Abstract
Bacteria growing in biofilms are responsible for a large number of persistent infections and are often more resistant to antibiotics than are free-floating bacteria. In a previous study, we identified a Pseudomonas aeruginosa gene, ndvB, which is important for the formation of periplasmic glucans. We established that these glucans function in biofilm-specific antibiotic resistance by sequestering antibiotic molecules away from their cellular targets. In this study, we investigate another function of ndvB in biofilm-specific antibiotic resistance. DNA microarray analysis identified 24 genes that were responsive to the presence of ndvB. A subset of 20 genes, including 8 ethanol oxidation genes (ercS′, erbR, exaA, exaB, eraR, pqqB, pqqC, and pqqE), was highly expressed in wild-type biofilm cells but not in ΔndvB biofilms, while 4 genes displayed the reciprocal expression pattern. Using quantitative real-time PCR, we confirmed the ndvB-dependent expression of the ethanol oxidation genes and additionally demonstrated that these genes were more highly expressed in biofilms than in planktonic cultures. Expression of erbR in ΔndvB biofilms was restored after the treatment of the biofilm with periplasmic extracts derived from wild-type biofilm cells. Inactivation of ethanol oxidation genes increased the sensitivity of biofilms to tobramycin. Together, these results reveal that ndvB affects the expression of multiple genes in biofilms and that ethanol oxidation genes are linked to biofilm-specific antibiotic resistance.
INTRODUCTION
Biofilms are the leading cause of hospital-acquired, implant-based infections and the basis of many persistent diseases, such as otitis media, periodontitis, and the chronic Pseudomonas aeruginosa lung infections that afflict patients with cystic fibrosis (CF) (4). The persistence of these infections is primarily attributed to the recalcitrance of biofilms to the immune system and antimicrobial agents. Bacteria growing in biofilms often show increased resistance to antibiotics (e.g., 10 to 100 times) compared to free-floating (planktonic) bacteria (20, 32). It is still not fully understood how biofilm cells become more resistant to antibiotics. Since the genetic makeup of the cells in the biofilm has not been altered, the increased resistance likely involves the altered expression of specific genes in the biofilm. It is well documented that the gene expression profile of biofilm cells is markedly different from that of planktonic cells (28, 34). Thus, a subset of these genes likely functions to protect biofilm cells from antibiotics.
We have identified several P. aeruginosa genes that contribute to biofilm-specific antibiotic resistance by screening for mutants with increased antibiotic sensitivity when growing in biofilms (21, 35, 36). One of these genes, ndvB, encodes a glucosyltransferase involved in the formation of cyclic glucans (21). The glucans are cyclic polymers of 12 to 15 β-(1→3)-linked glucose molecules with phosphoglycerol substitutions (27). Inactivation of ndvB blocked glucan production but did not affect growth, the kinetics of biofilm formation, or the architecture of the biofilms (21). However, biofilms of ndvB mutants exhibited increased sensitivity to the aminoglycosides tobramycin and gentamicin and the fluoroquinolone ciprofloxacin (21, 27). We and others have shown that antibiotics can physically interact with glucan-enriched periplasmic lysates and purified glucan preparations (21, 27). Thus, we proposed that glucans confer resistance to antibiotics by sequestering these antibiotics in the periplasm and away from their cytoplasmic targets (21).
