Abstract
The extensive use of antibiotics in medicine, the food industry, and agriculture has resulted in the frequent emergence of multidrug-resistant bacteria, which creates an urgent need for new antibiotics. It is now widely recognized that antimicrobial peptides (AMPs) could play a promising role in fighting multidrug-resistant bacteria. Antimicrobial peptide polybia-CP was purified from the venom of the social wasp Polybia paulista. In this study, we synthesized polybia-CP and studied its action mode of antibacterial activity. Our results revealed that polybia-CP has potent antibacterial activity against both Gram-positive and Gram-negative bacteria. The results from both the real bacterial membrane and the in vitro model membrane showed that polybia-CP is membrane active and that its action target is the membrane of bacteria. It is difficult for bacteria to develop resistance to polybia-CP, which may thus offer a new strategy for defending against resistant bacteria in medicine and the food and farming industries.
INTRODUCTION
In May 2011, an increased number of cases of enterohemorrhagic Escherichia coli (EHEC) infections with severe disease courses and fatalities occurred in Europe, which were caused by EHEC-contaminated food (e.g., vegetables, fruits, or meat). This event caused panic all over the world, for the isolated EHEC is resistant to antibiotics and caused severe economic losses in the agriculture industry. As we know, E. coli, one of the bacillus species, is widely distributed in nature and has a remarkable ability to survive under strong environmental stresses (18). In the food industry, bacillus contamination occurs often and always originates from soil, water, processing equipment, and the processing environment (1). Thus, the food safety situation is not so optimistic, and people should pay more attention to the problem of microbial contamination of food. In addition, nowadays the food industry suffers a growing pressure to reduce reliance on synthetic chemical preservatives. Consequently, alternative preservatives based on natural compounds and uncorrelated to the conventional multidrug-resistant mechanism are urgently needed.
At present, antimicrobial peptides (AMPs), which are heat tolerant and have rich sources (from prokaryotes [9], plants [14], insects [3], amphibians [17], and animals including humans [27]), have attracted more and more attention. AMPs have a broad antimicrobial spectrum and no cross-resistance compared with conventional antibiotics. Most conventional antibiotics target bacterial functions or growth processes (4), such as interfering with the synthesis of bacterial cell wall, essential bacterial enzymes, and proteins (8). The genome mutation of microbes could induce resistance to these antibiotics (12). Compared with conventional antibiotics, AMPs exhibit cytotoxity mainly by interacting with the lipid bilayers of membrane and destroying the integrity of cell membrane (23). It is difficult for microbes to develop resistance to them (24). In addition, AMPs could be degraded easily in vivo and their secondary metabolites are free amino acids, which prove that they have minimized side effects. Thus, AMPs may serve as a potentially significant group of food preservatives.
Polybia-CP belongs to a family of cationic antibacterial peptides and was originally isolated from the venom of the social wasp Polybia paulista (19). It is a typical α-helical, amphiphilic peptide and has limited toxicity against rat peritoneal mast cell and rat erythrocytes (19). In addition, its cytotoxity exhibition mode against tumor cells was membrane active (25). In the present study, we investigated the antimicrobial activity and the proposed mechanism of polybia-CP to support its potential to be used as a novel food preservative.
MATERIALS AND METHODS
Peptide synthesis.
Polybia-CP was synthesized by the stepwise solid-phase method using N-9-fluorenylmethoxy carbonyl (Fmoc) chemistry as previously reported (7). The peptides were purified by Sephadex gel column and reverse-phase high-performance liquid chromatography (HPLC; Waters) using a μBondapak C18 19-mm by 300-mm column with gradient elution of 20% to 75% CH3CN/H2O with 0.1% trifluoroacetic acid (TFA) at a flow rate of 8 ml/min. The atomic mass of the synthetic peptides was confirmed by electrospray ionization-mass spectrometry (ESI-MS).
Bacteria and reagents.
Bacterial strains used in this study were purchased from the American Type Culture Collection (ATCC): Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Bacillus subtilis (ATCC 23857), Staphylococcus aureus (ATCC 29213), and Staphylococcus epidermidis (ATCC 12228). All the bacteria were cultured in Mueller-Hinton (MH) broth. Prior to assays, the cells were grown overnight to stationary phase at 37°C in 5 ml MH broth. After incubation, 100 μl of bacteria was suspended in 5 ml of fresh MH broth (1/50 dilution) for an additional hour at 37°C to obtain a mid-log-phase culture.
