant from Proctor and Gamble Inc.
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson GG, O'Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 322:85–105 [DOI] [PubMed] [Google Scholar]
- 3. Andes D, Anon J, Jacobs MR, Craig WA. 2004. Application of pharmacokinetics and pharmacodynamics to antimicrobial therapy of respiratory tract infections. Clin. Lab. Med. 24:477–502 [DOI] [PubMed] [Google Scholar]
- 4. Andrew JH, Wale MC, Wale LJ, Greenwood D. 1987. The effect of cultural conditions on the activity of LY146032 against staphylococci and streptococci. J. Antimicrob. Chemother. 20:213–221 [DOI] [PubMed] [Google Scholar]
- 5. Antunes AL, et al. 2010. Application of a feasible method for determination of biofilm antimicrobial susceptibility in staphylococci. APMIS 118:873–877 [DOI] [PubMed] [Google Scholar]
- 6. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 7. Baudoux P, et al. 2007. Combined effect of pH and concentration on the activities of gentamicin and oxacillin against Staphylococcus aureus in pharmacodynamic models of extracellular and intracellular infections. J. Antimicrob. Chemother. 59:246–253 [DOI] [PubMed] [Google Scholar]
- 8. Bigger JW. 1944. Treatment of staphylococcal infections with penicillin—by intermittent sterilisation. Lancet ii:497–500 [Google Scholar]
- 9. Briandet R, et al. 2008. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl. Environ. Microbiol. 74:2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown MR, Collier PJ, Gilbert P. 1990. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 34:1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 46:1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ceri H, et al. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. CLSI 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI, Wayne, PA [Google Scholar]
- 14. Cozens RM, et al. 1986. Evaluation of the bactericidal activity of beta-lactam antibiotics on slowly growing bacteria cultured in the chemostat. Antimicrob. Agents Chemother. 29:797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cucarella C, et al. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donlan RM. 2000. Role of biofilms in antimicrobial resistance. ASAIO J. 46:S47–S52 [DOI] [PubMed] [Google Scholar]
- 17. Duguid IG, Evans E, Brown MR, Gilbert P. 1992. Effect of biofilm culture upon the susceptibility of Staphylococcus epidermidis to tobramycin. J. Antimicrob. Chemother. 30:803–810 [DOI] [PubMed] [Google Scholar]
- 18. Duguid IG, Evans E, Brown MR, Gilbert P. 1992. Growth-rate-independent killing by ciprofloxacin of biofilm-derived Staphylococcus epidermidis; evidence for cell-cycle dependency. J. Antimicrob. Chemother. 30:791–802 [DOI] [PubMed] [Google Scholar]
- 19. Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35:1824–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. EUCAST 2000. Determination of antimicrobial susceptibility test breakpoints. Clin. Microbiol. Infect. 6:570–572 [DOI] [PubMed] [Google Scholar]
- 21. Evans DJ, Allison DG, Brown MR, Gilbert P. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27:177–184 [DOI] [PubMed] [Google Scholar]
- 22. Fey PD. 2010. Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr. Opin. Microbiol. 13:610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Games PA, Keselman HJ, Rogan JC. 1981. Simultaneous pairwise multiple comparison procedures for means when sample sizes are unequal. Psychol. Bull. 90:594–598 [Google Scholar]
- 24. Gilbert P, Allison DG, Evans DJ, Handley PS, Brown MR. 1989. Growth rate control of adherent bacterial populations. Appl. Environ. Microbiol. 55:1308–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gillespie EL, Kuti JL, Nicolau DP. 2005. Pharmacodynamics of antimicrobials: treatment optimisation. Expert Opin. Drug Metab. Toxicol. 1:351–361 [DOI] [PubMed] [Google Scholar]
- 26. Gross M, Cramton SE, Gotz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69:3423–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harrison JJ, Ceri H, Stremick C, Turner RJ. 2004. Differences in biofilm and planktonic cell mediated reduction of metalloid oxyanions. FEMS Microbiol. Lett. 235:357–362 [DOI] [PubMed] [Google Scholar]
- 28. Hsiung TH, Olejnik S. 1994. Power of pairwise multiple comparisons in the unequal variance case. Commun. Stat. Simulat. 23:691–710 [Google Scholar]
- 29. Iordanescu S, Surdeanu M. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277–281 [DOI] [PubMed] [Google Scholar]