Cyclic glucans have been studied primarily in Rhizobium species, in which they have roles in hypo-osmotic adaptation and in plant infection (2). In Bradyrhizobium japonicum and Sinorhizobium meliloti, secreted glucans are important for the symbiotic relationship between these bacteria and their plant hosts (6, 7). ndvB mutants are unable to produce functional root nodules on plants, and addition of exogenous cyclic glucans can enhance nodule formation (8). Thus, it has been suggested that the glucans are involved in signaling between the bacteria and plants, although the mechanism has not been elucidated (2). These observations led us to propose that ndvB-derived glucans might function in signaling in P. aeruginosa. In this study, we explored this possibility by investigating gene expression differences between wild-type and ΔndvB bacteria growing in biofilms.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
All P. aeruginosa strains used in this study were derivatives of the PA14 wild-type strain and are listed in Table 1 (26). Transposon mutants were retrieved from the nonredundant PA14 transposon library (18). The ΔndvB mutant has been reported elsewhere (21). Additional unmarked deletion mutants were constructed in PA14 by allelic exchange with pEX18Gm derivatives as previously described (16, 21). Genes were cloned into the pJB866 vector as previously described (36), with open reading frames inserted downstream from the Pm promoter, which is capable of induction by m-toluic acid (1). Primers used for the construction of deletion mutants and pJB866 derivatives are listed in Table 2. Bacteria were grown at 37°C in rich medium (Luria-Bertani [LB]) or minimal medium. The minimal medium (M63-arginine) was M63 salts supplemented with arginine (0.4%) and MgSO4 (1 mM) (25). M63-ethanol was M63 salts supplemented with ethanol (0.5% [vol/vol]) and MgSO4 (1 mM). Tobramycin and ciprofloxacin were purchased from Research Production International (Mt. Prospect, IL) and MP Biomedicals (Solon, OH), respectively.
Table 1.
P. aeruginosa strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source |
|---|---|---|
| Strains | ||
| PA14 | P. aeruginosa burn wound isolate | 26 |
| TFM15 | PA14 ΔndvB | 21 |
| TFM215 | PA14 ΔexaA | This study |
| TFM212 | PA14 ΔpqqC | This study |
| TFM220 | PA14 ΔerbR | This study |
| TFM217 | PA14 ΔndvB ΔexaA | This study |
| TFM216 | PA14 ΔndvB ΔpqqC | This study |
| TFM221 | PA14 ΔndvB ΔerbR | This study |
| exaA::MrT7 | PA14_38860::MAR2 × T7 | 18 |
| exaC::MrT7 | PA14_38840::MAR2 × T7 | 18 |
| pqqC::MrT7 | PA14_38800::MAR2 × T7 | 18 |
| eraR::MrT7 | PA14_38900::MAR2 × T7 | 18 |
| Plasmids | ||
| pJB866 | Expression vector containing the m-toluic acid inducible promoter Pm; Tcr | 1 |
| pJB866-exaA | pJB866 with the PA14 exaA gene inserted downstream from the Pm promoter | This study |
| pJB866-pqqC | pJB866 with the PA14 pqqC gene inserted downstream from the Pm promoter | This study |