1-N-Phenylnaphthylamine (NPN) was purchased from J&K Scientific Ltd. Egg yolk l-α-phosphatidylethanolamine (EYPE) and egg yolk l-α-phosphatidyl-dl-glycerol (EYPG) were purchased from Avanti Polar. Plasmid DNA (PBR322) was purchased from Fermentas. Propidium iodide (PI) was purchased from Invitrogen.
Antimicrobial assay.
The MIC was determined using a standard serial dilution method described previously (5). In brief, 100-μl 2-fold serial dilutions of peptides in a range from 1 to 500 μM in MH broth were arranged in sterile 96-well plates, after which 10-μl aliquots of the cell suspension (1 × 106 CFU/ml) were added to each well and MH broth was added to a final volume of 200 μl. The antibacterial activity of polybia-CP was evaluated by visible turbidity in each well after incubation at 37°C for 18 h. The MICs were expressed as the minimum concentration of each sample required for a visible inhibition of growth. The experiment was conducted in triplicate.
Bactericidal activity.
The minimum bactericidal concentration (MBC) of polybia-CP was determined using the serial dilution assay described with modifications. Briefly, 100 μl of bacteria was incubated at 37°C overnight in the presence of different concentrations of polybia-CP (serial 2-fold dilutions) dissolved in phosphate-buffered saline (PBS) to a final volume of 200 μl. Following incubation, appropriate aliquots were plated on MH agar plates to accurately determine the 99.9% killing. The number of surviving bacteria was determined after overnight incubation at 37°C. Controls consisting of 100 μl PBS without peptide were added.
Temperature stability test.
In order to determine the effect of temperature on the stability of polybia-CP, polybia-CP was incubated in the water bath with different temperatures (20°C, 40°C, 60°C, 80°C, and 100°C) first; then, they were cooled down to room temperature and the MICs of polybia-CP toward E. coli were determined according to the antimicrobial assay described above.
Extracting E. coli genomic DNA.
Extraction of genomic DNA from E. coli ATCC 25922 by the N-cetyl-N,N,N-trimethyl ammonium bromide (CTAB) method was performed according to a procedure described previously (21).
OM permeabilization assay.
The outer membrane (OM) permeabilization of E. coli by peptides was evaluated by using the hydrophobic NPN (1-N-phenylnaphthylamine) fluorescent probe (11, 22). E. coli cells were harvested, washed, and resuspended in 5 mM HEPES, pH 7.2, with an optical density at 600 nm (OD600) value of 0.5 ± 0.02. The concentrations of peptides were the same as those described in the previous section. One hundred microliters of E. coli and 50 μl of peptides were mixed with 50 μl of 40 mM NPN in a 96-well black fluorotiter plate; the control was carried out with 5 mM HEPES alone. An increase in fluorescence due to partitioning of NPN into the OM was recorded as a function of time until no further increase in intensity was observed. Excitation and emission wavelengths were set at 350 nm and 420 nm, respectively.
Confocal fluorescence microscopy.
E. coli cells were incubated in the presence of polybia-CP (50 μM, final concentration) or in buffer alone. The mixture was incubated for 2 h at room temperature. Then, PI (100 μg/ml) was added and the suspension was further incubated for 5 min at room temperature in the dark. Microscopic analysis was done with laser confocal scanning microscope (Zeiss LSM 710 Meta).
Scanning electron microscopic (SEM) examination of bacterial membrane.
Mid-log-phase E. coli cells were resuspended at 1 × 108 CFU/ml in sodium-phosphate buffer, at a pH of 7.4, supplemented with 100 mM NaCl, and then incubated with polybia-CP at room temperature. Controls were run in the absence of peptide solution. After 30 min, the cells were fixed with an equal volume of 5% (vol/vol) glutaraldehyde in 0.2 M sodium-cacodylate buffer, at a pH of 7.4. After 2 h of fixation at 4°C, the samples were filtered on Isopore filters (0.2 μm pore size, Millipore, Bedford, MA) and washed extensively with 0.1 M cacodylate buffer, at a pH of 7.4. The filters were then treated with 1% (wt/vol) osmium tetroxide, washed in 5% (wt/vol) sucrose in cacodylate buffer, and subsequently dehydrated with a graded series of ethanol. After lyophilization and gold coating, the samples were evaluated on a scanning electron microscope (JSM-6380Lv).
Calcein leakage assay.
