Abstract
IncK plasmids encoding CTX-M-14 extended-spectrum β-lactamase (ESBL) and highly related to plasmid pCT were detected in 13 of 67 (19%) human clinical isolates of Escherichia coli with a group 9 CTX-M-type ESBL from the United Kingdom and in 2 quality assurance isolates. None of these E. coli strains was related to the cattle strain from which pCT was originally characterized.
TEXT
The ongoing horizontal spread of CTX-M extended-spectrum β-lactamases (ESBLs) among Escherichia coli strains from hospitals and the community is of great public health concern. In the United Kingdom, most ESBL-producing E. coli isolates from humans have the CTX-M-15 enzyme encoded by IncF plasmids, and many belong to the uropathogenic B2-ST131 lineage. However, approximately 4% of ESBL-positive clinical E. coli isolates in the United Kingdom instead produce a CTX-M group 9 ESBL.
Globally, after CTX-M-15, the CTX-M-14 ESBL (a member of the CTX-M group 9) is the second-most-prevalent CTX-M ESBL. Its dominance in Spain is thought to be due, in part, to the spread of an epidemic IncK plasmid (7). Recently, Cottell et al. (3) described an IncK plasmid, designated pCT (GenBank accession no. F868832.1), encoding CTX-M-14 ESBL, from an E. coli strain isolated from cattle in 2004 in the United Kingdom. Cottell et al. also detected pCT-like plasmids in Spanish, Australian, and Chinese isolates of E. coli (3).
We sought pCT-like plasmids among a United Kingdom collection of human clinical isolates of E. coli producing CTX-M group 9 ESBLs. Furthermore, we sought the presence of five pCT genetic regions to provide further insights into their distribution among pCT-like and other plasmids.
Sixty-nine E. coli isolates producing CTX-M group 9 ESBLs were recovered from the collection held by the United Kingdom Health Protection Agency's Antibiotic Resistance Monitoring and Reference Laboratory. All were United Kingdom isolates, and most were from human clinical samples obtained during 2003 to 2010, although two had been distributed as quality assurance specimens in the United Kingdom National External Quality Assessment Service (UK NEQAS) scheme. Identification of pCT-like plasmids was by PCR, as described by Cottell et al. (3). A bovine E. coli strain containing pCT (3) was used as a positive control. The method seeks five pCT genetic regions (pCT putative sigma factor, pCT shufflon recombinase, pCT pilN outer membrane protein, IncI complex nikB relaxase gene, and the pCT008-009 region). The CTX-M group 9 gene was sequenced by DNA sequencing to confirm the precise allele in selected isolates (3). IncK plasmid rep types were sought by PCR (1). The genetic relatedness of isolates found to produce CTX-M-14 ESBL and to harbor pCT-like IncK plasmids was examined by pulsed-field gel electrophoresis (PFGE) (8) and phylogenetic grouping (2).
Among the 69 E. coli isolates, 15 (22%) harbored plasmids that belonged to incompatibility group IncK as determined by rep PCR (1). These included 13 human clinical isolates and 2 quality assurance isolates; the remaining 54 (78%), all clinical isolates, did not harbor IncK plasmids. DNA sequencing confirmed that these 15 all carried blaCTX-M-14 and were positive by PCR for each of the five pCT genetic regions, indicating that they possessed pCT-like plasmids. The 13 clinical isolates were referred from nine different United Kingdom clinical laboratories, and the 2 quality assurance isolates were distributed by UK NEQAS, with their original source undisclosed to us. These isolates represented 11 different PFGE types at 85% banding similarity, and all were unrelated to the bovine strain of E. coli from which pCT was originally detected and characterized (Fig. 1). Three clusters were identified, three identical isolates from laboratory E and pairs of identical isolates from laboratory I and UK NEQAS (Fig. 1). The 15 isolates with pCT-like plasmids all belonged to commensal E. coli phylogenetic groups, specifically A and B1, rather than the virulent extraintestinal groups B2 and D, which commonly produce CTX-M-15 ESBL (6, 8).
Fig 1.
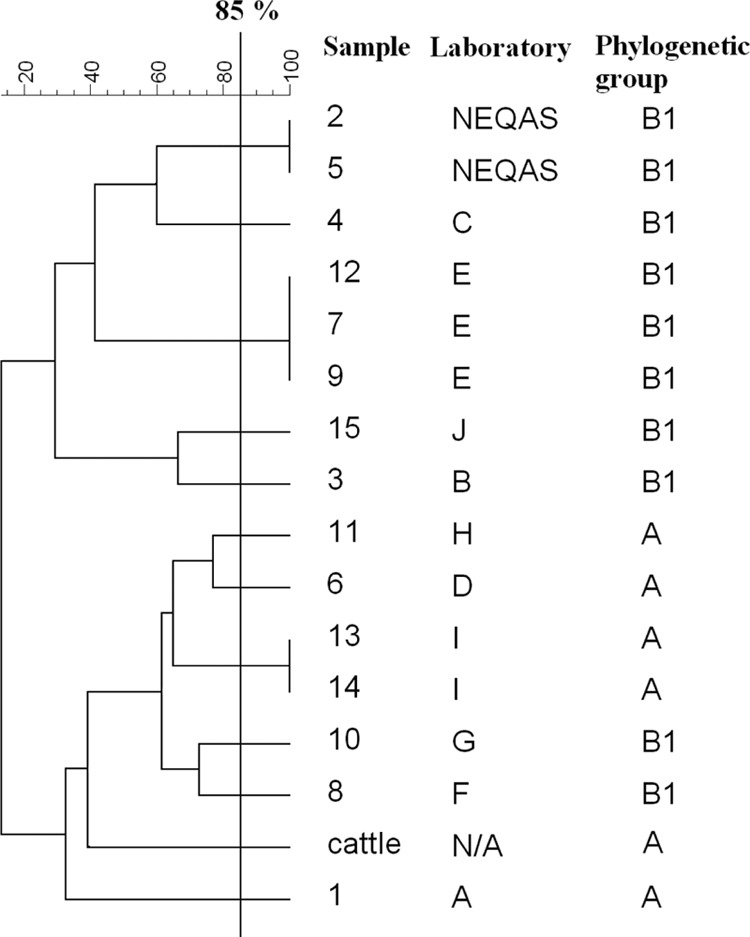
Dendrogram showing the genetic relatedness based on PFGE typing among the 15 E. coli producing CTX-M-14 ESBL and harboring pCT-like plasmids and a pCT-positive E. coli isolate from cattle (GenBank accession no. F868832.1).