| pJB866-erbR | pJB866 with the PA14 erbR gene inserted downstream from the Pm promoter | This study |
Tcr, tetracycline resistant.
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a | Function |
|---|---|---|
| exaAf1 | CCTAGAATTCGCGCAGTTCTGGTTGTAGGC | Deletion of exaA |
| exaAr2 | AACCGAGCTCCGTGGATCAGCAGGACCTTG | Deletion of exaA |
| exaAf3 | AATTGAGCTCTCCGGATCAAGCGCATGTACG | Deletion of exaA |
| exaAr4 | ATTGAAGCTTGAAGTTGCCGAAGCGCACCT | Deletion of exaA |
| pqqCf1 | AACTGAGCTCTAACTCGACCTGCCGGAATC | Deletion of pqqC |
| pqqCr2 | AACTGGTACCGTACATCGCGACATGGAAC | Deletion of pqqC |
| pqqCf3 | AACTGGTACCGACAGTTGGCCGCGCCACTA | Deletion of pqqC |
| pqqCr4 | ATCCAAGCTTCCGCTCGATGTTGTCGATG | Deletion of pqqC |
| agmR-StrEcoR Ib | GTCAGAATTCGCAACAGGCGGTAGAGATGG | Deletion of erbR |
| agmR-StrXbaI-R | CAACTCTAGACGATGACATTGTGGATGG | Deletion of erbR |
| agmR-XhaI-F | CAACTCTAGAAAGCTCAAGGTACACAACC | Deletion of erbR |
| agmR-HindIII Ib | GGCTAAGCTTGCTGATCGTCGACTATCAC | Deletion of erbR |
| ndvB JB-F | GTTGGAGCTCATGTCTTCACGCAAGATCGG | Cloning ndvB into pJB866 |
| ndvB JB-R | GTTGAAGCTTACCTCAACCGCCGATCTGCTC | Cloning ndvB into pJB866 |
| agmR JB-F2 | GTTCAAGCTTCATGTACAAGATCCTGATCGC | Cloning erbR into pJB866 |
| agmR JB-R2 | GTTCCTCGAGAGCATCGCCAGGACAGTGAG | Cloning erbR into pJB866 |
| exaA JB-F | ATCAGAGCTCATGACAACAAGAACCTCACC | Cloning exaA into pJB866 |
| exaA JB-F | GTTCAAGCTTATCGGGATACTCCGCAGGCG | Cloning exaA into pJB866 |
| pqqC JB-F | ATCAGACTCATGAGCCGTGCCGCCATGGA | Cloning pqqC into pJB866 |
| pqqC JB-R | GTTCAAGCTTATCGGCACGCTGTCGAGCGA | Cloning pqqC into pJB866 |
| rpoD-QF | CATCCGCATGATCAACGACA | rpoD qPCR primer |
| rpoD-QR | GATCGATATAGCCGCTGAGG | rpoD qPCR primer |
| ndvB-QF | GGCCTGAACATCTTCTTCACC | ndvB qPCR primer |
| ndvB-QR | GATCTTGCCGACCTTGAAGAC | ndvB qPCR primer |
| exaA-QF | AGACCAACACCATCATCGTC | exaA qPCR primer |
| exaA-QR | GGTGTGCTGGTAGAACCACT | exaA qPCR primer |
| exaB-QF | CTCCGCCTACAACCAGAACT | exaB qPCR primer |
| exaB-QR | CCTTCCTGGCTGATGAAGTC | exaB qPCR primer |
| exaC-QF | GGTCAAGGGCCAGTACTTCA | exaC qPCR primer |
| exaC-QR | GCGATCTTCAGCAGGATGTT | exaC qPCR primer |
| exaE-QF | AAATTCCTTCGCTGGTCATC | eraR qPCR primer |
| exaE-QR | AAGAACAGCACACGCAACTG | eraR qPCR primer |
| agmR-QF | CATTCCCGTGGTTATCGTCT | erbR qPCR primer |
| agmR-QR | AGGTAGACGTTGCCGTTGAG | erbR qPCR primer |
| pqqB-QF | GACGAGATGCTGGTCTGC | pqqB qPCR primer |
| pqqB-QR | ATTGGTGTTGTTGATGTGGA | pqqB qPCR primer |
| pqqC-QF | CGATAGTGCTCCAGGGTGAT | pqqC qPCR primer |
| pqqC-QR | GTCGACGCCTACGTCAACTT | pqqC qPCR primer |
| pqqE-QF | GGCACAACATCGACAACATC | pqqE qPCR primer |
| pqqE-QR | GTAGTAGTCGGGGGTGACGA | pqqE qPCR primer |
| ercS′-QF | CGAACTGAAGGCCAGCAACC | ercS′ qPCR primer |
| ercS′-QR | AGGACGAAGGCGTCGGAGAT | ercS′ qPCR primer |
Locations of restriction sites are underlined and in bold.
DNA microarray analysis.
