Abstract
Deletion of Bacillus subtilis spores' GerA germinant receptor (GR) had no effect on spore germination via the GerB plus GerK GRs, and loss of GerB plus GerK did not affect germination via GerA. Loss of one or two GRs also did not affect levels of GRs that were not deleted. Overexpression of GRs 5- to 18-fold increased rates of germination via the overexpressed GR and slowed germination by other GRs up to 15-fold. However, overexpression of one or two GRs had no effect on levels of GRs that were not overexpressed. These results suggest that either interaction between different GRs reduces the activity of GRs in triggering spore germination or all GRs compete for interaction with a limiting amount of a downstream signaling molecule in the germination pathway. Overexpression or deletion of GRs also had no effect on spores' levels of the GerD protein needed for normal GR-dependent germination or of the SpoVAD protein likely involved in dipicolinic acid release early in germination. Loss of GerD also had no effect on levels of GRs or SpoVAD. Spores of a strain lacking the only B. subtilis prelipoprotein diacylglycerol transferase, GerF, also had no detectable GerD or the GerA's C subunit, both of which are most likely lipoproteins; GerA's A subunit was also absent. However, levels of GerB's C subunit, also almost certainly a lipoprotein, and GerK's A subunit were normal in gerF spores. These results with gerF spores were consistent with effects of loss of GerF on spore germination by different GRs.
INTRODUCTION
While spores of various Bacillus species can remain dormant for extremely long periods, if nutrients return to their environment, spores can rapidly return to life via the process of germination (26, 31, 32). Nutrients trigger spore germination via their interaction with germinant receptors (GRs), proteins that recognize and respond to specific nutrients such as sugars and amino acids. GRs are located in the spore's inner membrane, where they are all colocalized in a small cluster (5, 9, 19, 29, 32). Binding of a specific nutrient germinant to a GR initiates the spore germination process by triggering the release of the spore core's large depot (∼20% of core dry weight) of pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]), although the mechanism whereby germinant binding triggers DPA release is not known.
The germination of Bacillus subtilis spores is the model system for studying spore germination. These spores contain three major GRs, termed GerA, GerB, and GerK, each of which contains A, B, and C subunits; all of these are required for GR function (32). The GerA GR alone triggers spore germination in response to either l-alanine or l-valine, while the GerB and GerK GRs are both required for germination with a mixture of l-asparagine, d-glucose, d-fructose, and K+ (AGFK). Loss of either GerB or GerK eliminates all response to the AGFK mixture. However, there are several GerB variants, termed GerB*, in which single amino acid changes in either the GerBA or GerBB subunit eliminate GerB's GerK dependence such that the GerB* GR alone can trigger spore germination with l-asparagine (23).
As noted above, all GRs are colocalized in a small cluster in B. subtilis spores' inner membrane (9), suggesting that there may be some GR-GR interactions, and there is also evidence for interactions between subunits of different GRs (4, 12). In addition, there is synergy between different B. subtilis GRs, including between the GerA and the GerB plus GerK GRs and between the GerA and the GerB* GRs, in particular at low concentrations of multiple germinants that activate the synergizing GRs (38). There are also reports that overexpression of one GR, for example, GerA, can increase the rate of spore germination with the GerA GR's cognate germinants, while decreasing rates of germination via other GRs (1, 3). The latter result is particularly striking, as it suggests that GRs may compete for a downstream signaling molecule in the germination pathway that could be present in limiting amounts such that overexpression of one GR reduces access of other GRs to this hypothetical signaling molecule. However, an equally plausible explanation for the inhibition of germination via one GR by overexpression of another is that overexpression of one GR drastically decreases the levels of other GRs. Indeed, decreases in levels of a particular GR or GRs result in decreased rates of spore germination via this GR or these GRs (7, 8, 24, 29). Recently, a battery of specific antisera have been generated that specifically recognize a number of B. subtilis germination proteins, including GR subunits, so it is now possible to determine levels of these proteins in spores (7, 16, 29, 34). Consequently, in the present work, we have measured rates of germination of spores of a variety of isogenic strains either with deletions of operons encoding GRs or with overexpressed GRs. We have also determined the levels of GR subunits and two other likely germination proteins, GerD and SpoVAD, in these spores, as well as in spores that lack GerD and the prelipoprotein diacylglycerol transferase, GerF, since GerD and GRs' C subunits are almost certainly lipoproteins (11, 31). The results indicate that while overexpression of one GR drastically reduced germination via other GRs, this had no effect on the level of the other GRs or levels of GerD or SpoVAD in the spore's inner membrane. These results thus suggest that either (i) there are GR-GR interactions that are needed for rapid spore germination, and formation of such productive interactions is blocked when levels of another GR are increased, or (ii) all GRs compete for binding to a hypothetical downstream signaling molecule needed for transduction of the GR signal to trigger DPA release. In addition, loss of GerD had no effect on spore levels of GR subunits or SpoVAD, loss of GerF reduced levels of GerD, GerAA, and GerAC in spores >20-fold, but loss of GerF had no discernible effect on spore levels of SpoVAD or GerB or GerK subunits.
MATERIALS AND METHODS
Spore preparation and purification.
The B. subtilis strains used in this work are isogenic derivatives of strain PS832, a prototrophic laboratory derivative of strain 168, and are listed in Table 1. Spores of these strains were prepared on rich 2× Schaeffer's medium-glucose agar plates at 37°C, and spores were harvested, purified, and stored as described previously (21, 22). All spore preparations used in this work were free (>98%) of growing or sporulating cells, germinated spores, and cell debris as determined by phase-contrast microscopy.
Table 1.
B. subtilis strains used
| Strain | Genotypea | Reference |
|---|---|---|
| FB10 | gerB* | 23 |
| FB20 | ΔgerA | 24 |
| FB58 | PsspB::gerB | 1 |
| FB60 | ΔgerB | 24 |
| FB61 | ΔgerA ΔgerB | 24 |
| FB62 | ΔgerD | 11 |
| FB68 | ΔgerK | 24 |
| FB72 | ΔgerA ΔgerB ΔgerK | 24 |
| FB87 | ΔgerB ΔgerK | 24 |
| PS533 | Wild type | 30 |
| PS3301 | ΔgerF | 11 |
| PS3407 | ΔgerD ΔgerF | 11 |
| PS3415 | PsspB::gerB* | 3 |
| PS3476 | PsspD::gerA | 3 |
| PS3477 | PsspD::gerB | 3 |
| PS3478 | PsspD::gerK | 3 |
| PS3499 | gerB* ΔgerK | 3 |
| PS3501 | PsspD::gerAgerB* | 3 |
| PS3502 | PsspD::gerB* | 3 |
| PS3521 | ΔgerAgerB* | 3 |
| PS3557 | PsspD::gerB PsspD::gerK | 3 |
| PS3665 | ΔgerA ΔgerKgerB* | 1 |
“PsspB::” or “PsspD::” indicates that the gene following is under the control of the promoter of the sspB or sspD gene, respectively.
Spore germination.
