Abstract
PSI-7977, a prodrug of 2′-F-2′-C-methyluridine monophosphate, is the purified diastereoisomer of PSI-7851 and is currently being investigated in phase 3 clinical trials for the treatment of hepatitis C. In this study, we profiled the activity of PSI-7977 and its ability to select for resistance using a number of different replicon cells. Results showed that PSI-7977 was active against genotype (GT) 1a, 1b, and 2a (strain JFH-1) replicons and chimeric replicons containing GT 2a (strain J6), 2b, and 3a NS5B polymerase. Cross-resistance studies using GT 1b replicons confirmed that the S282T change conferred resistance to PSI-7977. Subsequently, we evaluated the ability of PSI-7977 to select for resistance using GT 1a, 1b, and 2a (JFH-1) replicon cells. S282T was the common mutation selected among all three genotypes, but while it conferred resistance to PSI-7977 in GT 1a and 1b, JFH-1 GT 2a S282T showed only a very modest shift in 50% effective concentration (EC50) for PSI-7977. Sequence analysis of the JFH-1 NS5B region indicated that additional amino acid changes were selected both prior to and after the emergence of S282T. These include T179A, M289L, I293L, M434T, and H479P. Residues 179, 289, and 293 are located within the finger and palm domains, while 434 and 479 are located on the surface of the thumb domain. Data from the JFH-1 replicon variants showed that amino acid changes within the finger and palm domains together with S282T were required to confer resistance to PSI-7977, while the mutations on the thumb domain serve to enhance the replication capacity of the S282T replicons.
INTRODUCTION
Hepatitis C virus (HCV) currently infects more than 170 million people worldwide and is the leading cause of chronic liver disease and liver transplantation in the United States. HCV is a single plus-strand RNA virus about 9.6 kb in genome size and contains one open reading frame that encodes 10 structural and nonstructural (NS) proteins. The structural proteins (core, E1, and E2), which constitute the viral capsid and envelope proteins, are located at the amino terminus, followed by p7, which oligomerizes to form an ion channel critical for viral assembly, and the nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B), which are responsible for viral replication (35). Recently, two antiviral agents have been approved, in combination with pegylated alpha interferon and ribavirin (Peg-IFN/RBV), to treat patients infected with genotype (GT) 1 HCV. Both of these compounds, boceprevir (Victrelis) and telaprevir (Incivek), target the NS3/4A protease and inhibit HCV replication by blocking processing of NS3, NS4A/B, and NS5A/B (25, 33). These treatments showed improvement over Peg-IFN/RBV; however, patients may develop additional side effects corresponding to each of these antiviral compounds (4). Furthermore, viral resistance against these compounds has emerged both in vitro and in vivo as a result of the high genetic diversity of HCV and error-prone nature of the HCV NS5B RNA-dependent RNA polymerase (12). Therefore, research has focused on identifying therapies that are better tolerated, provide improved antiviral response, and have a higher barrier to resistance.
NS5B is an ideal antiviral target because this protein is essential in HCV replication. Two classes of inhibitors directed at the activity of NS5B are currently in clinical development: nucleoside/tide analogs, which target the active site, and nonnucleoside inhibitors (NNIs), which target other regions of the polymerase (39). Compared to NNIs, nucleoside/tide inhibitors have broader genotype coverage and a higher barrier to viral resistance (9, 24, 28). Previously, we reported the anti-HCV activity of PSI-7851, a phosphoramidate nucleotide prodrug of 2′-F-2′-C-methyluridine monophosphate (18). PSI-7851 is a 1:1 mixture of two diastereoisomers, PSI-7976 and PSI-7977, at the phosphorus center of the phosphoramidate moiety. PSI-7977, the Sp isomer, has better activity than PSI-7976 (Rp isomer) in replicon-based HCV inhibition assays (38) and is currently in phase 3 clinical trials. The 5′-triphosphate metabolite of PSI-7977, PSI-7409, inhibits HCV replication by serving as a nonobligate chain terminator (30). Cross-resistance studies have shown that PSI-7977 and PSI-7409 had reduced activity against GT 1b replicons and NS5B polymerase containing the S282T amino acid alteration, respectively (18, 38).
In this study, we further characterized the anti-HCV activity of PSI-7977 across various genotypes and subtypes and expanded the cross-resistance studies to include a panel of replicons containing resistant mutations selected by other classes of HCV inhibitors. Selection was performed with GT 1b (Con1 strain), 1a (H77 strain), and 2a (JFH-1 strain) replicon cells in order to examine for the emergence of any genotype- and/or subtype-dependent resistant variants associated with PSI-7977.
MATERIALS AND METHODS
Compounds.
PSI-7977 (prodrug of 2′-F-2′-C-methyluridine monophosphate) (38), PSI-352938 (prodrug of 2′-F-2′-C-methylguanosine monophosphate) (34), PSI-6130 (2′-F-2′-C-methylcytidine) (40), R1479 (4′-azidocytidine) (13), IDX-184 and INX-189 (prodrugs of 2′-C-methylguanosine monophosphate) (43, 48), NS5B nonnucleoside inhibitors NNI-1 (an indole analog), NNI-2 (a thiophene analog), NNI-3 (a benzothiadiazine analog), and HCV-796 (a benzofuran analog or NNI-4) (39), NS3 protease inhibitors telaprevir and RG7227 (ITMN-191) (21, 22), NS3/4a inhibitor ACH-806 (46), and NS5A inhibitor BMS-790052 (8) were synthesized at Pharmasset and shown to be >95 to 99% pure by proton nuclear magnetic resonance, mass spectroscopy, and high-performance liquid chromatography analysis. Ribavirin was obtained from Sigma-Aldrich.
Replicons and cells.
Plasmid DNA containing the GT 1a replicon (H77 strain; NCBI reference no. NC_004102.1) with adaptive mutations P1496L and S2204I (3) and the J6/JFH-1-derived GT 2a (JFH-1 GT 2a) were licensed from Apath. The JFH-1 GT 2a replicon contains a partial core (first 19 amino acids) and 3′ nontranslated region (NTR) from the J6 strain (GenBank accession no. AF177036) and 5′ NTR and the NS3 to NS5B region from the JFH-1 strain (GenBank accession no. AB047639). The JFH-1 replicon open reading frame also contains a Renilla luciferase reporter gene just upstream of the neomycin phosphotransferase gene (Neo). We generated a GT 1a replicon with a Renilla luciferase reporter gene (GT 1aRL) in between the 5′ NTR and Neo by molecular cloning and introduced an adaptive mutation (NS3 K583E) by site-directed mutagenesis to enhance its replication fitness. The Lunet cell line (14) and Con1-derived GT 1b ET replicon (GenBank accession no. AJ238799.1) with the firefly luciferase reporter gene and adaptative mutations E1202G, T1280I, and K1846T (23) were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). GT 1a, 1aRL, 1b, and JFH-1 2a replicon cell lines were each generated by electroporating the corresponding replicon RNA (10 μg) into the Lunet cells as described previously (17), followed by selection with G418. Plasmid containing the J6 GT 2a NS5B sequence (45) was synthesized at IDT DNA Technology (Coralville, IA). GT 2b and 3a NS5B sequences were isolated from human serum samples containing HCV RNA. The J6 GT 2a, GT 2b, and GT 3a NS5B sequences were each cloned into a Con1 GT 1b replicon that encodes a Renilla luciferase reporter gene using engineered PacI and AscI sites that framed the NS5B region to generate the chimeric replicons 1b/2a (J6), 1b/2b, and 1b/3a.
HCV replicon inhibition assays.