RNA was extracted from PA14 wild-type and ΔndvB biofilms grown in M63-arginine liquid medium using the Kadouri drip-fed method (23). Briefly, biofilms were grown in the wells of uncoated polystyrene 6-well plates for 48 h, washed, and detached through vigorous scraping. The cells were resuspended in fresh medium and centrifuged at 12,000 × g. The supernatant was removed, and the wash step was repeated. For both the wild-type and ΔndvB strains, three biological replicate samples were analyzed, each of which derived from two wells of biofilm-grown cells. RNA was purified using the PureLink Micro-to-Midi total RNA purification system (Invitrogen) according to the manufacturer's instructions. Cells were lysed with a 10-ml syringe and a 20-gauge needle. An on-column digestion was performed with DNase (Sigma), and RNA was eluted in RNase-free water (Invitrogen). RNA was analyzed for concentration and purity by an Agilent 2100 Bioanalyzer at the StemCore Microarray Facility (Ottawa Hospital Research Institute). cDNA synthesis, labeling, and hybridization to P. aeruginosa PAO1 GeneChips were performed at the StemCore Microarray Facility.
Quality assessment and analysis of microarray data were performed using an open-source R programming environment (5) in conjunction with Bioconductor software (10). Background adjustments, normalization, and selection of differentially expressed genes were obtained by robust microarray analysis (RMA) and MicroArray Suite 5.0 (MAS5.0) procedures. The overlapping genes (identified in both the RMA and MAS5.0 procedures) were selected for further study. t tests were used to identify genes that showed significant changes in transcript levels (P ≤ 0.001).
qPCR analysis.
RNA was extracted from planktonic cultures, drip-fed biofilms, or colony biofilms grown at 37°C. Planktonic cultures were grown in LB to early stationary phase or in M63-arginine to exponential phase. Kadouri drip-fed biofilms were grown as described in the microarray experiment. Colony biofilms on M63-arginine agar plates were prepared as previously described (36), with incubation for 24 h at 37°C followed by 16 h at room temperature. RNA extraction and cDNA synthesis were performed as previously described (36). cDNA was quantified using SYBR green detection of PCR products with the MyiQ single-color detection system (Bio-Rad). Each 20-μl quantitative real-time PCR (qPCR) mixture contained 2 μl cDNA (∼1.2 μg), 10 μl SYBR green PCR master mix (Applied BioSystems), and 100 pmol of each primer. The following thermal cycler conditions were used: 90 s at 95°C, followed by 45 cycles of 60 s at 95°C, 30 s at 56°C, and 30 s at 72°C. qPCR primers are listed in Table 2. RNAs isolated from at least two independent cell cultures were each tested in triplicate qPCRs, with expression of rpoD used as a reference standard. Statistical significance was determined by Student's t tests.
Planktonic growth in ethanol.
To examine growth of bacteria using ethanol as the sole carbon source, overnight cultures grown in M63-arginine were diluted in 50 ml of M63 containing 0.5% ethanol to equivalent optical densities at 600 nm. For growth in M63-arginine or LB broth, overnight LB cultures were diluted into the appropriate medium. Cultures were incubated with shaking at 37°C, and planktonic growth was monitored by absorbance readings at 600 nm. At least two biological replicates were performed for each condition.
Antibiotic resistance assays.
Minimum bactericidal concentrations (MBCs) were determined for biofilm (MBC-B) and planktonic (MBC-P) cultures as previously described (21, 36). Briefly, overnight cultures of bacteria were diluted (1:50) in M63-arginine in 96-well microtiter plates. For MBC-B assays, biofilms were allowed to form for 24 h, planktonic cells were removed, and the biofilms were exposed to serial dilutions of antibiotics. For MBC-P assays, antibiotics were added at the same time that the plates were inoculated. After 24 h of antibiotic exposure, bacterial survival was determined by spotting a small amount (ca. 3 μl) of culture on LB agar plates immediately after antibiotic treatment (MBC-P) or following a 24-h recovery period in which surviving cells can detach from biofilms into antibiotic-free growth medium (MBC-B). For MBC-B assays on ethanol minimal medium, M63 containing 0.5% ethanol as the carbon source was used.
For another measure of planktonic antibiotic resistance, MICs were determined in LB broth using the 2-fold broth dilution method (3). For strains carrying pJB866 derivatives, MICs were determined in LB or M63-arginine containing 2 mM m-toluic acid to induce expression of cloned genes.