Rates of B. subtilis spore germination were determined by measurement of the release of spores' large DPA depot by its fluorescence with Tb3+ using a multiwell fluorescence plate reader as described previously (39). Spore germination was routinely preceded by a heat shock of 30 min at 75°C followed by cooling on ice for ≥15 min. Germination was at 37°C, and conditions were as follows; all incubation mixtures contained 50 μM TbCl3 and spores at an optical density at 600 nm of 0.5 (i) with various concentrations of l-valine in 25 mM HEPES buffer (pH 7.4); (ii) with various concentrations of l-asparagine plus 10 mM d-glucose, 10 mM d-fructose, and 10 mM KCl (GFK) in 25 mM HEPES buffer (pH 7.4); and (iii) with various concentrations of l-asparagine in 25 mM HEPES buffer (pH 7.4). In these experiments, aliquots were also examined on a slide by phase-contrast microscopy to distinguish phase-bright (dormant) and phase-dark (germinated) spores at the end of germination incubations to determine the percentage of spores that had germinated. The relative rates of germination for different spore preparations were corrected for any slight differences in spore DPA content. Total spore DPA content was determined by boiling samples of dormant spores for 15 min, cooling on ice, centrifugation, and measurement of DPA in the supernatant fluid by its fluorescence with Tb3+ (39). All rates of spore germination shown in this work were determined at least in duplicate with two to four independent spore preparations. The significance of differences in rates of germination of spores of different strains was determined by a two-tailed Student t test.
Antibody production and purification.
The preparation of rabbit antisera against the B. subtilis GerAA, GerAC, GerBC, GerKA, and SpoVAD proteins was described previously, as was the source of all secondary antisera (7, 16, 29, 34). For production of antisera against the B. subtilis GerD protein, a truncated gerD gene lacking the signal sequence and the signal for diacylglycerol addition was amplified by PCR using genomic DNA from B. subtilis strain PS832 as the template; the 5′ primer introduced a NotI site and the 3′ primer introduced a KpnI site. The gerD PCR product was cloned into a modified pGEX plasmid containing a TEV protease cleavage site between an N-terminal glutathione S-transferase tag fused to the gerD gene. The GerD protein (residues 27 to 185) was expressed in Escherichia coli BL21 cells by induction with 1 mM isopropyl-β-d-thiogalactoside at 21°C for 16 h. The GerD protein was soluble and was purified by glutathione affinity chromatography followed by TEV protease cleavage and anion exchange and gel filtration chromatography.
The purified GerD protein was dialyzed against PBS (50 mM sodium phosphate, 150 mM NaCl [pH 7.2]), adjusted to a concentration of 1 mg/ml in PBS, and supplied in solution for polyclonal antibody production in rabbits (Pocono Rabbit Farm and Laboratory, Canadensis, PA). The antibody was detected in a bleed 2 months after initial injection, and the GerD antiserum was used without further treatment.
Determination of GR subunit, GerD, and SpoVAD levels in spores.
Levels of various GR subunits, GerD, and SpoVAD in spores were determined by Western blot analysis of equal aliquots of spores' inner membrane proteins that were isolated as described previously (25), since GRs, GerD, and SpoVAD are located in the spore's inner membrane (5, 7, 19, 28, 29, 34). The binding of the antisera to proteins on Western blots was detected with horseradish peroxidase coupled to goat anti-rabbit IgG, and binding of the secondary antibody was detected by chemiluminescence using X-ray film as described previously (29). To quantitate differences in GR levels between spores of different strains, the intensities of appropriate bands on X-ray film given by different amounts of appropriate inner membrane samples in Western blot analysis were compared on the same blot (see below). In a number of cases, blots were stripped and then reprobed with a different antiserum as described previously (29).
For all Western blot analyses, since recoveries of the inner membrane fraction could well vary between spore preparations, serial 2-fold dilutions of inner membrane fractions that were to be compared were run on SDS-polyacrylamide gel electrophoresis as described previously (16, 29), the gels were stained with Coomassie blue, and equivalent amounts of inner membrane protein were determined by inspection of the stained gels and further similar analyses if needed. This allowed quantitative comparison of levels of GR subunits, GerD, and SpoVAD in samples of inner membrane protein from different spore preparations. All quantitations of germination protein levels in spores of different strains were carried out at least twice on inner membrane fractions from at least two independent spore preparations using both visual inspection of Western blots and the program ImageJ, which gave extremely similar results. The significance of differences in levels of germination proteins in inner membrane fractions from spores of different strains was determined by a two-tailed Student t test.
RESULTS
Rates of AGFK and l-valine germination of spores with overexpressed or deleted GRs.
The overexpression of operons encoding GR subunits used promoters of several ssp genes that encode the abundant small, acid-soluble dormant spore proteins, either the moderately strong sspD promoter (PsspD) or the stronger sspB promoter (PsspB) (18, 31, 32). Like the operons encoding GRs, ssp promoters are expressed only in the developing forespore, and at the same time as the operons encoding GRs (18, 32, 33). Previous work showed that overexpression of the gerA operon from PsspD gives spores (↑gerA spores) that germinate faster with both saturating and subsaturating l-valine concentrations via the GerA GR than do wild-type spores (1, 3). This was also seen in the present work at both saturating and subsaturating l-valine concentrations, and the differences between rates of germination of wild-type and ↑gerA spores were significant (Fig. 1A). In contrast, deletion of the operon encoding the GerB or GerK GR or of both operons had no significant effect on germination via the GerA GR (Fig. 1A). l-Valine germination was also not affected by overexpression of the GerB GR from PsspD or PsspB (↑gerB and ↑↑gerB spores, respectively) (Fig. 1B and data not shown). However, overexpression of GerK from PsspD (↑gerK spores) or both GerB and GerK from PsspD (↑gerB ↑gerK spores) caused notable decreases in the rates of l-valine germination, and these differences were significant (Fig. 1B). It would have been valuable to examine the effects of overexpression of gerK from the stronger PsspB on l-valine germination. However, strains with gerK under PsspB control do not complete sporulation, as they appear to germinate and lyse within the developing sporangium, and this is also true of strains that carry gerA under PsspB control (3).
Fig 1.