The interaction and permeabilization of peptides against liposome were assayed by measuring calcein leakage. E. coli model cell membrane (EYPE-EYPG 70:30 mol %) was used in this study (16). Calcein-entrapped liposomes were prepared for use in dye leakage as described previously (26). First, the lipid was dissolved in chloroform. Then, the solvent was dried under a stream of nitrogen and the sample was kept under vacuum for at least 4 h to ensure complete solvent removal, and the obtained lipid films were hydrated in buffer (10 mM Tris, 154 mM NaCl, 0.1 mM EDTA in 18.2 MΩ Milli-Q water, pH 7.4) containing 70 mM calcein (fluorescein-bis-methyliminodiacetic acid; Sigma). Finally, the suspension was freeze-thawed in liquid nitrogen for nine cycles and extruded 30 times through polycarbonate filters (two stacked 0.1-μm-pore-size filters) using an Avanti Mini-Extruder (Avanti Polar Lipids Inc., Alabaster, AL). Calcein-entrapped vesicles were separated from free calcein by gel filtration chromatography on a Sephadex G-50 column. Entrapped large unilamellar vesicles (LUVs) in suspensions containing 100 μM lipids were incubated with various concentrations of the peptide (2 to 64 μM). The fluorescence of the released calcein was assessed with a spectrofluorometer (Tecan Infinite M200) at an excitation wavelength of 480 nm and an emission wavelength of 520 nm. Complete (100%) release was achieved via the addition of Triton X-100 to a final concentration of 1%. Spontaneous leakage was determined to be negligible at this time scale. The experiments were conducted at 25°C. The apparent percentage of calcein release was calculated in accordance with the following equation (28): release % = 100 × (F − F0/Ft − F0), in which F and Ft represent the fluorescence intensity prior to and after the addition of the detergent, respectively, and F0 represents the fluorescence of the intact vesicles.
Negative staining TEM.
Large unilamellar vesicles for transmission electron microscopy (TEM) analysis were prepared as described above for the calcein leakage assay. The liposomes in the presence or absence of polybia-CP were applied to glow-discharged carbon-coated copper grids and dried naturally. The grids were negative stained with (2% [wt/vol]) phosphotungstic acid for 1 min. Then, the additional phosphotungstic acid was soaked up by filter paper, and electron micrographs were then recorded (JEM1230) at nominal magnification (×20,000 to ×40,000) with an accelerating voltage of 120 kV.
DNA-binding assay.
Gel retardation experiments were performed for polybia-CP as described previously (2). Briefly, 300 ng of the plasmid DNA was mixed with increasing amounts of peptide in 30 μl of binding buffer (10 mM Tris-HCl, 1 mM EDTA buffer, pH 8.0). Reaction mixtures were incubated at room temperature for 30 min. Subsequently, native loading buffer was added (10% Ficoll 400, 10 mM Tris-HCl, pH 7.5, 50 mM EDTA, 0.25% bromophenol blue, and 0.25% xylene cyanol), and a 20-μl aliquot was subjected to 1.5% agarose gel electrophoresis. The migration of DNA was detected by the fluorescence of ethidium bromide (Bio-Rad).
Absorption titration.
DNA and polybia-CP were dissolved in 10 mM Tris-HCl buffer (pH 7.2) and then mixed to obtain various DNA/polybia-CP samples with constant DNA concentration and increasing polybia-CP concentrations (0, 5, 10, 25, and 50 μM). Transmittance of the mixed solutions was measured in the range of wavelengths from 220 to 320 nm (Nanodrop 2000). All measurements were made at a 10-mm optical path length at room temperature.
RESULTS
Antimicrobial activity and bactericidal activity.
To investigate the antibacterial activity of polybia-CP, it was synthesized by stepwise solid-phase assay, purified by HPLC, and confirmed by ESI-MS. The primary sequence of polybia-CP is ILGTILGLLKSL. The antimicrobial activities of the peptides against two Gram-negative and three Gram-positive species of bacteria were determined by the broth microdilution method. Polybia-CP displayed potent antibacterial activity against all the bacteria tested. The MICs of polybia-CP toward E. coli, P. aeruginosa, S. aureus, S. epidermidis, and B. subtilis were 16 μM, 128 μM, 4 μM, 16 μM, and 4 μM, respectively. The MBCs of polybia-CP against E. coli, S. aureus, and B. subtilis were 128 μM, 16 μM, and 8 μM. The bactericidal concentration is about 2- to 8-fold higher than its MICs. The temperature stability of polybia-CP at different temperatures was also determined. Our results after incubation in different temperatures ranging from 20°C to 100°C showed that the MICs were unaffected by temperature.
Outer membrane permeabilization assay.