Signaling experiment.
Periplasmic extracts were isolated from 750-ml PA14 planktonic cultures grown in M63-arginine, as previously described (21). The cells were pelleted, and the periplasmic contents were extracted with 70% ethanol at 70°C for 30 min. The insoluble fraction was removed by centrifugation, and the supernatant was precipitated by adding NaCl to 0.1 M and 10 volumes of 100% ethanol. After centrifugation for 30 min at 7,700 × g, the pellet was dried and suspended in 10 ml water. The presence of glucans in the periplasmic extracts was confirmed using the anthrone-sulfuric acid method of carbohydrate quantification (19, 21). Glucans were purified by gel filtration chromatography of periplasmic extracts with Sephadex G-75 resin, as previously described (21). The glucan-containing fractions were dried by Speed-Vac evaporation, and the pellets were suspended in water.
For the signaling experiment, the periplasmic extracts and purified glucans were added to wild-type and ΔndvB static biofilms that were pregrown for 48 h in 3 ml M63-arginine (with the medium refreshed at 24 h). Wild-type periplasmic extract (65 μg of anthrone-positive material), ΔndvB periplasmic extract (volume equivalent to that of the wild-type extract), purified glucans (65 μg of anthrone-positive material), or water was added to the biofilms. The volumes of the treatments were standardized to 144 μl with water. The biofilms were incubated for 8 h, RNA was isolated from the biofilms, and erbR expression was assayed by qPCR. The results represent two or three biological replicate samples, each tested in triplicate.
Microarray data accession number.
The microarray data have been deposited at the Gene Expression Omnibus (GEO) database (accession no. GSE32032).
RESULTS
ndvB inactivation results in differential transcription of 24 genes in biofilms.
To further investigate the function of P. aeruginosa glucans in biofilm-specific antibiotic resistance, we compared the global gene expression profiles of wild-type and ΔndvB biofilms. The biofilms were grown in M63-arginine for 48 h in a Kadouri drip-fed system. Microarray analysis of biofilm RNA led to the identification of 24 genes with differential expression in the ΔndvB biofilms (Table 2). Twenty of the genes were expressed preferentially in wild-type biofilms, and four of the genes were more highly expressed in ΔndvB biofilms.
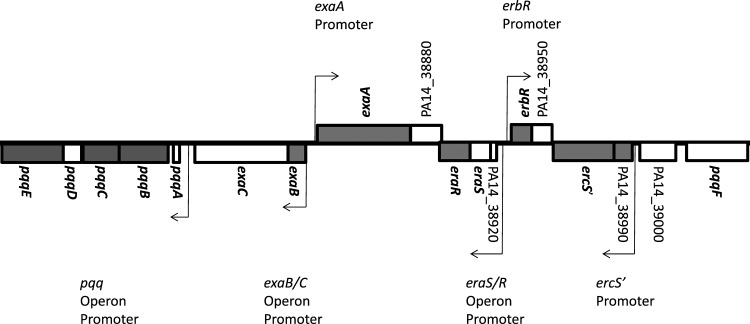
While most of the ndvB-responsive genes are predicted to encode proteins of unknown or unrelated functions, 8 of the genes are involved in ethanol oxidation (Table 3) (14, 22). These include the gene encoding a quinoprotein ethanol dehydrogenase (exaA) and genes involved in the synthesis of its cofactor pyrroloquinoline-quinone (PQQ) (pqqBCE). The PQQ-bound ethanol dehydrogenase is a periplasmic enzyme required for aerobic growth using ethanol as the sole source of carbon and energy (14). Another ndvB-responsive gene, exaB, encodes a cytochrome required for electron transport during growth on ethanol (29). Transcription of these genes is hierarchically controlled by at least six transcriptional regulators (22), three of which showed reduced expression in the ΔndvB biofilms (eraR, erbR, and ercS′). All of the ethanol oxidation genes discussed here, plus another uncharacterized gene identified in the microarray analysis (PA14_38990), are situated in the same genetic locus (Fig. 1). Since a large proportion of ndvB-responsive genes occupies this locus, we chose to investigate the genes further.