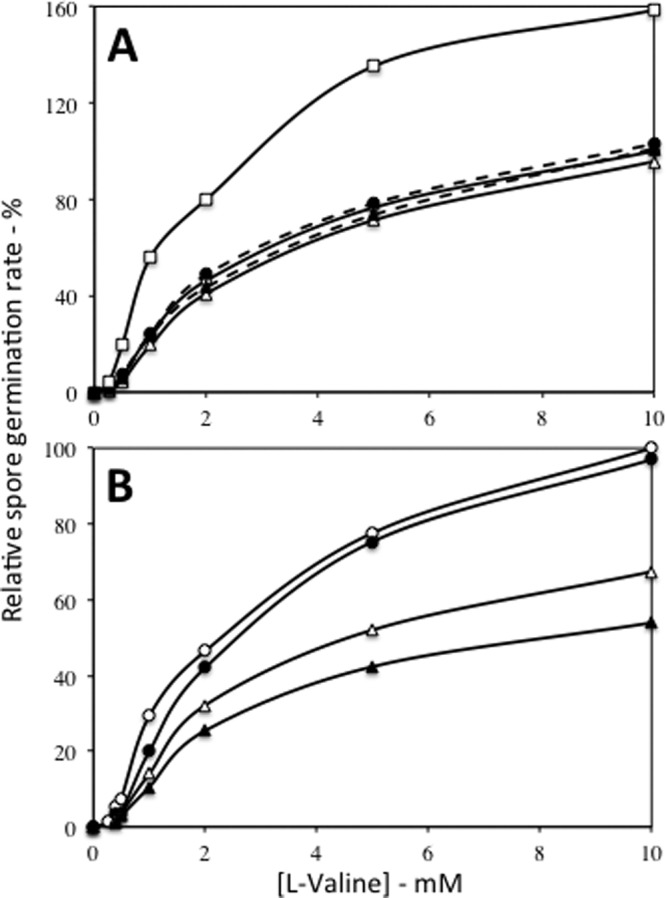
Effects of the absence of the GerB or GerK GRs or the overexpression of the GerA, GerB, or GerK GRs on the rate of l-valine germination via the GerA GR. Spores of various strains were prepared, purified, and germinated with various l-valine concentrations. Germination was measured by monitoring DPA release, and rates of DPA release were determined as described in Materials and Methods. The rate of germination of PS533 spores (wild-type) with 10 mM l-valine was defined as 100%. The symbols used for the spores of the various strains are as follows: (A) ○, PS533 (wild type); ●, FB60 (ΔgerB); ▵, FB68 (ΔgerK); ▲, FB87 (ΔgerB ΔgerK); and □, PS3476 (↑gerA); and (B) ○, PS533 (wild type); ●, PS3477 (↑gerB); ▵, PS3478 (↑gerK); and ▲, PS3557 (↑gerB ↑gerK). All rates shown are averages of results from at least duplicate measurements with at least two different spore preparations, and differences in relative germination rates were ≤15%. In panel A, the differences in the germination rates at ≥0.3 mM l-valine between wild-type spores and ↑gerA spores were significant (P < 0.001), but the differences between rates of germination of wild-type and ΔgerB, ΔgerK, or ΔgerB ΔgerK spores at different l-valine concentrations were not significant (P < 0.1 to P < 1). In panel B, the differences in the germination rates at ≥1 mM l-valine between wild-type spores and ↑gerK or ↑gerB ↑gerK spores were significant (P < 0.02), while differences between l-valine germination of wild-type and ↑gerB spores were not significant (P < 0.2). Note that in panel A, the open circles representing strain PS533 are largely obscured by the filled circles and triangles.
Analysis of AGFK germination via the GerB plus GerK GRs in strains with either overexpressed or missing GRs gave some similar results, in that overexpression of GerB, GerK, or GerB plus GerK under PsspD control increased rates of spore germination with AGFK significantly (Fig. 2), and similar results were obtained when the GerB GR was overexpressed from PsspB (data not shown). These differences were seen at asparagine concentrations of ≥0.5 mM in the AGFK mixture. Overexpression of GerA also inhibited AGFK germination >15-fold at low l-asparagine concentrations and ∼9-fold at saturating l-asparagine concentrations (Fig. 2). Deletion of the gerA operon also resulted in spores that germinated slightly faster with AGFK, but this difference was not significant (P < 0.5).
Fig 2.
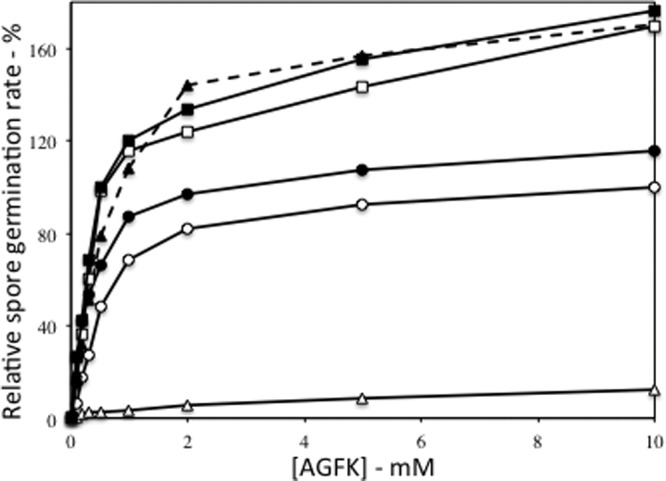
Effect of the absence of the GerA GR or the overexpression of the GerA, GerB, or GerK GRs on the rate of AGFK germination via the GerB plus GerK GRs. Spores of various strains were prepared, purified, and germinated with various concentrations of l-asparagine plus GFK. Germination was measured by monitoring DPA release, and rates of DPA release were determined as described in Materials and Methods. The rate of germination of PS533 (wild-type) spores with 10 mM l-asparagine plus GFK was defined as 100%. The symbols used for the spores of the various strains are as follows: ○, PS533 (wild type); ●, FB20 (ΔgerA); ▵, PS3476 (↑gerA); ▲, PS3477 (↑gerB); □, PS3478 (↑gerK); and ■, PS3577 (↑gerB ↑gerK). All rates shown are averages of results from at least duplicate measurements with at least two different spore preparations, and differences in relative germination rates were ≤15%. The differences in germination rates at ≥0.5 mM l-asparagine plus GFK between wild-type spores and ↑gerA, ↑gerB, ↑gerK, or ↑gerB ↑gerK spores were significant (P < 0.01), but the differences between the germination rates of wild-type and ΔgerA spores were not (P < 0.9).
Rates of l-asparagine germination of gerBB* spores with overexpressed or deleted GRs.
To further probe the effects of overexpression or deletion of GRs on spore germination, we examined spores of strains carrying the gerBB* mutation, resulting in a GerB* GR that can germinate in response to l-asparagine alone (23). The l-asparagine germination of spores carrying the GerB* GR was stimulated significantly by loss of both the GerK and GerA GRs, but not by loss of the GerA GR alone (Fig. 3A). GerB*-containing spores' germination with l-asparagine was also inhibited ∼9-fold by GerA overexpression at saturating l-asparagine levels, similar to the inhibition of AGFK germination via the GerB plus GerK GRs by GerA overexpression (Fig. 1 and 3A). As seen previously, overexpression of the GerB* GR from either PsspD or PsspB (↑gerB* or ↑↑gerB* spores, respectively) resulted in significant increases in the rate of l-asparagine germination (Fig. 3B), although with little effect on l-valine germination (data not shown) (1, 3). However, analysis of the effects of overexpression of the GerB* GR on l-valine germination via the GerA GR was complicated by the fact that the GerB* GR can also trigger spore germination with l-valine alone (X. Yi, B. Setlow, and P. Setlow, unpublished data).
Fig 3.