Inhibition of HCV replicon replication was determined by measuring the levels of luminescence expressed via the firefly or Renilla luciferase reporter gene using the Bright-Glo or Renilla-Glo reagent, respectively (Promega), or by real-time PCR (RT-PCR) using primers that anneal to the 5′ NTR. For the luciferase reporter assay, replicon cells were seeded at 3,000 cells/well in 96-well plates and incubated with 3-fold serially diluted compounds at 37°C in a humidified 5% CO2 atmosphere for 4 days. For transient assays, Lunet cells were transfected with 10 μg replicon RNA as described previously by electroporation (17). Replicon RNA was generated by in vitro transcription from linearized plasmids (restriction digested with ScaI for GT 1b and chimeric replicons, HpaI for GT 1a replicons, XbaI for JFH-1 GT 2a replicons) using a RiboMAX large-scale RNA T7 kit (Promega). Transfected cells were seeded at 8,000 cells/well in 96-well plates and incubated with serially diluted compounds for 4 days prior to performing the luciferase assay. For GT 1a replicons (wild type or F415Y), RT-PCR was used to measure HCV RNA levels. After incubation for 4 days, the supernatant was discarded and total RNA was extracted from cells using an RNeasy 96 kit as described by the manufacturer (Qiagen). The extracted RNA was reversed transcribed into cDNA, which was amplified as previously described (41). The threshold cycle (ΔΔCT) and percent inhibition for each sample were determined using the relative quantification method. The concentrations at which 50% inhibition was achieved (50% effective concentrations [EC50s]) were determined using the GraphPad Prism software (San Diego, CA).
Cross-resistance studies.
All replicon variants were generated using a QuikChange II XL site-directed mutagenesis kit (Agilent) with the appropriate primer sets. The non-NS5B panel of resistant replicons contained amino acid changes that conferred resistance to inhibitors targeting the NS3 protease (NS3 R155K, D168V), the NS3/4A binding surface (NS3 C16S, A39V), or the NS5A phosphoprotein (NS5A Y93H). The NS5B panel contained amino acid changes that conferred resistance to NS5B inhibitors targeting either the active sites (S96T/N142T, S282T, S15G/C223H/V321I, F415Y) or the other sites (C316Y, M414T, M423T, P495L). Most of the amino acid changes were generated using the Con1-derived GT 1b ET replicons, with the exception of the NS5B S15G/C223H/V321I mutations, which were generated in the JFH-1 replicons, and the NS5B F415Y mutation, which was generated in the GT 1a replicon. All mutations were verified by sequencing analysis. Plasmids containing replicon mutants were in vitro transcribed into RNA using the Ribomax large-scale RNA production system as recommended by the manufacturer (Promega, Madison, WI). RNA (10 μg) was electroporated into the highly permissive Lunet cells and selected using G418 to generate stable cells. Maintenance of the resistant mutation(s) was confirmed by sequencing of the corresponding nonstructural region and/or a reduction in activity of the appropriate reference compound. HCV inhibition assays were performed with PSI-7977 and the appropriate reference HCV inhibitors as described above.
Resistance selection.
Selection studies were conducted by culturing GT 1b, 1a, or JFH-1 GT 2a replicon cells in the presence of G418 and increasing concentrations of PSI-7977 starting at the approximate EC50 (for GT 1a and JFH-1 GT 2a replicons) or EC90 (for GT 1b replicons) values. Cells were passaged whenever they reached ∼80% confluence and replenished with G418 medium containing fresh compound. Replicon cells were also cultured in the presence of G418 and 0.2% dimethyl sulfoxide (DMSO) in parallel as a no-drug control. At various passages, both the PSI-7977-selected and no-drug control cells were tested for sensitivity to PSI-7977. For each assay, 3-fold serial dilutions of test compound were added to cells in duplicate and the cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 4 days. Inhibition of HCV replicon replication was determined as described above by luminescence (GT 1b and JFH-1 GT 2a) or RT-PCR (GT 1a). Aliquots of cells were also saved for RNA isolation, cDNA synthesis, and PCR amplification for sequencing.
Sequence analysis.
Total RNA was extracted from the selected cells and reverse transcribed into cDNA. The GT 1a and 1b NS5B region was amplified using a Titan one-tube RT-PCR kit (Roche Applied Science) and the appropriate primer sets (IDT DNA Technologies). Primers for GT 1b NS5B were 1bNS5B S1 (5′ CGT AAG CGA GGA GGC TAG T) and 1bNS5B A1 (5′ GTG TTT AGC TCC CCG TTC ATC). GT 1a NS5B primers consisted of 1aNS5B S1 (5′ GTT GAG TCC TAT TCT TC), 1aNS5B S2 (5′ GGC CGA CAC GGA AGA TGT C), 1aNS5B A1 (5′ GAG TGT TTA CCC CAA CCT TCA TCG G), and 1aNS5B A2 (5′ GGC CTA AGA GGC CGG AGT G). RNA extracted from the JFH-1 GT 2a replicon cells was reverse transcribed into cDNA using a Transcriptor first-strand cDNA synthesis kit (Roche), and the NS5B region was amplified with the Phusion polymerase (Thermo Fisher Scientific). Primers for JFH-1 GT 2a NS5B were 2aNS5B S1 (5′ CGA GGA GGA CGA TAC CAC CGT GTG CTG CTC C) and 2aNS5B A1 (5′ GTG TAC CTA GTG TGT GCC GCT CTA CCG AGC GG). All amplified products were gel purified and cloned into a TA TOPO pCR4 sequencing vector (Invitrogen) for subsequent clonal analysis. Sequence alignments were performed using Lasergene DNAStar software (Madison, WI).
Phenotypic assays.
Amino acid substitutions were introduced into the appropriate replicon plasmids using QuikChange II site-directed mutagenesis (Agilent) and primers (IDT DNA Technologies) with nucleotide substitutions that corresponded to changes at sites 179, 282, 289, 293, 434, and 479 as single changes or mutations in combination. A mutation(s) was confirmed by sequencing. Replicon RNA (10 μg) was electroporated into Lunet cells to evaluate for replication efficiency using a 4-day transient assay as described above. Replication fitness was determined by normalizing the luciferase expression at 96 h with that at 4 h and then dividing the normalized level of luciferase expression of the replicon mutant by that of the wild type. For susceptibility studies, cells transfected with the JFH-1 replicon variants were selected in G418 to generate stable cell lines prior to incubation with PSI-7977 or other control compounds. A 4-day transient assay was also used to evaluate GT 1aRL, 1b, 2a (JFH-1), 1b/2a (J6), 1b/2b, and 1b/3a replicons containing the S282T change, the GT 1aRL replicon containing the S282T/I434M changes, and GT 1b and 1b/2a (J6) replicons containing changes in residues 179, 289, and 282. All HCV inhibition assays were performed as described above.
RESULTS
Genotype coverage.
We have previously shown that PSI-7977 inhibited clone A (GT 1b) wild-type and S282T replicons with EC90 values of 0.42 and 7.8 μM, respectively (38). Here we evaluated the genotype coverage of PSI-7977 by using GT 1b (Con1)-, 1a (H77)-, and 2a (JFH-1)-derived replicons and GT 1b chimeric replicons containing the NS5B region from the J6 GT 2a isolate and from GT 2b and GT 3a patient isolates. As summarized in Table 1, PSI-7977 inhibited the replication of these replicons with similar EC50s (between 0.016 and 0.048 μM) and was especially active against the chimeric replicon containing the J6 NS5B (EC50 = 4.7 nM). We also assessed the activities of nonnucleoside inhibitors (NNIs) to compare them with the genotype coverage of PSI-7977. Among these, only the benzofuran compound HCV-796, which binds within the palm domain of the polymerase (10), was effective at inhibiting the replication of the various replicons, with EC50s ranging from 2 nM to 65 nM. The other three NNIs had limited genotype coverage. The thiophene and benzothiadiazine compounds (20, 31) were most active against the replication of GT 1b replicons but were ∼10-fold less active against GT 1a replicons. The benzothiadiazine analog showed weak activity against chimeric replicons containing GT 2a (J6) and 2b NS5B and was inactive against the JFH-1 GT 2a replicons and chimeric replicons with NS5B from GT 3a. The thiophene inhibitor failed to inhibit replicons containing GT 2 or 3 NS5B. The indole analog, which targets the alternative GTP binding site on the surface of the thumb domain (2), inhibited the replication of replicons containing NS5B from GT 1 and 3a but was much less active against replicons with GT 2 NS5B polymerase.
Table 1.