Table 3.
Genes differentially expressed in wild-type and ΔndvB biofilmsa
| PA14 geneb | P. aeruginosa PAO1 orthologue | Fold changec | P value (× 10−5) | Gene name | Functiond |
|---|---|---|---|---|---|
| PA14_03900 | PA0299 | −2.36 | 4.48 | spuC | Putrescine-pyruvate aminotransferase |
| PA14_09320 | PA4222 | −2.76 | 8.57 | pchI | ATP-binding component of an ABC transporter |
| PA14_11210 | PA4072 | 2.19 | 7.48 | Putative amino acid permease | |
| PA14_24720 | PA3044 | 2.76 | 1.07 | Putative two-component sensor | |
| PA14_29040 | PA2715 | 2.10 | 1.04 | Putative ferredoxin | |
| PA14_29220 | PA2700 | 2.43 | 1.41 | opdB | Putative porin |
| PA14_36120 | PA2210 | 2.99 | 1.33 | Putative MFS transporter | |
| PA14_38780 | PA1989 | 2.82 | 1.11 | pqqE | PQQ biosynthesis protein E |
| PA14_38800 | PA1987 | 2.11 | 1.09 | pqqC | PQQ biosynthesis protein C |
| PA14_38820 | PA1986 | 3.75 | 3.86 | pqqB | PQQ biosynthesis protein B |
| PA14_38850 | PA1983 | 2.95 | 2.51 | exaB | Cytochromec550 |
| PA14_38860 | PA1982 | 3.75 | 5.67 | exaA | Ethanol dehydrogenase |
| PA14_38900 | PA1980 | 3.36 | 3.37 | eraR | Response regulator (formerlyexaE) |
| PA14_38930 | PA1978 | 2.89 | 1.89 | erbR | Response regulator (formerlyagmR) |
| PA14_38970 | PA1976 | 3.89 | 2.89 | ercS′ | Sensor kinase |
| PA14_38990 | PA1975 | 3.01 | 2.03 | Hypothetical | |
| PA14_51205 | PA1015 | 2.10 | 6.51 | Putative transcriptional regulator | |
| PA14_52230 | PA0931 | 2.13 | 2.98 | pirA | Ferric enterobactin receptor |
| PA14_57275 | PA4407 | −2.30 | 5.14 | ftsZ | Cell division protein |
| PA14_58890 | PA4540 | 2.53 | 6.15 | Hypothetical | |
| PA14_64240 | PA4857 | 5.10 | 4.73 | MarC family protein | |
| PA14_64680 | PA4894 | 2.74 | 7.54 | Hypothetical | |
| PA14_70140 | PA5312 | −2.90 | 2.41 | aldH | Putative aldehyde dehydrogenase |
| PA14_71250 | PA5397 | 2.14 | 8.46 | Hypothetical |
This list includes all genes differentially expressed by at least 2-fold in a DNA microarray analysis comparing wild-type and ΔndvB biofilms (P < 10−4).
Genes involved in ethanol oxidation are indicated in bold. A gene coregulated with ethanol oxidation genes is underlined.
Values indicate the fold change in gene expression between wild-type and ΔndvB biofilms. Positive values indicate greater expression in wild-type biofilms. Negative values indicate greater expression in ΔndvB biofilms.
MFS, major facilitator superfamily.
Fig 1.
Chromosomal arrangement of ethanol oxidation genes in P. aeruginosa strain PA14. ndvB-responsive genes identified by microarray analysis are shown in gray. Genes previously reported to be involved in ethanol oxidation are in bold (14, 22). The confirmed promoters for several operons of ethanol oxidation genes are indicated by arrows (12, 13, 22).