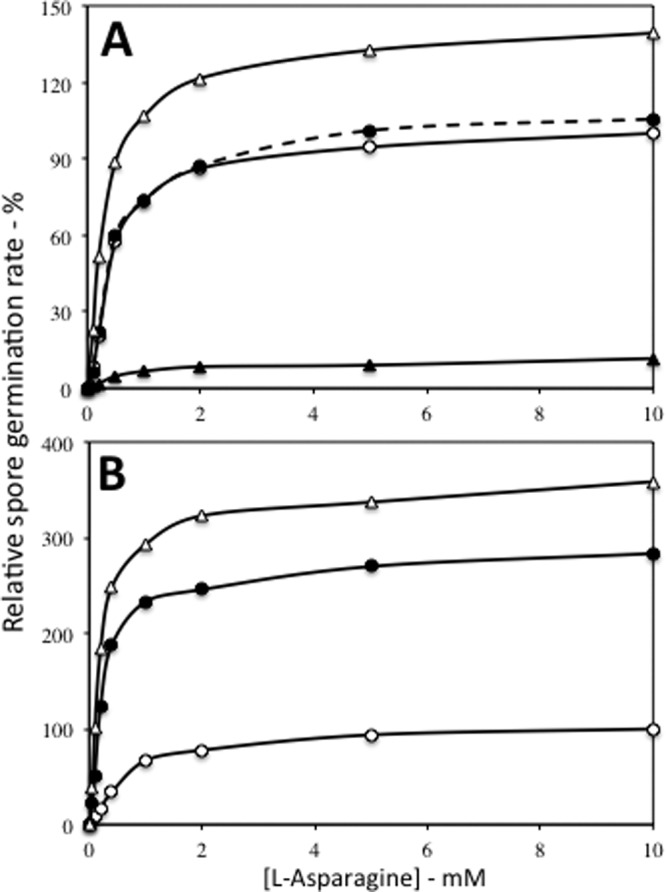
Effects of the absence of the GerA or GerK GRs and overexpression of the GerA or GerB* GRs on the rate of l-asparagine germination via the GerB* GR. Spores of various strains were prepared, purified, and germinated with various l-asparagine concentrations. Germination was measured by monitoring DPA release, and rates of DPA release were determined as described in Materials and Methods. The rate of germination of FB10 spores (gerB*) with 10 mM l-asparagine was defined as 100%. The symbols used for the spores of the various strains are as follows: (A) ○, FB10 (gerB*); ●, PS3521 (ΔgerA gerB*); ▵, PS3665 (ΔgerA gerB* ΔgerK); and ▲, PS3501 (↑gerA gerB*); and (B) ○, FB10 (gerB*); ●, PS3502 (↑gerB*); and ▵, PS3415 (↑↑gerB*). All rates shown are averages of results from at least duplicate measurements with at least two different spore preparations, and differences in relative germination rates were ≤15%. In panel A, the differences in germination rates at ≥0.3 mM l-asparagine between FB10 spores and ↑gerA and ΔgerA ΔgerK spores were significant (P < 0.01), but the differences between the germination rates of FB10 and ΔgerA spores were not (P < 1). In panel B, the differences in germination rates at ≥0.5 mM l-asparagine between FB10 spores and ↑gerB* or ↑↑gerB* spores were significant (P < 0.008).
Levels of GRs in spores of strains with operons encoding GRs overexpressed or deleted.
The data presented above indicated that changes in levels of various GRs due to overexpression from PsspD or PsspB increased rates of germination via the overexpressed GR or GRs and decreased germination via those GRs that were not overexpressed. The latter effect was most evident in the large inhibition of AGFK and l-asparagine germination via the GerB plus GerK GRs and the GerB* GR, respectively, by overexpression of GerA. There was also a much smaller inhibition of l-valine germination via GerA by overexpression of GerK or GerB plus GerK. Deletion of various GRs generally had only small effects on germination via remaining GRs, although removal of GerA did appear to result in some stimulation in GerB*- or GerB-plus-GerK-dependent germination.
An obvious question about the inhibitory effects of GR overexpression on germination via GRs that are not overexpressed is whether these effects are on the activity of the GRs not overexpressed or on these GRs' levels. To answer this question, we turned to Western blot analysis of levels of a number of germination proteins in the spores of the various strains (Fig. 4 and 5; Table 2). These analyses showed first that levels of SpoVAD, and by inference levels of all of the proteins encoded by the heptacistronic spoVA operon, were similar in spores of all strains analyzed. The SpoVA proteins are thought to play a major role in DPA uptake into developing spores in sporulation and appear likely to play a role in DPA release in spore germination as well (17, 32, 34, 35). Levels of the auxiliary germination protein GerD, which is essential for normal GR-dependent germination (27), were also similar in spores of all strains with absent or overexpressed GRs.
Fig 4.
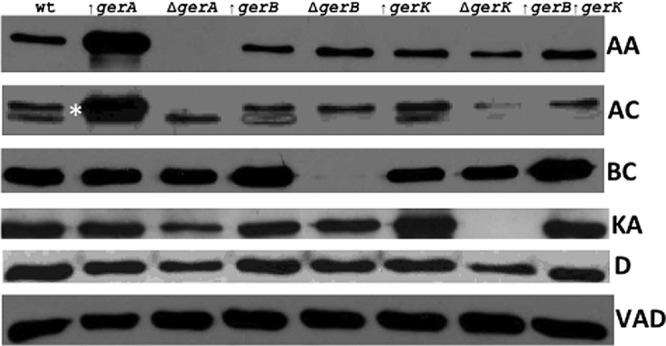
Western blot analysis of levels of germination proteins in dormant spores of various B. subtilis strains. Dormant spores of various strains were prepared and purified and the inner membrane fractions were isolated as described in Materials and Methods. Aliquots of equal amounts of inner membrane protein from the different initial dormant spores (∼0.5 mg [dry weight]) were then used for Western blot analysis using different antisera as described in Materials and Methods. The genotypes of the spores used are given above the GerAA strip, and “wt” denotes wild-type (PS533) spores. The GerAA (AA), GerAC (AC), and GerBC (BC) strips are from the same Western blot probed first with anti-GerAA serum, then stripped and reprobed with anti-GerBC serum, and then stripped and reprobed with anti-GerAC serum. The SpoVAD (VAD), GerD (D), and GerKA (KA) strips are from different Western blots and from either the same spore preparation as used for analysis of GerAA, GerAC, and GerBC (SpoVAD) or from different spore preparations (GerD and GerKA). In the GerAC strip, GerAC is the upper band in the doublet denoted by the white asterisk to the right of the wild-type lane; note that this upper band is absent in the ΔgerA spores. The lower band is GerBC that was detected earlier by anti-GerBC serum that was not removed well when the blot was stripped. Note that the apparently lower levels of GerAC in ΔgerK spores and GerKA in ΔgerA spores were not seen in other comparable Western blots.
Fig 5.