Activities of PSI-7977 and representative nonnucleoside inhibitors in various genotypes and subtypes of HCVa
| Inhibitor | GT 1b (Con1) |
GT 1a (H77) |
GT 2a (JFH-1) |
GT 1b/2a NS5B (J6) |
GT 1b/2b NS5B (patient isolate) |
GT 1b/3a NS5B (patient isolate) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | EC50 (μM) | CC50 (μM) | |
| PSI-7977 NI | 0.048 ± 0.013 | >20 | 0.044 ± 0.0047 | >20 | 0.037 ± 0.0036 | >20 | 0.0047 ± 0.0017 | >20 | 0.020 ± 0.0044 | >20 | 0.016 ± 0.0034 | >20 |
| Indole NNI-1 | 0.33 ± 0.056 | 25.6 ± 7.5 | 0.51 ± 0.030 | 32.2 ± 8.4 | 16.3 ± 4.5 | 33.3 ± 5.6 | 26.4 ± 2.4 | 36.5 ± 10.4 | 20.2 ± 4.2 | 38.3 ± 11.4 | 0.039 ± 0.01 | 22.3 ± 4.1 |
| Thiophene NNI-2 | 0.14 ± 0.083 | 30.2 ± 4.0 | 1.2 ± 0.21 | 34. 6 ± 3.0 | 22.8 ± 5.0 | 34.1 ± 7.6 | >50 | 29.8 ± 10.0 | >50 | >50 | >50 | 38.0 ± 3.3 |
| Benzothiadiazine NNI-3 | 0.15 ± 0.086 | >50 | 2.0 ± 0.73 | >50 | >50 | >50 | 28.5 ± 2.5 | >50 | 32.8 ± 1.6 | >50 | >50 | >50 |
| HCV-796 NNI-4 | 0.0081 ± 0.0022 | >5 | 0.043 ± 0.0079 | >5 | 0.065 ± 0.014 | >5 | 0.020 ± 0.0093 | >5 | 0.011 ± 0.0010 | >5 | 0.0020 ± 0.00061 | >5 |
NI, nucleotide inhibitor; NNI, nonnucleoside inhibitor; CC50, 50% cytotoxic concentration. Transient assays were performed for GT 1b/2a, 1b/2b, and 1b/3a chimeric replicons, which were the GT 1b replicon with its NS5B region swapped with that isolated from J6 GT 2a, 2b, and 3a, respectively. Results are average ± standard deviation from at least three independent experiments performed in duplicates.
Cross-resistance studies.
Cross-resistance studies were performed using replicon mutants generated by site-directed mutagenesis with mutations within the nonstructural region of GT 1b replicons, unless indicated otherwise. Two different panels of resistant replicons were evaluated: the first panel contained resistant mutations within the NS5B region, while the second panel contained resistant mutations outside NS5B.
Among the various NS5B mutations, we included representative amino acid changes (C316Y, M414T, M423T, P495L) that conferred resistance to the four main classes of NNIs (12), the S96T/N142T substitutions that conferred resistance to the 4′-azidocytidine nucleoside R1479 (1), the S282T substitution which conferred resistance to 2′-C-methylnucleosides/tides (29) and 2′-F-2′-C-methylpyrimidines (18, 40), the S15G/C223H/V321I (JFH-1 GT 2a) changes that in combination conferred resistance to 2′-F-2′-C-methylguanosine analogs (15), and the F415Y (GT 1a) alteration that has previously been found in patients treated with ribavirin (47). As summarized in Table 2, the replicons with the C316Y, M414T, M423T, and P495L changes remained fully susceptible to the inhibition of PSI-7977, while showing a 26- to 70-fold reduction in sensitivity for the corresponding class of NNIs. Among the amino acid alterations that conferred resistance to nucleoside/tide inhibitors, PSI-7977 showed a 9.5-fold increase in EC50 against the S282T replicons. The S96T/N142T, S15G/C223H/V321I (GT 2a), and F415Y (GT 1a) replicons remained susceptible to PSI-7977. As expected, the reference compounds R1479 and PSI-352938 were 5.4- and 12.6-fold less active against the S96T/N142T replicons and the JFH-1 GT 2a S15G/C223H/V321I replicons, respectively. However, our data showed that the activity of ribavirin against the GT 1a F415Y replicons was similar to that of wild type.
Table 2.
Cross-resistance studies of PSI-7977 using replicons with NS5B mutationsa
| Resistant mutation | HCV inhibitor | EC50 (μM) |
Fold change in EC50 | |
|---|---|---|---|---|
| WT replicon | Mutant replicon | |||
| C316Y | PSI-7977 | 0.035 ± 0.0037 | 0.021 ± 0.0021 | 0.61 ± 0.057 |
| HCV-796 | 0.0063 ± 0.0010 | 0.31 ± 0.19 | 47.2 ± 6.0 | |
| M414T | PSI-7977 | 0.044 ± 0.014 | 0.036 ± 0.016 | 0.81 ± 0.22 |
| Benzothiadiazine | 0.13 ± 0.047 | 3.6 ± 1.5 | 26.7 ± 4.4 | |
| M423T | PSI-7977 | 0.035 ± 0.0037 | 0.031 ± 0.0035 | 0.89 ± 0.094 |
| Thiophene | 0.14 ± 0.043 | 5.9 ± 1.7 | 42.7 ± 6.0 | |
| P495L | PSI-7977 | 0.040 ± 0.0047 | 0.040 ± 0.0097 | 1.0 ± 0.31 |
| Indole | 0.41 ± 0.12 | 29.2 ± 11.1 | 70.4 ± 9.6 | |
| S96T | PSI-7977 | 0.035 ± 0.014 | 0.028 ± 0.011 | 0.94 ± 0.53 |
| R1479 | 3.24 ± 0.77 | 17.6 ± 5.6 | 5.4 ± 0.54 | |
| S282T | PSI-7977 | 0.059 ± 0.0073 | 0.56 ± 0.10 | 9.5 ± 2.3 |
| PSI-352938 | 0.033 ± 0.0093 | 0.061 ± 0.022 | 1.8 ± 0.24 | |
| S15G/C223H/V321Ib | PSI-7977 | 0.15 ± 0.027 | 0.13 ± 0.072 | 0.84 ± 0.20 |
| PSI-352938 | 0.12 ± 0.048 | 1.5 ± 0.47 | 12.6 ± 1.1 | |
| F415Yc | PSI-7977 | 0.053 ± 0.031 | 0.063 ± 0.030 | 1.3 ± 0.25 |
| Ribavirin | 38.2 ± 8.0 | 25.1 ± 4.8 | 0.66 ± 0.017 | |
Mutations were generated in the GT 1b replicon NS5B region unless indicated otherwise. EC50 fold change was determined by normalizing the values of the replicon mutants with that of the wild type (WT). Results are reported as average ± standard deviation from at least three independent experiments performed in duplicate.
Mutations were generated in JFH-1 GT 2a replicons.
The mutation was generated in GT 1a replicons.
Table S1 in the supplemental material summarizes the data for PSI-7977 and the appropriate reference inhibitors against replicons with resistant mutations within the NS3/4A protease and NS5A phosphoprotein. Results showed that PSI-7977 was active in replicons with the NS3 R155K or D168V alteration. Consistent with previous findings (12, 19, 21), the R155K mutation was cross resistant to both RG7227 (a macrocyclic compound) and telaprevir (a linear ketoamide compound), while D168V conferred resistance only to RG7227 and not telaprevir. PSI-7977 also retained its activity against replicons with amino acid changes of NS3 C16S, A39V (resistant to the NS3/4A inhibitor ACH-806), or NS5A Y93H (resistant to the NS5A inhibitor BMS-790052).
Selection studies using GT 1a, 1b, and 2a replicon cells.
Emergence of resistance has become a concern for antiviral compounds because it may lead to viral breakthrough and treatment failures. In order to evaluate the ability of PSI-7977 to select for resistance, selection studies were performed using GT 1a, 1b, and the JFH-1 GT 2a replicon cells. Performing the studies with all three replicon cell lines also allowed us to determine if there was any genotype- and/or subtype-dependent resistance associated with PSI-7977. Subtype-dependent resistance has previously been reported for the NS3 protease and nonnucleoside NS5B inhibitors (27). Recently, we reported that nucleotide prodrugs of 2′-F-2′-C-methylguanosine monophosphate, PSI-352938 and PSI-353661, were able to select for resistant mutations in the JFH-1-derived replicons but not in GT 1a and 1b replicons due to differences in fitness caused by the amino acid changes (15).
Replicon cells were cultured in the presence of increasing concentrations of PSI-7977, and emergence of resistance was monitored at various times during the selection studies. Results of the susceptibility tests and NS5B sequencing are summarized in Table 3. For GT 1b and 1a replicons, the S282T change was selected when cells were treated with PSI-7977 at approximately 20- to 40-fold over its EC50. The emergence of S282T coincided with a 5- to 6-fold reduction in activity of PSI-7977. While S282T was the only change selected in GT 1b NS5B, an additional amino acid change (I434M) was selected in GT 1a replicons. Clonal analysis showed that I434M was observed in combination with S282T in 50% of the cDNA clones sequenced.