The ability to permeabilize the outer membrane of E. coli was determined by using NPN. NPN is a hydrophobic fluorescent probe that fluoresces weakly in an aqueous environment and strongly when it enters a hydrophobic environment such as the interior of membrane (13). Normally, NPN is excluded from Gram-negative bacterial cells. In this study, the maximum excitation wavelength is 350 nm, and the maximum emission wavelength is 420 nm, similar to the maxima observed with NPN added to aqueous solution. Compared to cells with NPN only, polybia-CP induced a rapid increase in fluorescence in a dose-dependent manner (Fig. 1). This indicated that polybia-CP could disrupt the bacterial outer membrane and permeabilize the outer membrane.
Fig 1.
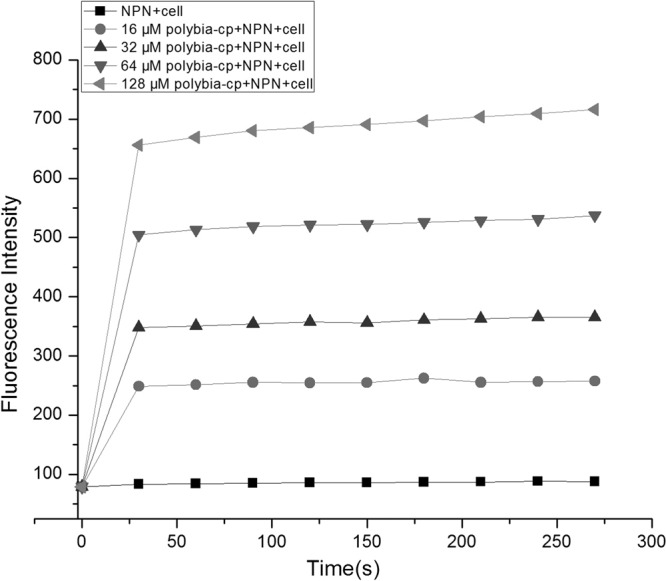
Effects of polybia-CP on the permeabilization of the outer membrane of E. coli.
Permeabilization of membrane of viable bacteria.
To determine the effect of polybia-CP to the integrity of the membrane of bacteria, DNA-intercalating fluorescent dye PI (15) was used to incubate with E. coli in the presence or absence of polybia-CP. PI could pass through the damaged membrane and intercalate into DNA and then be visualized by confocal laser-scanning microscopy. As shown in Fig. 2, the membrane of control bacteria was integrated and unstained by PI. However, most of bacteria incubated with PI in the presence of polybia-CP were stained by PI. That is to say, polybia-CP could disrupt the integrity of bacterial membrane and induce the uptake of PI. Thus, we could conclude that the cytoplasm also could leak out of the bacteria and lead to the death of bacteria.
Fig 2.
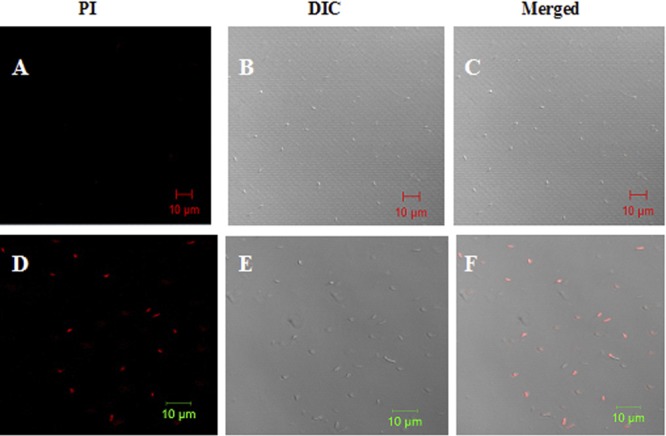
Effect of polybia-CP on bacterial membrane permeabilization. E. coli cells were incubated with PBS control (A to C) or polybia-CP (50 μM) (D to F) for 90 min at room temperature. Bacteria were stained with PI and analyzed by confocal laser-scanning microscopy to assess the integrity of the bacterial membrane. Results are the representative of three separate experiments. Polybia-CP could induce the uptake of PI. Bar, 10 μm.
Morphology changes of bacteria after treating by polybia-CP.
The PI uptake assay determined that polybia-CP could disrupt the integrity of bacterial membrane indirectly. To determine the influence of polybia-CP on the membrane directly, SEM was used to examine the morphological changes on the bacteria induced by polybia-CP. As shown in Fig. 3, polybia-CP could induce the morphological change dramatically. The untreated E. coli showed a normal smooth surface, while E. coli treated with 50 μM polybia-CP revealed a swollen, beruffled surface likely to represent pore formation. Taking this together with the results of the PI uptake assay and the OM permeation assay, we cautiously conclude that the membrane was disrupted. PI could enter into the bacteria through the disrupted membrane. Correspondingly, the cell content also could leak out from these pores.
Fig 3.