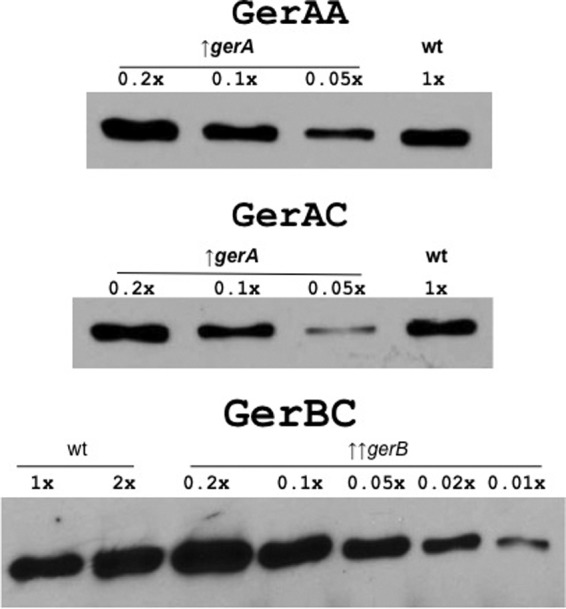
Western blot analysis of levels of overexpression of GerAA, GerAC, and GerBC from PsspD and PsspB. Spores of strains PS533 (wild-type [wt]), PS3476 (↑gerA), and FB58 (↑↑gerB) were prepared and purified, and the inner membrane fractions were isolated as described in Materials and Methods. The protein concentrations in the inner membrane fractions were determined such that the 1× amount of each one was identical and represented the inner membrane protein from ∼0.5 mg dry spores. Various amounts of inner membrane protein were analyzed on the same Western blot as probed with anti-GerAA serum and then stripped and reprobed with anti-GerAC serum to determine levels of overexpression of GerAA and GerAC in spores of the ↑gerA strain in which the gerA operon is under PsspD control. Various amounts of inner membrane protein were also analyzed on another Western blot probed with anti-GerBC serum to determine the level of overexpression of GerBC in spores of the ↑↑gerB strain, in which the gerB operon is under PsspB control.
Table 2.
Relative levels of germination proteins in spores of different strainsa
| Strain (genotype) | Relative germination protein level |
|||||
|---|---|---|---|---|---|---|
| SpoVAD | GerAA | GerAC | GerBC | GerKA | GerD | |
| PS533 (wt) c | 1d | 1d | 1d | 1d | 1d | 1d |
| PS3477 (↑gerB)e | 1 | 1 | 1 | 3.3b | 1 | 1 |
| FB58 (↑↑gerB)e | 1 | 1 | 1 | 18b | 1 | 1 |
| PS3478 (↑gerK)e | 1 | 1 | 1 | 1 | 4.4b | 1 |
| PS3557 (↑gerB ↑gerK)e | 1 | 1 | 1 | 2.3b | 3b | 1 |
| PS3476 (↑gerA)e | 1 | 8b | 8b | 1 | 1 | 1 |
| FB62 (ΔgerD) | 1 | 1 | 1 | 1 | 1 | -f |
| PS3301 (ΔgerF)g | 1 | <0.05 | <0.05 | 1 | 1 | <0.05 |
| FB10 (gerB*)h | 1 | 1 | 1 | 1 | 1 | 1 |
| PS3502 (↑gerB*)e | 1 | 1 | 1 | 8b | 1 | 1 |
| PS3415 (↑↑gerB*)e | 1 | 1 | 1 | 12b | 1 | 1 |
| PS3501 (↑gerA gerB*)e | 1 | 9b | 4.4b | 1 | 1 | 1 |
Levels of various germination proteins in spores of various strains were determined as described in Materials and Methods. Values for individual proteins are expressed relative to the levels of this same protein in PS533 (wild-type [wt]) spores that were set at 1. Values in bold were significantly different from values for wild-type spores.
The differences between the level of this protein from that in wild-type spores was always significant, with P values of <0.05 and most often even lower.
The same results were obtained with spores of strains FB20 (ΔgerA), FB60 (ΔgerB), FB61 (ΔgerA ΔgerB), FB68 (ΔgerK), and FB87 (ΔgerB ΔgerK), except for the absence of the subunit(s) of the deleted GR.
Value for this individual protein was set at 1.
The symbols ↑ and ↑↑ indicate that the operon following was under the control of the moderately strong PsspD promoter and the stronger PsspB promoter, respectively.
—, no protein seen, as expected due to the deletion of the gene encoding this protein.
GerAA and GerAC were also not detected in spores of strain PS3407 (ΔgerD ΔgerF).
The same results were obtained with spores of strains PS3499 (ΔgerK gerB*), PS3521 (ΔgerA gerB*), and PS3665 (ΔgerA ΔgerK gerB*), except for the absence of the subunit(s) of the deleted GR.
Examination of spores lacking various GRs showed that levels of other GRs were not noticeably affected by loss of one or more other GRs (Fig. 4; Table 2). However, overexpression of GRs from PsspD or PsspB did result in elevation of the overexpressed GR's level up to ∼18-fold, with overexpression under PsspB control giving a higher level of overexpression (Fig. 5; Table 2). In addition, overexpression of the gerA operon under PsspD control in the wild-type and gerB* backgrounds gave ∼8-fold overexpression of GerAA and 8- or 4-fold overexpression of GerAC in the wild-type and gerB* backgrounds, respectively (Fig. 5; Table 2). However, in spores with an overexpressed GR or GRs, there were no significant effects on levels of the GRs encoded by the operons that were not under PsspD or PsspB control (Fig. 4; Table 2).
Effects of loss of GerD and GerF on germination protein levels.
Two other proteins that have major effects on rates of GR-dependent spore germination are GerD and GerF (11, 27, 32). The precise function of GerD is not known, but it is required for normal nutrient germination of B. subtilis spores. Indeed, spores lacking GerD germinated extremely poorly with l-valine via GerA or AGFK via GerB plus GerK (Fig. 6). GerF is the only prelipoprotein diacylglycerol transferase in B. subtilis, and a number of germination proteins, including GerAC, GerBC, GerKC, and GerD, are lipoproteins that are thought to be anchored in the spore's inner membrane by a diacylglycerol moiety linked to a cysteine residue near these proteins' N termini (11, 32). This modification appears to be essential for GerAC function, at least partially required for GerBC function but perhaps not for GerKC function, and likely to be essential for GerD function, since spores of a strain with a gerD gene encoding a protein in which the diacylglycerylated cysteine has been replaced by alanine do not accumulate GerD (11, 19). Indeed, gerF B. subtilis spores did not germinate with l-valine via GerA but did germinate, albeit slowly, with AGFK via GerB plus GerK (Fig. 6), as found previously (11). While it is thought that the effects of GerD and GerF on GR-dependent spore germination are via effects on GR function, it is certainly possible that these two proteins also have effects on GR levels. Consequently, levels of various germination proteins were determined in wild-type, gerD, and gerF spores (Fig. 7; Table 2). Loss of GerD had minimal, if any, effects on levels of GR subunits or SpoVAD, and loss of GerF had little to no effect on levels of GerBC, GerKA, or SpoVAD. However, levels of GerD, GerAA, and GerAC were reduced >20-fold in ΔgerF spores, and these proteins were also not found in any other fraction of ΔgerF spores (data not shown). This effect of a gerF deletion is certainly consistent with the absence of GerA-dependent germination in gerF spores as well as the disruption of the GR cluster in these spores (9, 11).