Table 3.
Selection studies using PSI-7977 in GT 1b, 1a, and JFH-1 2a repliconsa
| Replicon cell | Day of selection | PSI-7977 concnb (μM) | Fold change in EC50 | NS5B amino acid substitution(s) |
|---|---|---|---|---|
| GT 1b | 36 | 0.6 | 3.4 | None |
| 56 | 0.9 | 5.0 | S282T | |
| GT 1a | 86 | 0.5 | 1.1 | None |
| 139 | 2.0 | 6.1 | S282T, I434 M | |
| GT 2a | 105 | 1.2 | 5.9 | T179A, M289L, I293L |
| 137 | 3.0 | 21.9 | T179A, S282T, M289L, I293L | |
| 149 | 3.0 | 13.5 | T179A, S282T, M289L, I293L, M434T, H479P |
Lunet cells stably expressing GT 1b, 1a, or JFH-1 2a replicons were cultured in the presence of increasing concentrations of PSI-7977. The starting concentration for PSI-7977 was 0.3 μM in GT 1b replicon cells and 0.05 μM in GT 1a and JFH-1 2a replicon cells. Susceptibility studies and NS5B sequencing were performed at various days of selection to monitor for resistance. EC50 fold change (single-point determinations performed in duplicate on the particular day of selection) was determined by normalizing the EC50 of the PSI-7977-selected cells with that of the no-drug control cells.
Concentrations of PSI-7977 in which replicons cells were cultured and when the mutation(s) was identified.
In contrast to GT 1a and 1b replicons, the amino acid alterations selected by PSI-7977 in the JFH-1 GT 2a replicons appeared to be more complex. Additional mutations appeared both prior to and after the selection of S282T, which emerged when cells were treated with PSI-7977 at concentrations about 70-fold over its EC50. In total, we observed five other amino acid changes: T179A, M289L, I293L, M434T, and H479P. Prior to the emergence of S282T, the EC50 fold change for PSI-7977 was about 6, and cDNA clones sequenced contained the T179A, M289L, and/or I293L alteration. Emergence of S282T appeared to further increase the EC50 of PSI-7977 to about 20-fold. Further incubation of JFH-1 replicon cells with PSI-7977 selected two more amino acid substitutions: M434T and H479P.
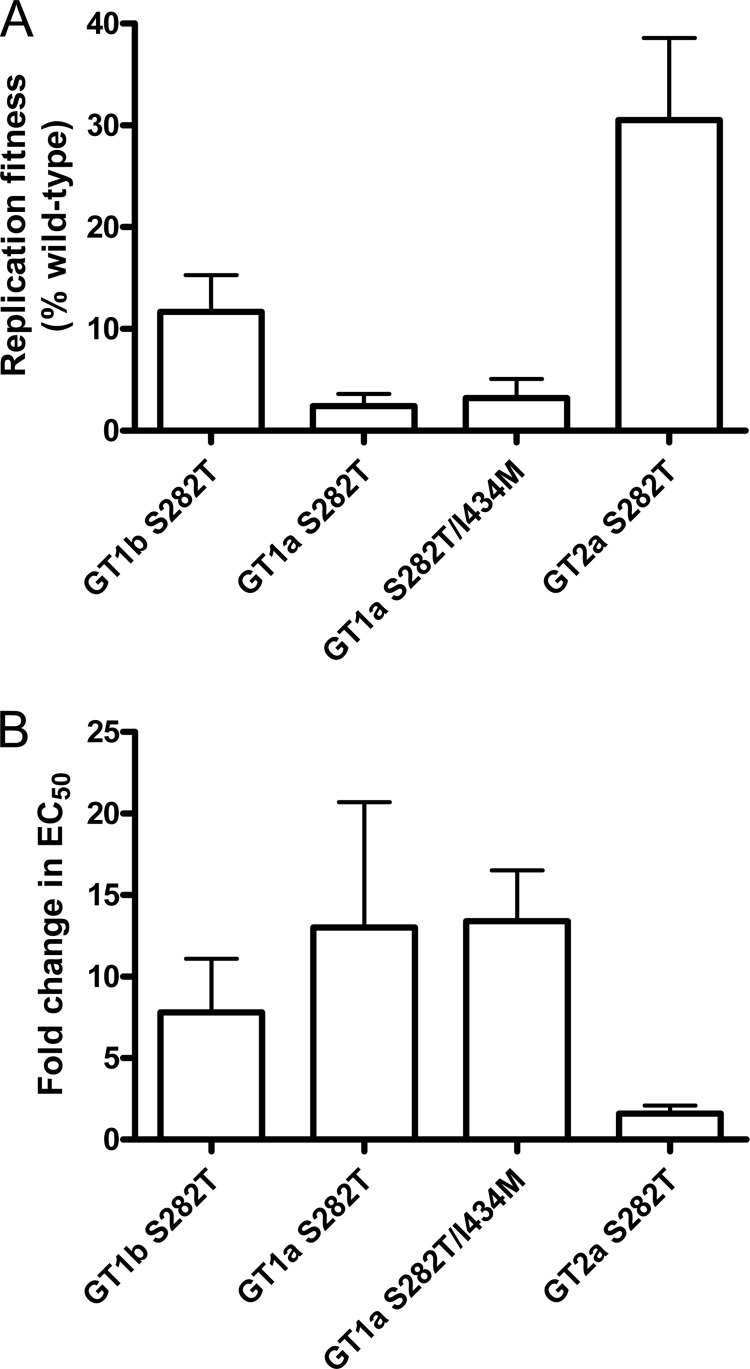
Since S282T was the common alteration selected by PSI-7977 in all three genotypes, the Thr substitution at position 282 was introduced into each replicon and transient assays were performed to examine the effect of S282T on PSI-7977 activity and replication fitness. Results from our fitness analysis showed that the GT 1a S282T replicon was the most unfit replicon (∼3%) among the three genotypes (Fig. 1A). Consistent with previous findings (1, 24, 29), our results showed that the GT 1b S282T replicon replicated at about 12% of its wild type. Compared to GT 1a and 1b, the JFH-1-derived GT 2a S282T replicon was the most fit (∼30%), though it was still significantly less efficient than its wild-type replicon. The more striking contrast between GT 1 and the JFH-1 S282T replicons was the effect on PSI-7977 activity. As shown in Fig. 1B, the S282T change in GT 1b and 1a replicons conferred resistance to PSI-7977, with EC50 increases of 7.8- and 13-fold, respectively. Combining the S282T mutation with I434M in GT 1a did not further increase the EC50 fold change of PSI-7977 over that for S282T alone. Interestingly, the JFH-1 GT 2a S282T replicons remained quite susceptible to PSI-7977 (EC50 fold change, ∼2). These data suggest that the additional mutations within JFH-1 NS5B may play a role in conferring resistance to PSI-7977.
Fig 1.
Phenotypic analysis of the NS5B S282T mutation in GT 1b, 1a, and JFH-1 2a replicons. Replicons containing S282T were generated by site-directed mutagenesis and tested for susceptibility to PSI-7977 using a 4-day transient assay. Cells containing GT 1a S282T/I434M replicons were also evaluated. (A) Replication fitness of S282T replicons from GT 1b, 1a, and JFH-1 2a relative to the corresponding wild type. (B) Effect of S282T on PSI-7977 activity. EC50 fold change was determined by normalizing the EC50s from the S282T replicons with that of the corresponding wild type. Results are reported as average ± SD from at least two independent experiments performed in duplicate.
Sequence analyses of PSI-7977-selected JFH-1 replicons.
To further examine these additional amino acid changes selected by PSI-7977, we first performed clonal sequencing of the NS5B region from various passages of the selection study to determine the progression and frequency of these mutations (Table 4). Subsequently, replicon mutants were constructed and susceptibility studies were performed to determine if these additional mutations would affect the activity of PSI-7977 (Table 5).
Table 4.