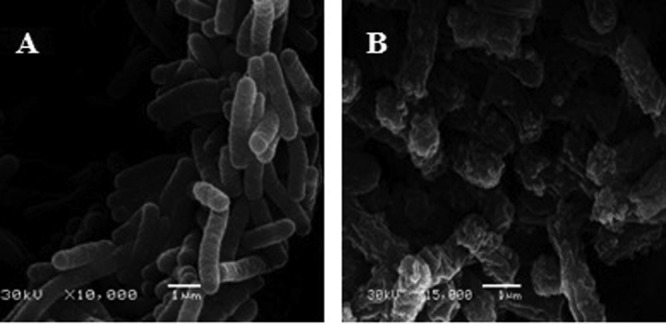
Scanning electron micrographs of E. coli in the absence (A) or presence (B) of polybia-CP after 30 min of exposure to polybia-CP. Bar, 1 μm.
Peptide-induced dye leakage from calcein-loaded LUVs.
As we know, the true membrane of bacteria contains not only lipids but also a wide variety of other biological molecules, such as proteins, carbohydrates, and cholesterol. To determine the action target of polybia-CP, the pure lipid membrane model should be involved. In this study, we studied the ability of polybia-CP to cause membrane leakage by testing their ability to evoke calcein released from LUVs made of negatively charged phospholipids. The percentage of calcein leakage at 2 min after exposure to the polybia-CP was used as a measurement of membrane permeability. The dose-response curves of peptide-induced calcein release are shown in Fig. 4.
Fig 4.
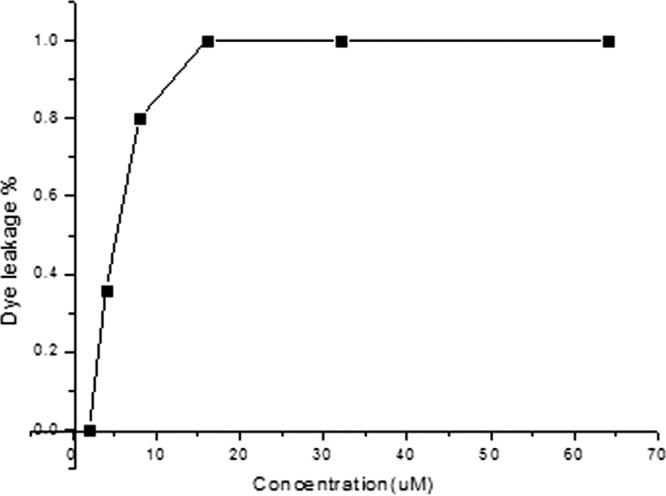
Dose-dependent peptide-induced calcein release from negatively charged EYPE-EYPG (7:3) LUVs. The percentage of dye leakage caused by polybia-CP was calculated as follows: % dye leakage = 100 × [(F − F0)/(Ft − F0)], where F is the leaked fluorescence intensity achieved 2 min after addition of polybia-CP, and F0 and Ft are the intensities seen without the peptide and with Triton X-100 (full leakage; positive control), respectively.
Morphology changes of LUVs after treating by polybia-CP.
In order to directly observe the effect of polybia-CP on the model membrane, TEM was used to monitor the morphological change of LUVs. As shown in Fig. 5, after a short time exposure to polybia-CP, the integrity of LUVs was disrupted thoroughly. The smooth, regular surface of LUVs became loose and fragmented. This result was consistent with the calcein leakage assay. While exposed to polybia-CP, the integrity of LUVs was disrupted, and then the entrapped calcein leaked out of LUVs and made the fluorescence intensity rise remarkably.
Fig 5.
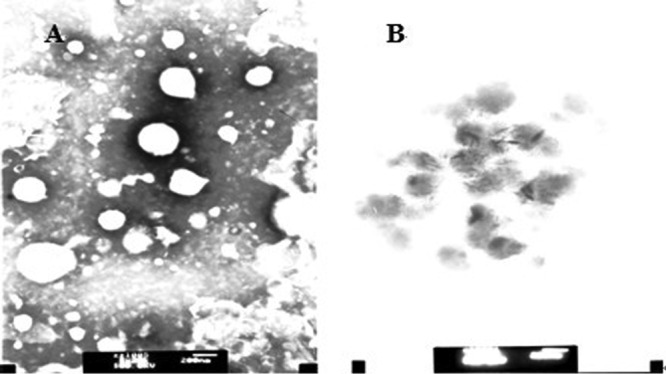
Transmission electron microscopy of polybia-CP-treated LUVs. Transmission electron micrographs showing the morphological changes in liposomes composed of EYPE-EYPG (7:3) in the absence (A) and presence (B) of polybia-CP. The inset showed that polybia-CP could disrupt the integrity of liposome. Bar, 200 nm.