Fig 6.
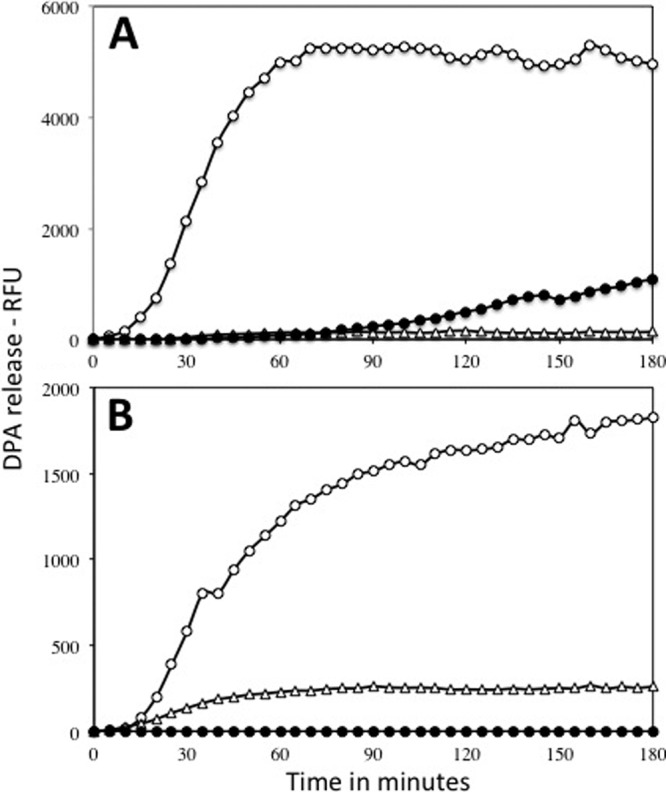
Germination of wild-type, gerD, and gerF spores with l-valine (A) or AGFK (B). Spores of strains PS533 (wild-type) (○), FB62 (ΔgerD) (●), and PS3301 (ΔgerF) (▵) were prepared and purified, and spore germination with either 10 mM l-valine (A) or 10 mM l-asparagine plus GFK (B) was followed by measuring DPA release fluorometrically in relative fluorescence units (RFU) as described in Materials and Methods.
Fig 7.
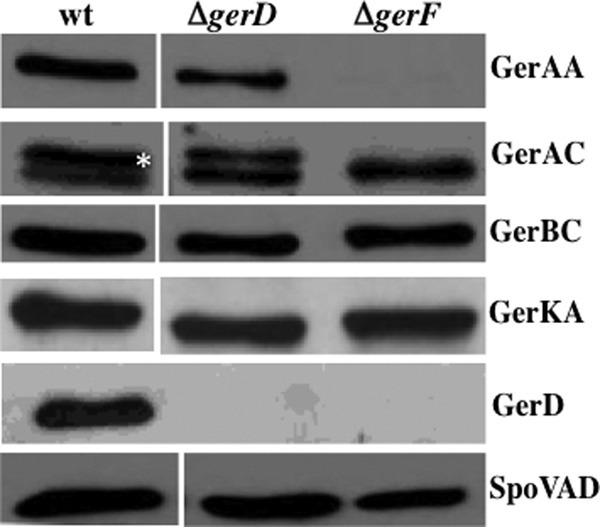
Western blot analysis of levels of germination proteins in spores lacking GerD or the prelipoprotein diacylglyceroltransferase, GerF. Spores of strains PS533 (wild type [wt]), FB62 (ΔgerD), and PS3301 (ΔgerF) were prepared and purified and inner membrane fractions were isolated as described in Materials and Methods. Equal amounts of inner membrane protein from ∼0.5 mg dry spores (defined as 1×) were analyzed on Western blots by probing with antisera against various germination proteins as described in Materials and Methods. The GerAA, GerAC, and GerBC strips are from the same Western blot probed first with anti-GerAA serum, then stripped and reprobed with anti-GerBC serum, and then stripped and reprobed with anti-GerAC serum. The SpoVAD, GerD, and GerKA strips are from different Western blots but from either the same spore preparation as used for analysis of GerAA, GerAC, and GerBC (SpoVAD) or different spore preparations (GerD and GerKA). Note that while the wt, ΔgerD, and ΔgerF lanes treated with the different antisera were from the same Western blot, intervening lanes have been removed for simplicity (except in the GerD strip), and the wt and the ΔgerD and ΔgerF lanes (the latter two lanes were run adjacent to each other) were moved adjacent to each other using PowerPoint. In the GerD strip, all three samples were run adjacent to each other. In the GerAC strip, GerAC is the upper band in the doublet denoted by the white asterisk adjacent to the upper band in the wt lane. The lower band is GerBC that was detected earlier by anti-GerBC serum that was not removed well when the blot was stripped. The levels of GerAA, GerAC, and GerD in ΔgerF spores were >20-fold lower than in wild-type spores, as determined by running Western blots with 0.1× the wild-type spore extract and 2× the ΔgerF spore extract and comparing the intensities of the appropriate bands (data not shown). The slightly lower level of SpoVAD in the ΔgerF strain was not seen in other comparable Western blots.
DISCUSSION
The work in this communication has led to a number of conclusions about rates of germination and germination protein levels with spores of B. subtilis. Some of these conclusions reinforce ones made previously (1, 3, 24). These include that (i) deletion of operons encoding GRs has very little to no effect on rates of germination via other GRs; (ii) overexpression of operons encoding GRs increases rates of germination of spores by these GRs (1, 3); and (iii) overexpression of operons encoding GRs, in particular GerA, decreases rates of spore germination by other GRs (1, 3).