- 30. Jackson DW, et al. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khan W, et al. 2010. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int. Microbiol. 13:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koike M, Iida K, Matsuo T. 1969. Electron microscopic studies on mode of action of polymyxin. J. Bacteriol. 97:448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kristian SA, et al. 2004. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb. Pathog. 36:237–245 [DOI] [PubMed] [Google Scholar]
- 34. Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34:426–444 [DOI] [PubMed] [Google Scholar]
- 35. Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556–562 [DOI] [PubMed] [Google Scholar]
- 36. Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 37. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 38. MacKenzie FM, Gould IM. 1993. The post-antibiotic effect. J. Antimicrob. Chemother. 32:519–537 [DOI] [PubMed] [Google Scholar]
- 39. Mah TF, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 40. Martinez JL, Rojo F. 2011. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 35:768–789 [DOI] [PubMed] [Google Scholar]
- 41. McLaughlin K, et al. 2000. Increased risk for posttransplant lymphoproliferative disease in recipients of liver transplants with hepatitis C. Liver Transpl. 6:570–574 [DOI] [PubMed] [Google Scholar]
- 42. Mouton JW. 2002. Breakpoints: current practice and future perspectives. Int. J. Antimicrob. Agents 19:323–331 [DOI] [PubMed] [Google Scholar]
- 43. Naves P, Del Prado G, Ponte C, Soriano F. 2010. Differences in the in vitro susceptibility of planktonic and biofilm-associated Escherichia coli strains to antimicrobial agents. J. Chemother. 22:312–317 [DOI] [PubMed] [Google Scholar]
- 44. Nguyen D, et al. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ooi N, et al. 2010. XF-70 and XF-73, novel antibacterial agents active against slow-growing and nondividing cultures of Staphylococcus aureus including biofilms. J. Antimicrob. Chemother. 65:72–78 [DOI] [PubMed] [Google Scholar]
- 46. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 47. Qu Y, Daley AJ, Istivan TS, Rouch DA, Deighton MA. 2010. Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of Staphylococcus epidermidis biofilms to antibiotics. J. Antimicrob. Chemother. 65:1405–1411 [DOI] [PubMed] [Google Scholar]
- 48. Regoes RR, et al. 2004. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents Chemother. 48:3670–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rivardo F, Martinotti MG, Turner RJ, Ceri H. 2011. Synergistic effect of lipopeptide biosurfactant with antibiotics against Escherichia coli CFT073 biofilm. Int. J. Antimicrob. Agents 37:324–331 [DOI] [PubMed] [Google Scholar]
- 50. Sandoe JA, Wysome J, West AP, Heritage J, Wilcox MH. 2006. Measurement of ampicillin, vancomycin, linezolid and gentamicin activity against enterococcal biofilms. J. Antimicrob. Chemother. 57:767–770 [DOI] [PubMed] [Google Scholar]
- 51. Sepandj F, Ceri H, Gibb A, Read R, Olson M. 2004. Minimum inhibitory concentration (MIC) versus minimum biofilm eliminating concentration (MBEC) in evaluation of antibiotic sensitivity of Gram-negative bacilli causing peritonitis. Perit. Dial. Int. 24:65–67 [PubMed] [Google Scholar]
- 52. Shapiro JA, Nguyen VL, Chamberlain NR. 2011. Evidence for persisters in Staphylococcus epidermidis RP62a planktonic cultures and biofilms. J. Med. Microbiol. 60:950–960 [DOI] [PubMed] [Google Scholar]
- 53. Soriano F, Ponte C. 2009. Comment on: functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63:1301. [DOI] [PubMed] [Google Scholar]
- 54. Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Theilacker C, et al. 2011. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BMC Microbiol. 11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J. Gen. Microbiol. 132:1297–1304 [DOI] [PubMed] [Google Scholar]
- 57. Udekwu KI, Parrish N, Ankomah P, Baquero F, Levin BR. 2009. Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63:745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang X, et al. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56:1648–1663 [DOI] [PubMed] [Google Scholar]
- 59. Wiuff C, et al. 2005. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 49:1483–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. 2003. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob. Agents Chemother. 47:3407–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]