DNA-binding activity.
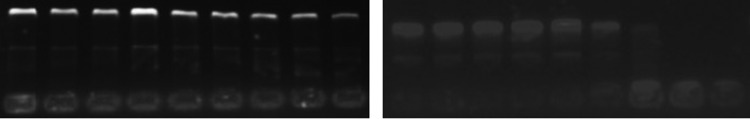
Together with the results of PI uptake assay, SEM, calcein leakage assay, and TEM microscopy, we can conclude that polybia-CP could disrupt the integrity of membrane, leading to the leakage of cell content of bacteria. At the same time, during the process of interaction with membrane, polybia-CP inevitably could enter into cytoplasm, but the effect of polybia-CP in the cytoplasm is unknown. Thus, we examined the DNA-binding properties of polybia-CP to attempt to determine the target of action. The DNA-binding affinities of the peptides were examined by analyzing the maximum absorption intensity shifts and the electrophoretic mobilities of plasmid DNA bands at various peptide/DNA weight ratios. Electronic absorption spectroscopy is usually to determine the binding of complexes with the DNA, for a complex bound to DNA through intercalation could be characterized by the change of blue shift in wavelength (20). As shown in Fig. 6, our results showed that polybia-CP could not induce the shift of maximum absorption intensity of the genome DNA of bacteria. Compared with LL-37, polybia-CP could not inhibit the migration of plasmid DNA at concentrations even above 30 μM, while LL-37 inhibited the migration of DNA at concentrations above 16 μM (Fig. 7). Then we could cautiously conclude that polybia-CP did not interact with the genome of bacteria and that the membrane may be its only target.
Fig 6.
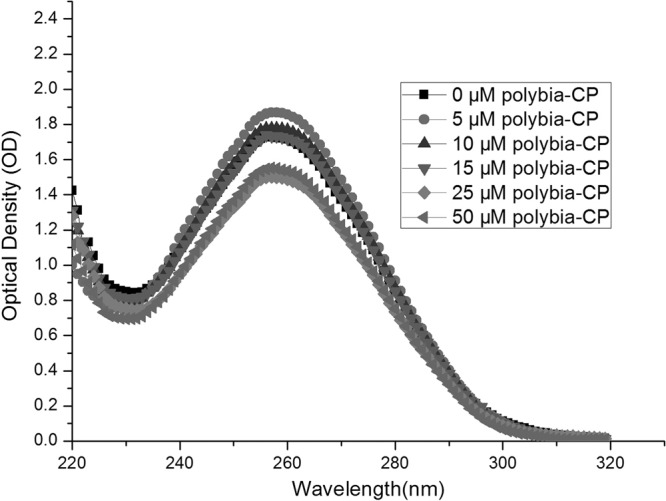
UV spectra of E. coli genomic DNA in the presence of increasing amounts of polybia-CP. E. coli genomic DNA was treated with increasing amounts of polybia-CP (peptide concentrations were 0 μM, 5 μM, 10 μM, 15 μM, 25 μM, and 50 μM). UV spectra were measured in the range of wavelengths from 220 to 320 nm. All measurements were made at a 10-mm optical path length at room temperature.
Fig 7.
Interaction of the peptides with plasmid DNA. Binding was assayed by measuring the inhibition of migration by plasmid DNA. The interaction of polybia-CP (left; lanes 1 to 9, 0 μM, 2 μM, 4 μM, 6 μM, 8 μM, 10 μM, 15 μM, 20 μM, and 30 μM, respectively) and LL-37 (right; lanes 1 to 9, 0 μM, 0.5 μM, 1 μM, 2 μM, 4 μM, 8 μM, 16 μM, 32 μM, and 64 μM, respectively) with plasmid DNA were examined by electrophoresis assay. Polybia-CP and LL-33 were coincubated with plasmid DNA for 30 min at room temperature before electrophoresis on a 1.5% agarose gel.
DISCUSSION
The extensive use of antibiotics in medicine and agriculture has resulted in the frequent emergence of resistant bacteria, like those termed “superbacteria” and EHEC, which put people in the potentially dangerous situation of having no usable antibiotics forthcoming in the near future. In addition, the use of chemical food preservatives makes this situation worse and is often associated with deleterious side effects, such as cumulative toxicity and degradation problems. Also, more and more people are paying attention to food preservatives, and safer food preservatives have been required in recent years. The amphipathic α-helical cationic antibacterial peptides, which lack the toxicity of conventional chemotherapeutic agents, are degraded easily in vivo, and are unaffected by common mechanism of chemoresistance, would be ideal food preservative candidates and attract great interest all over the world.