There are also a number of new conclusions from the present work, including the following. (i) Levels of overexpression of GerAA and GerAC when the gerA operon was expressed under the control of PsspD were essentially identical in the otherwise wild-type background, although overexpression of GerAC was lower in the gerB* background (Table 2). This is consistent with the cotranscription of gerAA and gerAC in the gerA operon and suggests that translational coupling of the gerAC gene is relatively efficient. (ii) Levels of GerBC overexpression under PsspD and PsspB control were between 3- and 8-fold (PsspD) and between 12- and 18-fold (PsspB). Previously GerBA was found to be overexpressed ∼20-fold from PsspD and ∼200-fold from PsspB, much higher levels of overexpression than for GerBC. The reason for this difference in levels of overexpression of GerBA, encoded by the first gene in the gerB operon, or GerBC, encoded by the last gene in the operon, is not clear, but perhaps the translational coupling in the gerB operon is not as efficient as in the gerA operon. Indeed, there is an 8-nucleotide (nt) gap between the gerBA and gerBB coding regions, while there is a 30-nt overlap between the coding regions of gerAA and gerAB, although there are 1-nt overlaps between coding regions of gerAB and gerAC and those of gerBB and gerBC. (iii) The levels of GerAA, GerAC, and GerB* overexpression from PsspD or PsspB, 4- to 12-fold, are higher than the increased maximal rates of germination of these spores with l-valine or l-asparagine, respectively. These maximal rates increased only ∼1.6-fold for ↑gerA spores and ∼ 3.5-fold for ↑↑gerB* spores. However, at subsaturating l-valine and l-asparagine concentrations, for example, 0.5 mM l-valine, there was an ∼5-fold-higher germination rate with ↑gerA spores and an ∼10-fold-higher germination rate with ↑↑gerB* spores. These values are relatively close to the elevated GerAA/GerAC levels in ↑gerA spores and the elevated GerBC levels in ↑↑gerB spores. This much higher increase in germination rates of spores with overexpressed GRs at subsaturating germinant concentrations was also seen previously (1, 3). Presumably, at saturating germinant levels, some factor other than GR level becomes rate limiting for spore germination. (iv) Overexpression or deletion of GRs had no effect on levels of SpoVAD, a protein likely involved in DPA efflux during spore germination. Previous results have indicated that B. subtilis sporulation in a poor medium results in ∼3-fold decreases in spore levels of GRs but also no changes in SpoVAD levels (29). Presumably, there is some coregulation of operons encoding GRs that does not extend to the spoVA operon. While this GR coregulation mechanism is not known, one candidate for a possible regulatory protein in B. subtilis is SpoVT, which represses expression of operons encoding GRs but has no effect on the spoVA operon (2, 37). Another candidate for a protein that might regulate GR levels is PrpE, which has been suggested to modulate levels of at least the GerA and GerK GRs (10), although neither the effects of this protein on GerD and SpoVA protein levels in spores nor how this protein might exert effects on GerA and GerK levels is known. (v) Overexpression or deletion of GRs also had no effect on GerD levels in spores. In contrast to the results with SpoVAD noted above, GerD levels are decreased ∼3-fold in spores prepared in a poor medium (29), yet SpoVT is not thought to affect gerD expression (2, 37). However, the lack of effect of GR overexpression on GerD levels is consistent with the fact that germination via at least some of the GRs not overexpressed does not go down. In addition, GerD overexpression does not increase GR-dependent spore germination (27). (vi) The absence of GerD had no notable effect on GR levels, even though all GR-dependent spore germination is greatly decreased (27). Thus, alterations in GR levels are not the cause of the slow GR-dependent germination with gerD spores, although the GR clustering in the dormant spore's inner membrane is disrupted in such spores (9). (vii) Lack of the prelipoprotein diacylglycerol transferase, GerF, resulted in the absence of GerD, GerAA, and GerAC from spores, and not just from the inner membrane. Presumably, GerD and GerAC are degraded if they are not diacylglycerylated, and without GerAC, GerAA does not assemble into a GR and is also degraded. Previous work has shown that GerD that cannot be deacylglycerylated due to replacement of the modified Cys residue by Ala is also absent from spores even if GerF is present (20). A GerAC protein with a similar change also results in the lack of GerA-dependent germination, as does a gerF null mutation (11), consistent with the absence of GerAA and GerAC in gerF spores seen in the present work. (viii) In contrast to the absence of GerAA, GerAC, and GerD, gerF spores had relatively normal levels of GerBC and GerKA. This was not unexpected, because gerF spores still exhibit significant germination via the GerB plus GerK GRs, and spores that contain GerF still exhibit significant germination via the GerB plus GerK GRs even if the GerBC or GerKC proteins lack the diacylglycerylated Cys residue (11). However, it is not clear why the absence of a diacylglycerylated Cys residue on GerAC has such a large effect on GerA GR assembly in the spore's inner membrane yet has essentially no such effect on assembly of the GerB or GerK GRs. One possibility is that GerAC and GerD actually have different modifications in their N-terminal regions, as several structures other than S-diacylglycerylation of an N-terminal Cys residue have been recently identified in lipoproteins in Bacillus species (15). These additional structures include acetylation of the N-terminal Cys residue's amino group in addition to its S-diacylglycerylation found in B. subtilis and related species, and an N-terminal Cys residue with a fatty acid acylating the amino group and only a single fatty acid attached to the S-glycerol backbone in at least Bacillus cereus (15). However, which of the germination lipoproteins have these various structures and the sequence elements that determine which structure is formed are not known. It is also notable that at least one other B. subtilis membrane lipoprotein, the OpuAC protein, which is involved in glycine betaine transport, also remains in the cell membrane if not lipidated and is functional (13).
It is also possible that overexpression of a GR overloads either the system for protein insertion into membranes or the prelipoprotein diacylglycerol transferase that adds lipid to GerD, GerAC, GerBC, and perhaps GerKC (11). However, while these are certainly logical possibilities, both seem unlikely for at least two reasons. (i) Overexpression of the GerB GR in which GerBC is lipidated (11) has no significant effects either on germination via the GerA GR, as shown in the present work and previously (3), or on GerAC levels, as shown in the present work, and in the absence of lipidation, GerAC was not found in spores. (ii) While GRs are overexpressed significantly from either the PsspD or PsspB promoter, the levels of GRs are normally ≤100 molecules/spore (25; K.-A.V. Stewart and P. Setlow, unpublished data), not high at all, and even overexpressed levels seem unlikely to saturate the system for protein insertion into membranes. Indeed, at least overexpressed GerBA is found only in the inner membrane (25).
The conclusion that appears to have the most significance with respect to our understanding of the mechanisms of spore germination is that overexpression of one GR, in particular GerA, inhibits the rates of germination via other GRs up to ∼15-fold. While this inhibitory effect could have been due to elevated levels of one GR resulting in lower levels of other GRs, this was not the case, as up to 18-fold-elevated GR levels had no significant effect on levels of other GRs. Consequently, the effects of GR overexpression on germination via other GRs must be due to an effect on the activity or functionality of these other GRs. Much recent work has shown that the major variability in rates of germination between individual spores of different strains with different GR levels or without GerD, and at different germinant concentrations, is in the length of the lag period (Tlag) between mixing spores with a germinant and the initiation of the rapid release of the spore's DPA depot (14, 36, 40). Thus, lower germinant concentrations or the lack of GerD results in longer average Tlags, while higher germinant concentrations or elevated GR levels result in shorter average Tlags. Consequently, it appears most likely that the inhibitory effect of GerA overexpression on spores' germination rates via other GRs will be to greatly increase average Tlags with the other GRs.
We envisage two possible mechanisms for the inhibitory effect of GerA overexpression on germination via other GRs, one involving inhibitory GR-GR interactions and the other involving GRs competing for an essential downstream signaling molecule in the spore germination pathway. In examining these two possible mechanisms, it is important to keep in mind that the components of the dormant spore's inner membrane most likely have minimal mobility (6). Consequently, interactions of GRs in the spore's inner membrane probably do not change during dormancy but, rather, are those that were in place when the spore became dormant. It is also notable that all GRs are colocalized in a small cluster in the spore's inner membrane (9).