Progression of amino acid changes within JFH-1 GT 2a NS5B under selective pressure with PSI-7977a
| Day | No. of cDNA clones sequenced | Single change (n) | Double changes (n) | Multiple changes (n) |
|---|---|---|---|---|
| 57 | 10 | M289L (7) | ||
| 68 | 10 | M289L (6) | ||
| I293L (1) | ||||
| 85 | 10 | S282T (1) | ||
| M289L (4) | ||||
| 105 | 11 | T179A (2) | M289L/I293L (2) | |
| M289L (2) | M289I/I293L (1) | |||
| 127 | 10 | M289L (4) | T179A/M289L (1) | T179A/S282T/I293L (1) |
| S282T/M289L (1) | T179A/M289L/I293L (2) | |||
| M289L/I293L (1) | ||||
| 137 | 10 | S282T/M289L (8) | T179A/S282T/M289L/I293L (1) | |
| 149 | 10 | S282T/M289L (1) | T179A/S282T/I293L (2) | |
| S282T/M289L/I293L (2) | ||||
| S282T/M289L/I293L/M434T (1) | ||||
| T179A/S282T/M289L/I293L (1) | ||||
| T179A/S282T/M289L/I293L/H479P (1) | ||||
| T179A/S282T/M289L/I293L/M434T(2) | ||||
| 158 | 11 | S282T/M289L/I293L (1) | ||
| S282T/M289L/H479P (1) | ||||
| T179A/S282T/M289L/I293L (2) | ||||
| T179A/S282T/I293L/H479P (5) | ||||
| S282T/M289L/I293L/H479P (1) | ||||
| T179A/S282T/M289L/H479P (1) |
Total RNA was extracted from PSI-7977-selected cells from various passages, starting on day 57. The JFH-1 NS5B region was reverse transcribed into cDNA and amplified for clonal analysis. Changes within the finger domain (T179A and S282T) are in bold, of which the S282T change is also underlined. Changes within the palm domain (M289L and I293L) are in italic. Changes within the thumb domain (M434T and H479P) are normal text. n, number of cDNA clones with the indicated amino acid substitution(s).
Table 5.
Phenotypic analysis of JFH-1 GT 2a replicon variantsa
| Change and mutation | Fold change in EC50 | Fitness (% relative to WT) |
|---|---|---|
| Single-residue changes | ||
| T179A | 1.1 ± 0.2 | 89.0 ± 11.2 |
| S282T | 2.8 ± 0.7 | 30.5 ± 8.1 |
| M289L | 2.9 ± 0.9 | 99.9 ± 9.8 |
| I293L | 1.3 ± 0.3 | 93.9 ± 5.4 |
| M434T | 0.9 ± 0.2 | 104.7 ± 13.7 |
| H479P | 1.0 ± 0.3 | 126.0 ± 37.1 |
| Combination of residue changes in finger and palm domains | ||
| T179A/M289L | 2.1 ± 0.2 | 44.8 ± 10.6 |
| M289L/I293L | 2.5 ± 0.5 | 150.4 ± 16.7 |
| T179A/M289L/I293L | 3.5 ± 0.6 | 95.6 ± 26 |
| S282T/M289L | 6.3 ± 1.0 | 32.3 ± 4.9 |
| S282T/M289L/I293L | 5.6 ± 1.9 | 45.2 ± 9.3 |
| T179A/S282T/I293L | 8.3 ± 0.7 | 1.8 ± 0.6 |
| T179A/S282T/M289L/I293L | 9.5 ± 2.5 | 6.6 ± 2.2 |
| Combination of residue changes in finger, palm, and thumb domains | ||
| S282T/M289L/H479P | 7.4 ± 0.6 | 72.5 ± 12.6 |
| S282T/M289L/I293L/M434T | 7.1 ± 0.1 | 83.0 ± 9.3 |
| S282T/M289L/I293L/H479P | 8.4 ± 0.5 | 126.0 ± 6.9 |
| T179A/S282T/I293L/H479P | 9.0 ± 1.5 | 85.5 ± 11.3 |
| T179A/S282T/M289L/I293L/M434T | 10.6 ± 1.9 | 45.1 ± 6.5 |
| T179A/S282T/M289L/I293L/H479P | 11.8 ± 3.0 | 38.8 ± 5.3 |
| T179A/S282T/M289L/H479P | 11.4 ± 1.2 | 0.26 ± 0.13 |
Mutations in JFH-1 NS5B were generated by mutagenesis, and replicons were electroporated into Lunet cells, which were subjected to G418 selection prior to the susceptibility studies. Changes within the finger domain (T179A and S282T) are in bold, of which S282T is also underlined. Changes within the palm domain (M289L and I293L) are in italic. Changes within the thumb domain (M434T and H479P) are normal text. EC50 fold change was determined by normalizing the EC50s for PSI-7977 from the replicon variants with that of the wild type (WT). Replication fitness was determined by measuring the expression of luciferase at 4 h and 96 h posttransfection and normalizing the luminescence levels with that of the wild type. All values are reported as average ± standard deviation from at least three independent experiments performed in duplicate.
Our clonal analysis showed that M289L was the first and most predominant single amino acid change selected by PSI-7977. Among the 27 cDNA clones with a single mutation, 23 clones contained the M289L change, while only 1 cDNA clone contained S282T. Two other amino acid changes were selected by PSI-7977 at a relatively lower frequency prior to the appearance of S282T: T179A and I293L appeared either as single changes or in combination with M289L. By day 127, T179A and I293L existed as double or multiple mutations in combination with M289L and/or S282T. S282T did not become the predominant mutation until day 137, when cells were treated with 3 μM PSI-7977, and was mostly associated with the M289L change either as double mutations or in combinations with the other amino acid changes. Also by day 137, the replicons no longer contained just a single mutation in NS5B. Most replicons contained more than two mutations by day 149, and all replicons contained three or more mutations by day 158. The last two substitutions, M434T and H479P, did not appear until day 149. None of these amino acid changes were observed in cells treated with DMSO, with the exception of T179A, which appeared on day 158 as a low-frequency change (1 out of 11 cDNA clones sequenced) in the DMSO-treated replicons compared to PSI-7977-selected replicons, in which 8 out of 11 cDNA clones sequenced contained the T179A change.
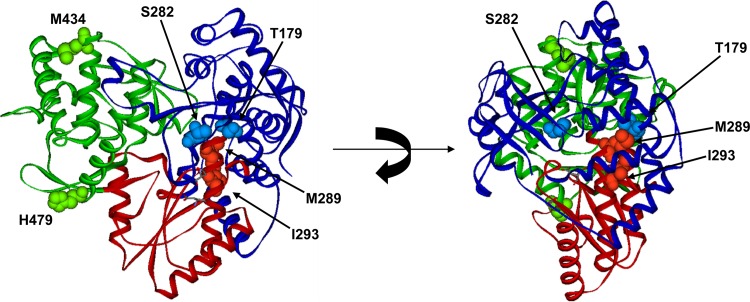
We mapped the locations of these amino acids using the JFH-1 NS5B crystal structure (37). As previously shown (6, 29), residue 282 is located close to the NS5B polymerase active site. Both M289 and I293 are located on the same alpha helix in the palm domain, while T179 is directly across from M289 about 3.8 Å away on an alpha helix within the finger domain (Fig. 2). Among these changes, T179 is closest to S282, but they are still 12.8 Å away from each other. The other two mutations, M434T and H479P, which were selected after the emergence of S282T, are located on the other side of the polymerase on the surface of the thumb domain.
Fig 2.
Locations of the PSI-7977-selected amino acid changes using JFH-1 NS5B (Protein Data Bank accession number 3I5K). The finger domain is colored blue, the thumb domain is colored green, and the palm domain is colored red. The two catalytic residues D220 and D318 are shown as sticks to show the proposed active site of RNA synthesis. The PSI-7977-selected amino acid changes (residues 179, 282, 289, 293, 434, and 479) are shown as spheres. The image on the right is a 90°C rotation of the image from the left to show the proximity of the mutations within finger and palm domains.
Phenotypic analysis of PSI-7977-selected amino acid substitutions.