Among the 13 human clinical isolates with pCT-like plasmids, 12 dated from 2004 to 2005 and the remaining clinical isolate dated from 2006. This is not statistically significant (P = 0.16, Fisher's exact test) but is noteworthy since the strain hosting pCT was originally isolated in 2004. The two NEQAS isolates were distributed in 2004. It is possible that CTX-M-14 ESBL encoded on IncK pCT-like plasmids was disseminated in the United Kingdom in both humans and animal isolates in 2004 to 2006 but that this plasmid type has subsequently become less prevalent, at least among human clinical isolates; this hypothesis requires further investigation.
The 54 E. coli isolates not harboring IncK rep-type plasmids were all negative by PCR for the pCT shufflon recombinase element and the pCT008-009 region element, both of which are thought to be unique to pCT-like plasmids. However, nine (13%) isolates were positive for the other three pCT elements sought, i.e., the IncI complex nikB relaxase element, the pCT pilN outer membrane protein, and the pCT putative sigma factor (profile 1). A further 10 (15%) were positive only for the IncI complex nikB relaxase element (profile 2). The nikB genes of pCT-like plasmids show little sequence divergence, in contrast to the nikB genes of non-pCT like plasmids (3), and the core genes of pCT related to replication or transfer are similar to those found on plasmids of rep type IncI (3). The remaining 35 (51%) isolates were negative for all pCT elements (profile 3), definitively showing that their group 9 CTX-M ESBLs were not encoded by pCT-like plasmids. pCT-like plasmids are IncK rep-type plasmids harboring blaCTX-M-14 which are positive by PCR for the five following pCT genetic regions: pCT putative sigma factor, pCT shufflon recombinase, pCT pilN outer membrane protein, IncI complex nikB relaxase gene, and the pCT008-009 region (3). The blaCTX-M alleles were amplified and sequenced from 12 isolates not harboring IncK plasmids and selected to represent the three different PCR profiles (4 isolates of each); all harbored blaCTX-M-14. Thus, we confirmed the findings of other studies (4, 5), which have established that IncK pCT-like plasmids are not a unique vehicle of CTX-M-14 ESBL dissemination in the United Kingdom.
The PCR method described by Cottell et al. (3) is useful for identifying pCT-like plasmids rapidly and for tracing their spread. Nevertheless, the present data show that three of the five genetic regions used as markers were not unique either to pCT or to IncK plasmids, confirming the findings of Cottell et al. (3). Our data suggest that a more cost-effective approach would be to screen isolates for the pCT shufflon recombinase element and the pCT008-009 region element (both of which were only present in E. coli isolates with incompatibility group IncK plasmids and were always associated with the other three regions—the IncI complex nikB relaxase element, pCT pilN outer membrane protein, and pCT putative sigma factor), followed by screening only positive isolates for the remaining three pCT regions.
In summary, pCT-like plasmids encoding CTX-M-14 ESBL were detected in diverse clinical strains of E. coli from different parts of the United Kingdom. However, these plasmids, which resembled an element first characterized from a bovine strain (3), were found in only one-fifth of the isolates with group 9 CTX-M ESBLs, and none of the host E. coli strains was related to the original bovine host strain. This study illustrates the complex biogeography of horizontally acquired antimicrobial resistance elements, which includes plasmids that are shared by distinct E. coli strains of animal and human origin.
ACKNOWLEDGMENTS
This work was supported by funding from the European Community (TROCAR contract HEALTH-F3-2008-223031).
N.W. and D.M.L. have both accepted research grants and speaking invitations from various pharmaceutical companies. D.M.L. has shares in GlaxoSmithKline, Merck, Pfizer, Dechra, and Eco Animal Health within diversified portfolios. All other authors have no interests to declare.
Footnotes
Published ahead of print 26 March 2012
REFERENCES
- 1. Carattoli A, et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 2. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cottell JL, et al. 2011. Complete sequence and molecular epidemiology of an IncK epidemic plasmid encoding blaCTX-M-14 widely disseminated in humans and animals. Emerg. Infect. Dis. 17:645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dhanji H, et al. 2011. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum beta-lactamase from UK river water. J. Antimicrob. Chemother. 66:512–516 [DOI] [PubMed] [Google Scholar]
- 5. Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 67:878–885 [DOI] [PubMed] [Google Scholar]
- 6. Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multi-resistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 7. Valverde A, et al. 2009. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53:5204–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woodford N, et al. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54:735–743 [DOI] [PubMed] [Google Scholar]