The results presented in this study showed that polybia-CP has potent antimicrobial activity against both Gram-positive and -negative bacteria. However, the mechanism of the action is unknown. Thus, SEM was used to visualize the morphological changes of bacteria treated by polybia-CP. The treated bacteria showed a disrupted surface likely to represent pore formation, whereas the untreated bacteria were characterized by a normal smooth surface. In our previous study of the cytotoxity exhibition mode of polybia-CP, our results demonstrated that polybia-CP could disrupt the integrity of cell membrane and exhibited its cytotoxity against tumor cells (25). Thus, we wanted to know if it also targets the plasma membrane of bacteria, disrupts the integrity of the membrane, and kills the bacteria. In the present study, we also examined the effect of polybia-CP on the pure lipid LUVs, which was comprised of EYPE-EYPG and used as the model of bacterial membrane. The TEM results showed that polybia-CP could disrupt the integrity of LUVs after a short-term incubation. In addition, a DNA electrophoresis experiment was employed to evaluate whether polybia-CP could kill the bacteria through disrupting DNA genome of bacteria. Our results showed that polybia-CP has no effect on the plasmid DNA. Taking this together with the previous results, we can cautiously speculate that the target of polybia-CP is the plasma membrane of bacteria.
As we know, both Gram-positive and Gram-negative bacteria have cytoplasma membrane surrounding the cell, and their membranes are comprised of different phospholipids, with lipoteichoic acids (LTAs) embedded in Gram-positive bacterial membrane and lipopolysaccharide (LPS) embedded in Gram-negative bacterial membrane (6). The bacterial resistance was mainly attributable to single genes or different genes encoding some activity (such as DNA damage repair, membrane permeability and efflux, rRNA mutation, aminoglycoside modification, etc.) involved with detoxification or related tasks (10). Cationic and amphipathic polybia-CP can enter into the membrane bilayers and takes an α-helical conformation. Then, the amino acid residue can interact with lipid molecules and disrupt the lipids' orderly alignment. Finally, water channels form, leading to bacterial death (25). This process occurs solely due to the physical action of water flow through the channels, and there is not any other protein or enzyme involved. In addition, bacterial survival depends on membrane lipid homeostasis and an ability to adjust lipid composition to acclimatize the bacterial cell to optimize growth in diverse environments (29). Also, bacterial mechanisms to control the expression of the genes solely responsible for the formation of fatty acids and the modification of existing fatty acyl chains to adjust the biophysical properties of the fatty acids to maintain a stable membrane are very sophisticated (30). Thus, it is difficult for bacteria to develop resistance to polybia-CP unless the nature of the bacterial membrane changes.
In conclusion, our results indicate that polybia-CP could kill the bacteria by disrupting the membrane and is not affected by the common resistance mechanism. Our findings also underscore that the antimicrobial activity of polybia-CP is unaffected by high temperature. Furthermore, the fact that polybia-CP could be degraded quickly in vivo and that their secondary metabolites are free amino acids proves that it has minimal side effects compared with conventional chemotherapeutic agents. Thus, with the increasing resistance to conventional antibiotics, there is no doubt that polybia-CP could offer a new strategy for defending against resistant bacteria in medicine and the food and farming industries. Also, polybia-CP has desirable and novel antibiotic and food preservative features that can be developed.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (nos. 90813012 and 20932003), the Key National S&T Program “Major New Drug Development” of the Ministry of Science and Technology (2012ZX09504001-003), the Chunhui Program of Ministry of Education of China (no. 2010082), and the Fundamental Research Funds for the Central Universities (lzujbky-2010-129).