Based on the sequence analysis, we generated JFH-1 replicons with each of these mutations as single or multiple changes and evaluated them for their effect on replication fitness using transient assays and susceptibility to PSI-7977 using cells that expressed the transfected replicons. Among those with single amino acid substitutions, replicons containing the S282T change were the most unfit, while those containing T179A, I293L, M289L, M434T, and H479P replicated with very similar efficiency as wild type (Table 5). No significant effect on the activity of PSI-7977 was observed with T179A, I293L, M434T, or H479P, while S282T and M289L replicons both showed slight increases (∼3-fold) in EC50s. Next, we combined amino acid substitutions within the finger and palm domains of the NS5B polymerase. Among these, three replicons contained mutations without S282T, while the other four contained mutations combined with S282T. As shown in Table 5, replicons that did not have the S282T change showed a 2.1- to 3.5-fold shift in EC50s, while replicons that contained S282T in combination with the finger and/or palm domain mutations reduced the activity of PSI-7977 by 5.6- to 9.5-fold. Combining S282T with the palm domain mutations (S282T/M289L and S282T/M289L/I293L) resulted in EC50 fold shifts of about 6, while combining S282T with mutations from both the finger and palm domains (T179A/S282T/I293L, T179A/S282T/M289L/I293L) resulted in EC50 fold changes of about 9. Addition of the thumb domain M434T or H479P change to the various S282T replicons with multiple mutations did not appear to further enhance the EC50 fold shifts for PSI-7977 (Table 5).
Analysis of the replication capacity of replicons containing multiple mutations also showed an interesting profile. While combining the S282T change with mutations from both the finger and palm domains enhanced the EC50 fold shifts of PSI-7977, these combinations did not appear to improve the fitness of the S282T replicon (Table 5). For example, adding the palm mutation M289L or the M289L/I293L mutations to S282T did not significantly increase the replication over that of the S282T replicon alone. Adding both finger and palm mutations (T179A/I293L or T179A/M289L/I293L) to S282T actually reduced the replication capacity to below 10% of that of the wild type. In contrast, the two amino acid changes in the thumb domain appeared to serve as compensatory mutations for the S282T variants (Table 5). Specifically, adding H479P to the S282T/M289L replicons improved the replication capacity from 32% to 73%; adding M434T or H479P to the S282T/M289L/I293L replicons improved the replication capacity from 45% to 83% and 126%, respectively; adding H479P to T179A/S282T/I293L improved the replication capacity from 1.8% to 86%; and adding M434T or H479P to T179A/S282T/M289L/I293L improved the replication capacity from 6.6% to 45% and 39%, respectively. Interestingly, the S282T/M289L/I293L/H479P replicons, which had the highest level of fitness (126%), had a low frequency in the selected replicons (Table 4). Among replicons with multiple mutations, the combination with the highest frequency was T179A/S282T/I293L/H479P. On the basis of the EC50 fold shift data, the fold shift for T179A/S282T/I293L (8.3-fold) is higher than that for S282T/M289L/I293L (5.6-fold). We speculate that the HCV replicon would favor adding the H479P change to the highest-shift mutant in order to more effectively evade the inhibition by PSI-7977.
The one exception to a relatively fit variant, despite the presence of the thumb mutation, was T179A/S282T/M289L/H479P, which replicated with the poorest fitness of only 0.26%. However, we did not observe the clone with the corresponding triple mutation without the H479P change in any cDNA clones sequenced (Table 4), suggesting that replicons with that particular combination may have impaired replication capacity.
Cross-resistance studies using JFH-1 S282T replicons.
Our phenotypic analysis of JFH-1 GT 2a replicons indicates that combination of S282T with additional amino acid changes conferred resistance to PSI-7977. To determine if this was also a requirement for other classes of nucleoside/tide analogs, representative JFH-1 GT 2a replicon variants were examined for their susceptibility to PSI-6130 (2′-F-2′-C-methylcytidine nucleoside analog of RG-7128) and to INX-189 and IDX-184 (both prodrugs of 2′-C-methylguanosine monophosphate). It has previously been reported that PSI-6130, INX-189, and IDX-184 were less active against GT 1b S282T replicons (26, 40, 43). In addition, we included in these studies PSI-352938, a prodrug of 2′-F-2′-C-methylguanosine monophosphate that remains active against GT 1b replicons with the S282T change (16). Results are summarized in Table 6. Similar to PSI-7977, our data showed that the JFH-1 GT 2a replicons containing the S282T change alone were still susceptible to PSI-6130 (EC50 fold change, 1.4). Interestingly, the JFH-1 GT 2a S282T replicons displayed 3.8- and 7.4-fold reductions in sensitivity to the 2′-C-methylnucleotide prodrugs IDX-184 and INX-189, respectively. Addition of the other finger, palm, and thumb mutations did further increase the EC50 fold changes for PSI-7977, PSI-6130, INX-189, and IDX-184. The greatest fold shifts were associated with the 2′-C-methylnucleotide prodrugs (14- to 19-fold), followed by PSI-7977 (9-fold) and PSI-6130 (5-fold). All JFH-1 GT 2a S282T replicon variants, either a single mutation or mutations in combination, remained susceptible to PSI-352938.
Table 6.
Cross-resistance studies of nucleoside/tide inhibitors against JFH-1 GT 2a S282T replicon variantsa
| Replicon | Fold change in EC50 |
||||
|---|---|---|---|---|---|
| PSI-7977 | PSI-6130 | INX-189 | IDX-184 | PSI-352938 | |
| S282T | 2.8 ± 0.7 | 1.4 ± 0.2 | 7.4 ± 3.8 | 3.8 ± 0.8 | 0.5 ± 0.07 |
| S282T/M289L | 6.3 ± 1.0 | 3.7 ± 0.7 | 22.4 ± 6.4 | 6.4 ± 2.0 | 1.2 ± 0.1 |
| T179A/S282T/M289L/I293L | 9.5 ± 2.5 | 5.3 ± 1.9 | 18.7 ± 8.1 | 15.2 ± 3.9 | 1.5 ± 0.9 |
| T179A/S282T/I293L/H479P | 9.0 ± 1.5 | 4.8 ± 1.1 | 19.0 ± 5.0 | 14.1 ± 3.2 | 1.1 ± 0.3 |
Lunet cells expressing the JFH-1 GT 2a replicon variants were tested for cross-resistance among PSI-7977, PSI-6130, INX-189, IDX-184, and PSI-352938. EC50 fold change was determined by normalizing the EC50 from the replicon variants with that of the wild type. Results are reported as average ± standard deviation from at least three independent experiments performed in duplicate.
Effect of S282T in other NS5B isolates.