The basis for the GR-GR interaction model is as follows. There is much evidence that GRs can cooperate in spore germination, with the requirement for both GerB and GerK GRs for AGFK germination being a prime example (32). The GerK receptor can also stimulate l-valine germination via the GerA GR (1). While none of these interactions have been shown to be direct, this seems likely. Consequently, it is possible that elevation of levels of one or more GRs would alter the pattern of GR-GR interactions in the spore's germination protein cluster. If this is indeed the case and if GR-GR interactions can also inhibit germination by one of the interacting GR partners, this could explain the inhibition of germination of one GR's function by an overexpressed GR. However, given the much larger effects of GerA overexpression on AGFK or l-asparagine germination than the effects of GerB plus GerK overexpression on l-valine germination, GerA must be either more effective in forming partners with heterologous GRs or more inhibitory when it does so. One concern about this model is, however, that while some types of GR-GR interactions that stimulate GR function have been well documented, as described above, there is yet no direct evidence for an inhibitory GR-GR interaction.
The model involving different GRs competing for a common downstream signaling molecule in the germination pathway again involves a major unknown—the precise identity of the downstream signaling molecule, which has previously been called a signal integrator (SI) (1). There is no direct evidence for such an SI, although the strong synergy between different GRs in stimulating spore germination, in particular at low concentrations of nutrient combinations, is certainly consistent with there being an SI that sums signals from multiple GRs (38). In this model, all GR signals would be fed into the SI, with the SI then triggering a downstream germination event such as DPA release. Again, GerA must be the most effective GR in capturing the SI and preventing signals from the GerB plus GerK GRs or the GerB* GR from being transmitted. This would likely require that levels of the SI molecule be relatively low so that there is only an amount no higher than normal GR levels. Consequently, when one or more GRs are overexpressed, the GR that bound best to the SI, presumably GerA, would displace other GRs from the germination signal transduction pathway. At present, we cannot decide conclusively between these two models, but it should be possible to design experiments to do this, and this work is currently in progress.
ACKNOWLEDGMENTS
This communication is based upon work supported by a Department of Defense Multidisciplinary University Research Initiative through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286.
We are grateful to Yunefeng Li, Bing Hao, and George Korza for assistance in preparing the antiserum against the B. subtilis GerD protein.
Footnotes
Published ahead of print 6 April 2012
REFERENCES
- 1. Atluri S, Ragkousi K, Cortezzo D, Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagyan I, Hobot J, Cutting SM. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cabrera-Martinez R-M, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christie G, Lowe CR. 2007. Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J. Bacteriol. 189:4375–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooper GR, Moir A. 2011. Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis. J. Bacteriol. 193:2261–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowan AE, et al. 2004. Lipids in the inner membrane of dormant spores of Bacillus species are immobile. Proc. Natl. Acad. Sci. U. S. A. 101:7733–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh S, Scotland M, Setlow P. 2012. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J. Bacteriol. 194:2221–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh S, Setlow P. 2009. Isolation and characterization of superdormant spores of Bacillus species. J. Bacteriol. 191:1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81:1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hinc K, et al. 2006. Expression of genes coding for GerA and GerK spore germination receptors is dependent on the protein phosphatase PrpE. J. Bacteriol. 188:4373–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Igarashi T, Setlow B, Paidhungat M, Setlow P. 2004. Analysis of the effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 186:2984–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Igarashi T, Setlow P. 2005. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J. Bacteriol. 187:2513–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kempf B, Gade J, Bremer E. 1997. Lipoproteins from the osmoregulated ABC transport system OpuA of Bacillus subtilis: purification of the glycine betaine binding protein and characterization of a functional lipidless mutant. J. Bacteriol. 179:6213–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong L, et al. 2011. Phase contrast microscopy, fluorescence microscopy, Raman spectroscopy and optical tweezers to characterize the germination of individual bacterial spores. Nat. Protoc. 6:625–639 [DOI] [PubMed] [Google Scholar]
- 15. Kurokawa K, Ryu, et al. 2 February 2012. Novel bacterial lipoprotein structures conserved in low-GC content Gram-positive bacteria are recognized by Toll-like receptors. J. Biol. Chem. doi:10.1074/jbc.M111.292235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, et al. 2011. Structure-based functional studies of the effects of amino acid substitutions in GerBC, the C subunit of the Bacillus subtilis GerB spore germinant receptor. J. Bacteriol. 193:4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, et al. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194:1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mason JM, Hackett RH, Setlow P. 1988. Regulation of expression of small, acid-soluble proteins in Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mongkolthanaruk W, Cooper GR, Mawer JS, Allan RN, Moir A. 2011. Effect of amino acid substitutions in the GerAA protein on the function of the alanine-responsive germinant receptor of Bacillus subtilis spores. J. Bacteriol. 193:2268–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mongkolthanaruk W, Robinson C, Moir A. 2009. Localization of the GerD spore germination protein in the Bacillus subtilis spore. Microbiology 155:1146–1151 [DOI] [PubMed] [Google Scholar]
- 21. Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- 22. Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paidhungat M, Setlow P. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant D-alanine. J. Bacteriol. 181:3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paidhungat M, Setlow P. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:4886–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19:85–94 [DOI] [PubMed] [Google Scholar]
- 27. Pelczar PL, Igarashi T, Setlow B, Setlow P. 2007. The role of GerD in the germination of Bacillus subtilis spores. J. Bacteriol. 189:1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pelczar PL, Setlow P. 2008. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J. Bacteriol. 190:5635–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramirez-Peralta A, Zhang P, Li Y-Q, Setlow P. 2012. Effects of sporulation conditions on the germination and germination protein levels of spores of Bacillus subtilis. Appl. Environ. Microbiol. 78:2689–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Setlow P, Johnson EA. Spores and their significance. In Doyle MP, Buchanan R. (ed), Food microbiology: fundamentals and frontiers, 4th ed, in press. ASM Press, Washington, DC [Google Scholar]
- 32. Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 33. Sun D, et al. 1991. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EσF: identification of features of good EσF-dependent promoters. J. Bacteriol. 173:7867–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187:5677–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vepachedu VR, Setlow P. 2007. Analysis of interactions between nutrient germinant receptors and SpoVA proteins of Bacillus subtilis spores. FEMS Microbiol. Lett. 274:42–47 [DOI] [PubMed] [Google Scholar]
- 36. Wang G, Yi X, Li Y-Q, Setlow P. 2011. Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins. J. Bacteriol. 193:2301–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang ST, et al. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16–37 [DOI] [PubMed] [Google Scholar]
- 38. Yi X, Liu J, Faeder JR, Setlow P. 2011. Synergism between different germinant receptors in the germination of Bacillus subtilis spores. J. Bacteriol. 193:4664–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yi X, Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192:3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang P, et al. 2010. Factors affecting the variability in the time between addition of nutrient germinants and rapid DPA release during germination of spores of Bacillus species. J. Bacteriol. 192:3608–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]