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Abdou AM, Higashiguchi S, Aboueleinin AM, Kim M, Ibrahim HI. 2007. Antimicrobial peptides derived from hen egg lysozyme with inhibitory effect against Bacillus species. Food Control 18:173–178 [Google Scholar]
- 2. Beckloff N, et al. 2007. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob. Agents Chemother. 51:4125–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boman HG, Hultmark D. 1987. Cell free immunity in insects. Annu. Rev. Microbiol. 41:103–126 [DOI] [PubMed] [Google Scholar]
- 4. Calderon CB, Sabundayo BP. Antimicrobial classifications: drugs for bugs. In Schwalbe R, Steele-Moore L, Goodwin AC. (ed), Antimicrobial susceptibility testing protocols. CRC Press, Boca Raton, FL [Google Scholar]
- 5. Chou HT, et al. 2008. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. Int. J. Antimicrob. Agents 32:130–138 [DOI] [PubMed] [Google Scholar]
- 6. Epand RM, Epand RF. 2009. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta 1788:289–294 [DOI] [PubMed] [Google Scholar]
- 7. Fields GB, Noble RL. 1990. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 35:161–214 [DOI] [PubMed] [Google Scholar]
- 8. Finberg RW, et al. 2004. The importance of bactericidal drugs: future directions in infectious disease. Clin. Infect. Dis. 39:1314–1320 [DOI] [PubMed] [Google Scholar]
- 9. Fitton JE, Dell A, Shaw WV. 1980. The amino acid sequence of the delta haemolysin of Staphylococcus aureus. FEBS Lett. 115:209–212 [DOI] [PubMed] [Google Scholar]
- 10. Heinemann JA, Ankenbauer RG, Carlos F, Amábile-Cuevas CF. 2000. Do antibiotics maintain antibiotic resistance? Drug Discov. Today 5:195–204 [DOI] [PubMed] [Google Scholar]
- 11. Helander IM, Mattila-Sandholm T. 2000. Fluorometric assessment of Gram-negative bacterial permeabilization. J. Appl. Microbiol. 88:213–219 [DOI] [PubMed] [Google Scholar]
- 12. Henderson-Begg SK, et al. 2010. Mutation frequency in antibiotic-resistant and -susceptible isolates of Streptococcus pneumoniae. Int. J. Antimicrob. Agents 35:342–346 [DOI] [PubMed] [Google Scholar]
- 13. Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morassutti C, Amicis FD, Skerlavaj B, Zanetti M, Marchetti S. 2002. Production of a recombinant antimicrobial peptide in transgenic plants using a modified VMA intein expression system. FEBS Lett. 519:141–146 [DOI] [PubMed] [Google Scholar]
- 15. Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. 1991. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 139:271–279 [DOI] [PubMed] [Google Scholar]
- 16. Oren Z, Hong J, Shai Y. 1997. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J. Biol. Chem. 272:14643–14649 [DOI] [PubMed] [Google Scholar]
- 17. Park S, et al. 2001. Structural study of novel antimicrobial peptides, nigrocins, isolated from Rana nigromaculata. FEBS Lett. 507:95–100 [DOI] [PubMed] [Google Scholar]
- 18. Sheath PHA, Mair NS, Sharpe ME, Holt JG. 1886. Bergey's manual of systematic bacteriology, vol 2 Williams and Wilkins, Baltimore, MD [Google Scholar]
- 19. Souza BM, et al. 2005. Structural and functional characterization of two novel peptide toxins isolated from the venom of the social wasp Polybia paulista. Peptides 26:2157–2164 [DOI] [PubMed] [Google Scholar]
- 20. Tang YL, Shi YH, Zhao W, Hao G, Le GW. 2009. Interaction of MDpep9, a novel antimicrobial peptide from Chinese traditional edible larvae of housefly, with Escherichia coli genomic DNA. Food Chem. 115:867–872 [Google Scholar]
- 21. Van Soolingen D, Hermans PE, de Haas PE, Soll DR, Van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang B, et al. 2010. Inhibition of bacterial growth and intramniotic infection in a guinea pig model of chorioamnionitis using PAMAM dendrimers. Int. J. Pharm. 395:298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang KR, et al. 2008. Antitumor effects, cell selectivity and structure–activity relationship of a novel antimicrobial peptide polybia-MPI. Peptides 29:963–968 [DOI] [PubMed] [Google Scholar]
- 24. Wang KR, et al. 2009. Novel mode of action of polybia-MPI, a novel antimicrobial peptide, in multi-drug resistant leukemic cells. Cancer Lett. 278:65–72 [DOI] [PubMed] [Google Scholar]
- 25. Wang KR, et al. 2011. Novel cytotoxity exhibition mode of polybia-CP, a novel antimicrobial peptide from the venom of the social wasp Polybia paulista. Toxicology 288:27–33 [DOI] [PubMed] [Google Scholar]
- 26. Won A, Ianoul A. 2009. Interactions of antimicrobial peptide from C-terminus of myotoxin II with phospholipid mono- and bilayers. Biochim. Biophys. Acta 1788:2277–2283 [DOI] [PubMed] [Google Scholar]
- 27. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 28. Zhang LJ, Rozek A, Hancock RWW. 2001. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 276:35714–35722 [DOI] [PubMed] [Google Scholar]
- 29. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 [DOI] [PubMed] [Google Scholar]
- 30. Zhang YM, Rock CO. 2009. Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 50:S115–S119 [DOI] [PMC free article] [PubMed] [Google Scholar]