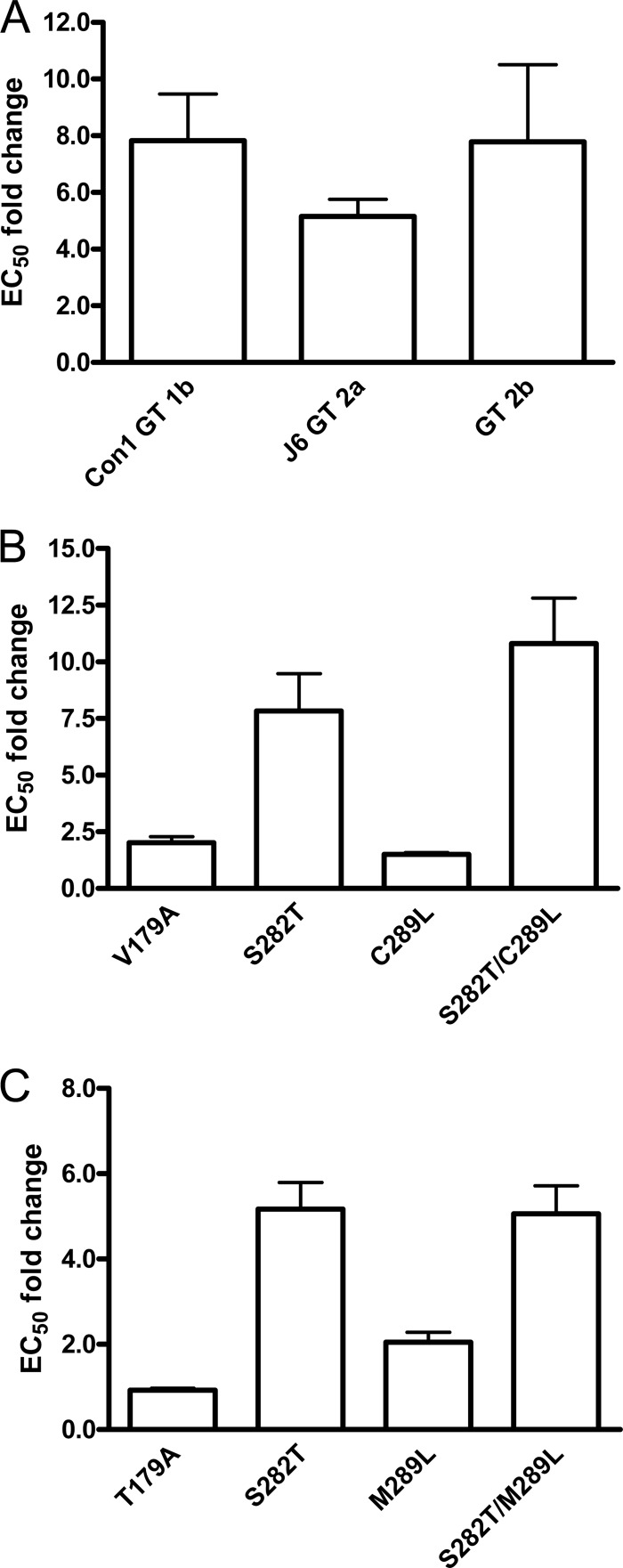
Examination of the various HCV NS5B isolates from the Los Alamos database indicates that with the exception of residues 282 and 479, positions 179, 289, 293, and 414 are not highly conserved among the various HCV genotypes. Interestingly, while position 282 is mostly a Ser, position 479 is mostly a Pro in HCV NS5B, including other GT 2a isolates. We first introduced the S282T change alone into chimeric replicons containing NS5B from J6 GT 2a, GT 2b, and GT 3a in order to determine if S282T was sufficient to confer resistance to PSI-7977. Data from the transient assays showed that the activity of PSI-7977 was reduced by 5.2- and 7.8-fold in J6 GT 2a and GT 2b S282T chimeric replicons, respectively, quite comparable to that in GT 1b S282T replicons (Fig. 3A). We were not able to determine an EC50 for PSI-7977 using GT 3a S282T because this substitution was highly unfit (the wild type itself was the least efficient in replication among the various replicons). The effect of substituting amino acids 179 and 289 with Ala and Leu, respectively, in Con1 GT 1b and J6 GT 2a NS5B was also examined. Residue 179 is a Val and residue 289 is a Cys in Con1 GT 1b, while J6 GT 2a NS5B contains the same amino acids at positions 179 and 289 as JFH-1. Similar to JFH-1, the mutation in GT 1b and J6 GT 2a replicons with single substitution of V/T179A or C/M289L did not significantly affect the activity of PSI-7977 (Fig. 3B and C). Addition of C289L to S282T in Con1 GT 1b showed a slight increase in EC50 fold shifts, from 7.8 in the S282T replicon to 10.8 in the double mutant (Fig. 3B), but the replication fitness of the C289L/S282T replicons was about 10-fold lower than that of the S282T replicons (data not shown). In contrast to JFH-1, adding M289L to S282T in the J6 GT 2a replicons did not appear to further enhance the resistance mediated by the S282T change alone (Fig. 3C).
Fig 3.
Effect of residue 282, 179, and 289 alterations in replicons from other genotypes. (A) S282T change in GT 1b replicons and chimeric replicons with J6 GT 2a or GT 2b NS5B showed reduced susceptibility to PSI-7977. (B) Effect of V179A, S282T, C289L, and S282T/C289L in Con1 GT 1b replicons on PSI-7977 activity. (C) Effect of T179A, S282T, M289L, and S282T/M289L in J6 GT 2a chimeric replicons on PSI-7977 activity. Susceptibility studies of PSI-7977 were performed using a 4-day transient assay in Lunet cells transfected with replicon RNA. Results are reported as average ± SD from at least three independent experiments performed in duplicate.
DISCUSSION
The genetic diversity of HCV, which classifies the virus into six major genotypes, multiple subtypes, and numerous quasispecies, presents the virus with opportunities to naturally select for the genetic isolate that is best fit for replication. With the development of antiviral compounds and the monitoring of resistance, it has become apparent that mutations which would allow the virus to escape inhibition by drugs will be enriched over the baseline genomic sequences. These studies have also led to the identification of subtype-dependent resistant mutations for NS3 protease, NS5A, and NS5B nonnucleoside inhibitors in both replicons and clinical isolates (7, 21, 27, 32, 42). Compared to these three classes of HCV inhibitors, relatively less is known about genotype- and subtype-dependent resistance for nucleoside/tide analogs targeting the active site of NS5B. Herein we present anti-HCV results, cross-resistance analysis, and resistance selection studies of the phosphoramidate nucleotide prodrug PSI-7977 using replicons from various genotypes and subtypes.
Results from studies using GT 1a (H77)-, 1b (Con1)-, and 2a (JFH-1)-derived replicons and chimeric replicons with the NS5B region from GT 2a (J6), 2b, and 3a clearly showed that PSI-7977 is a potent HCV inhibitor across NS5B proteins from different isolates. Our previous work with PSI-7409, the active 5′-triphosphate metabolite of PSI-7977, also showed that it inhibited the enzymatic activity of NS5B polymerase from GTs 1 to 4 with similar 50% inhibitory concentrations (18). In comparison, nonnucleoside inhibitors are more selective in their targets and exhibit a genotype- and/or subtype-dependent range of activities. Currently, we are working on generating replicative chimeric replicons with NS5B from GTs 4 to 6, but as the 5′-triphosphate metabolite of PSI-7977 targets the active conserved site of the polymerase, we expect that PSI-7977 would retain its activity in these constructs.
Cross-resistance studies using the panels of replicons with mutations in NS3/4A protease, NS5A, and NS5B showed that with the exception of GT 1b S282T, these replicon variants remained fully susceptible to PSI-7977. An efficacious antiviral therapy for hepatitis C patients will likely require the combination of two or more HCV inhibitors to prevent the emergence of resistant variants. Our data therefore indicate that PSI-7977 can be combined with different classes of HCV inhibitors, including ribavirin and the 2′-F-2′-C-methylguanosine analogs, which exhibit a different resistance profile. It was interesting to note that the F415Y change, which was previously observed in GT 1a-infected patients treated with ribavirin (47), did not confer resistance to ribavirin in our hands. As GT 1a replicons were not available at that time, the cross-resistance data from the Young et al. study were generated using GT 1b replicons (which naturally contained a Tyr at position 415) and reverse substitution to generate the Y415F change (47). It is possible that the F415Y change alone in GT 1a replicons is not sufficient to reduce the activity of ribavirin.
Unlike the NS3 protease, NS5A, and NS5B nonnucleoside inhibitors, PSI-7977 did not appear to exhibit genotype- or subtype-dependent resistance, as it selected the S282T change in replicons from H77 GT 1a, Con1 GT 1b, and JFH-1 GT 2a. While selection was not performed with the chimeric replicons, variants containing NS5B from the J6 GT 2a and GT 2b isolates confirmed that S282T did confer resistance to PSI-7977, comparable to the effect observed in GT 1b and 1a S282T replicons. Interestingly, analysis of the PSI-7977-selected JFH-1 GT 2a replicons indicates that JFH-1 was capable of enriching for amino acid changes in addition to S282T and that reduction of PSI-7977 activity was associated with additional mutations in combination with S282T. In particular, S282T together with mutations from both the finger (T179A) and palm (M289L and I293L) domains was essential to conferring resistance to PSI-7977. The two changes on the surface of the thumb domain, M434T and H479P, each showed a compensatory effect that improved the fitness of the S282T variants. Using the JFH-1 infectious virus system, Cheng et al. have recently reported that PSI-7851 (a diastereoisomer mixture that contains PSI-7977), but not 2′-C-methyladenosine or 2′-C-methylcytidine analogs, selected the M289L, S282T, and R543H changes (5). M289L and S282T were the dominant amino acid changes according to our genotypic analysis. Together the data support the suggestion that similar amino acid changes which at least include S282T and M289L could be selected by PSI-7851 and its pure diastereoisomer PSI-7977 using both the JFH-1 replicon and infectious virus systems.