Expression of ethanol oxidation genes in biofilms involves NdvB and ErbR.
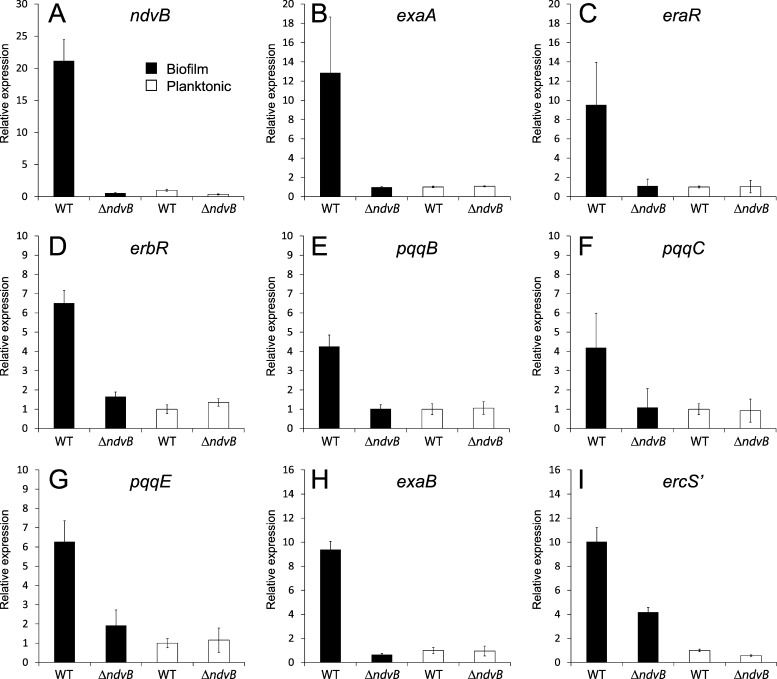
The microarray experiment indicated that the expression of ethanol oxidation genes in P. aeruginosa biofilms requires ndvB. We used quantitative real-time PCR (qPCR) of RNA isolated from biofilms to validate the ndvB-dependent expression of the eight ethanol oxidation genes identified in the microarray analysis. As shown in Fig. 2, expression of each gene was reduced in ΔndvB biofilms relative to that in wild-type biofilms, confirming the microarray results. In addition, the genes were more highly expressed in wild-type biofilms than in planktonic cells (Fig. 2). These data suggest that the ethanol oxidation genes are induced in P. aeruginosa biofilms and that ndvB is required for this induction (21).
Fig 2.
qPCR analysis of ethanol oxidation genes in biofilms and planktonic cultures of wild-type and ΔndvB strains. RNA was extracted from drip-fed biofilms grown in M63-arginine and planktonic cultures grown in LB (A to G) or from colony biofilms and planktonic cultures that were both grown in M63-arginine (H and I). Gene expression of the wild type and the ΔndvB mutant under each condition is given relative to that of the wild-type planktonic cultures. The error bars indicate standard deviations for two biological replicates tested in triplicate qPCRs. Tested genes include ndvB (A), exaA (B), eraR (C), erbR (D), pqqB (E), pqqC (F), pqqE (G), exaB (H), and ercS′ (I). All of the genes showed a statistically significant (P < 0.05) increase in expression in wild-type biofilms compared to that in ΔndvB biofilms. WT, wild type.