The locations of these residues on the polymerase suggest that they could be involved in regulating the conformation of NS5B. Residues 179, 289, and 293 are within two alpha helices that are across from each other in the finger and palm domains. In particular, T179 is on top of M289, which is in turn stacked directly above I293. S282 is an extension from the helix containing residues 289 and 293. Mutations in 179, 289, and 293 could affect the interaction between the two helices, which might in turn impact the strand containing S282. Residues 434 and 479 are located on the surface of the thumb domain far away from the active site of RNA synthesis and thus are unlikely to directly interfere with the nucleotide substrates. Schmitt et al. have suggested that extensive hydrophobic interaction within the thumb domain of JFH-1 NS5B could provide stabilization of this enzyme (36). It is possible that altering amino acid 434 or 479 could compensate for conformation changes induced by residue 282, which is close to the active site, and residues 179, 289, and 293, which are along the alpha helices within the finger and palm domains.
The susceptibility of the JFH-1 S282T replicons appeared to vary among the different classes of nucleoside/tide inhibitors (Table 6). JFH-1 replicons with the single S282T mutation showed 4- and 7-fold increases in EC50s for IDX-184 and INX-189, respectively, but were less resistant to PSI-7977 and PSI-6130. Among JFH-1 S282T replicons with multiple mutations, PSI-6130 (a 2′-F-2′-C-methylcytidine) showed the lowest EC50 fold changes (∼5-fold). PSI-7977 (a 2′-F-2′-C-methyluridine prodrug), which shares a similar modified sugar as PSI-6130 but a different base, showed ∼9-fold increases in EC50s. INX-189 and IDX-184 (prodrugs of 2′-C-methylguanosine), which contain a different modified sugar and a different base, had the highest increases in EC50s (19- and 15-fold, respectively). In contrast, PSI-352938 (a prodrug of 2′-F-2′-C-methylguanosine monophosphate), which shares the same 2′-F-2′-C-methyl-modified sugar as PSI-7977 and PSI-6130 but carries a different base, remained active against the S282T replicon variants. These data suggest that the Thr substitution at residue 282 and possibly its nearby amino acids could affect the interaction with the sugar and base moieties of the nucleotides. Work is under way using molecular modeling, crystallization, and mutagenesis of residue 282 within both the replicons and NS5B polymerase to determine how this would affect nucleotide interaction.
In conclusion, PSI-7977 is a potent HCV inhibitor with broad genotype coverage. Cross-resistance and selection studies showed that S282T is likely the amino acid change that will be selected by PSI-7977 across various genotypes and subtypes. JFH-1 is a highly unique strain capable of efficient replication and infection (11, 44), and this particular isolate appeared to require additional amino acid changes together with S282T to reduce the activity of PSI-7977. Our studies presented here further suggest that it is possible for the diverse genome of HCV to evolve a mechanism to compensate for the poor fitness as a result of the S282T amino acid alteration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ralph Mosley for helpful discussion.
All authors were past employees of Pharmasset, Inc.
Footnotes
Published ahead of print 19 March 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Ali S, et al. 2008. Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479. Antimicrob. Agents Chemother. 52:4356–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beaulieu PL, et al. 2011. From benzimidazole to indole-5-carboxamide thumb pocket I inhibitors of HCV NS5B polymerase. Part 1. Indole C-2 SAR and discovery of diamide derivatives with nanomolar potency in cell-based subgenomic replicons. Bioorg. Med. Chem. Lett. 21:3658–3663 [DOI] [PubMed] [Google Scholar]
- 3. Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butt AA, Kanwal F. 2012. Boceprevir and telaprevir in the management of hepatitis C virus-infected patients. Clin. Infect. Dis. 54:96–104 [DOI] [PubMed] [Google Scholar]
- 5. Cheng G, et al. 2011. Nucleotide inhibitor resistance selections using GT2a infectious virus: PSI-7851 and GS-6620 select for a novel resistance pathway, including substitutions of M289V/L followed by S282T. J. Hepatol. 54:S473 [Google Scholar]
- 6. Dutartre H, Bussetta C, Boretto J, Canard B. 2006. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50:4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fridell RA, et al. 2011. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935 [DOI] [PubMed] [Google Scholar]
- 8. Gao M, et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herlihy KJ, et al. 2008. Development of intergenotypic chimeric replicons to determine the broad-spectrum antiviral activities of hepatitis C virus polymerase inhibitors. Antimicrob. Agents Chemother. 52:3523–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howe AY, et al. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kato T, et al. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817 [DOI] [PubMed] [Google Scholar]
- 12. Kieffer TL, Kwong AD, Picchio GR. 2010. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J. Antimicrob. Chemother. 65:202–212 [DOI] [PubMed] [Google Scholar]
- 13. Klumpp K, et al. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793–3799 [DOI] [PubMed] [Google Scholar]
- 14. Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lam AM, et al. 2011. Hepatitis C virus nucleotide inhibitors PSI-352938 and PSI-353661 exhibit a novel mechanism of resistance requiring multiple mutations within replicon RNA. J. Virol. 85:12334–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam AM, et al. 2011. Inhibition of hepatitis C virus replicon RNA synthesis by PSI-352938, a cyclic phosphate prodrug of β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine. Antimicrob. Agents Chemother. 55:2566–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lam AM, Frick DN. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lam AM, et al. 2010. PSI-7851, a pronucleotide of beta-d-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob. Agents Chemother. 54:3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lenz O, et al. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Pogam S, et al. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim SR, et al. 2012. Virologic escape during danoprevir (ITMN-191/RG7227) monotherapy is hepatitis C virus subtype dependent and associated with R155K substitution. Antimicrob. Agents Chemother. 56:271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin K, Perni RB, Kwong AD, Lin C. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ludmerer SW, et al. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malcolm BA, et al. 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob. Agents Chemother. 50:1013–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarville JF, et al. Abstr. 5th Int. Workshop Hepatitis C-Resistance New Compounds, abstr. 9; Boston, MA. 2010. [Google Scholar]
- 27. McCown MF, Rajyaguru S, Kular S, Cammack N, Najera I. 2009. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob. Agents Chemother. 53:2129–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCown MF, et al. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 52:1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Migliaccio G, et al. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164–49170 [DOI] [PubMed] [Google Scholar]
- 30. Murakami E, et al. 2010. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 285:34337–34347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen TT, et al. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nyanguile O, et al. 2010. 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus. J. Virol. 84:2923–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perni RB, et al. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reddy PG, et al. 2010. 2′-Deoxy-2′-alpha-fluoro-2′-beta-C-methyl 3′,5′-cyclic phosphate nucleotide prodrug analogs as inhibitors of HCV NS5B polymerase: discovery of PSI-352938. Bioorg. Med. Chem. Lett. 20:7376–7380 [DOI] [PubMed] [Google Scholar]
- 35. Reed KE, Rice CM. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55–84 [DOI] [PubMed] [Google Scholar]
- 36. Schmitt M, et al. 2011. A comprehensive structure-function comparison of hepatitis C virus strain JFH1 and J6 polymerases reveals a key residue stimulating replication in cell culture across genotypes. J. Virol. 85:2565–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simister P, et al. 2009. Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J. Virol. 83:11926–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sofia MJ, et al. 2010. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 53:7202–7218 [DOI] [PubMed] [Google Scholar]
- 39. Sofia MJ, Chang W, Furman PA, Mosley RT, Ross BS. 2012. Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J. Med. Chem. 55:2481–2531 [DOI] [PubMed] [Google Scholar]
- 40. Stuyver LJ, et al. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79–87 [DOI] [PubMed] [Google Scholar]
- 41. Stuyver LJ, et al. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sullivan JC, et al. 2011. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. J. Hepatol. 54:S4. [DOI] [PubMed] [Google Scholar]
- 43. Vernachio JH, et al. 2011. INX-08189, a phosphoramidate prodrug of 6-O-methyl-2′-C-methyl guanosine, is a potent inhibitor of hepatitis C virus replication with excellent pharmacokinetic and pharmacodynamic properties. Antimicrob. Agents Chemother. 55:1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yanagi M, Purcell RH, Emerson SU, Bukh J. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250–263 [DOI] [PubMed] [Google Scholar]
- 46. Yang W, et al. 2008. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob. Agents Chemother. 52:2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young KC, et al. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869–878 [DOI] [PubMed] [Google Scholar]
- 48. Zhou XJ, et al. 2011. Safety and pharmacokinetics of IDX184, a liver-targeted nucleotide polymerase inhibitor of hepatitis C virus, in healthy subjects. Antimicrob. Agents Chemother. 55:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.