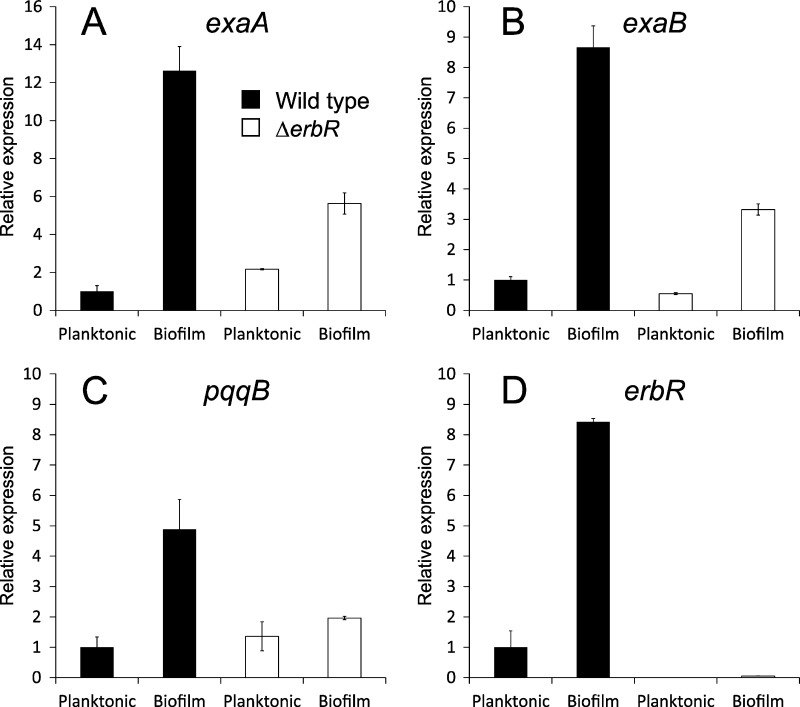
The genes encoding the P. aeruginosa ethanol oxidation system are controlled by a hierarchical signal transduction network (13, 22). Previous work with P. aeruginosa strain ATCC 17933 found that the response regulator ErbR (formerly AgmR) is a key member of this network (13). In the presence of ethanol, ErbR was shown to directly activate transcription of the exaBC, eraSR, and pqqABCDE operons and indirectly activate transcription of exaA (13). We suspected that the ndvB-dependent induction of the ethanol oxidation genes in biofilms also requires erbR. Therefore, we examined expression of three genes with different promoters (exaA, exaB, and pqqB) in wild-type and ΔerbR strains. Each gene was induced in wild-type biofilms relative to its expression in planktonic cells (Fig. 3), and gene expression was diminished in the ΔerbR biofilms, indicating that erbR is involved in the induction (Fig. 3). However, expression was not completely abolished in the ΔerbR biofilms, suggesting that other regulatory mechanisms contribute to the induction of these genes in biofilms. Overall, these results indicate that the induction of ethanol oxidation genes in P. aeruginosa biofilms involves both known (erbR) and novel (ndvB) regulatory elements.
Fig 3.
Induction of exaA, exaB, and pqqB in biofilms involves the response regulator ErbR. Colony biofilms and planktonic cultures of wild-type and ΔerbR strains were grown in M63-arginine. RNA was assayed by qPCR for expression of exaA (A), exaB (B), pqqB (C), and erbR (D). Gene expression is given relative to that of the wild-type planktonic cultures. For each condition, two biological replicate samples were tested in triplicate qPCRs.
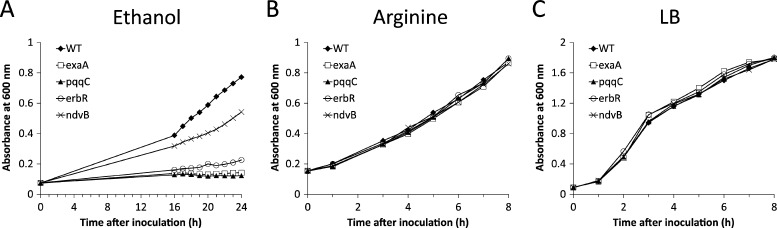
Planktonic growth in ethanol is compromised in the ΔndvB mutant.
A potential consequence of the reduced expression of the ethanol oxidation genes in the ΔndvB mutant is suboptimal growth on ethanol. We tested this possibility by comparing planktonic growth on ethanol minimal medium of the ΔndvB mutant, the wild type, and three mutants lacking ethanol oxidation genes (ΔexaA, ΔpqqC, and ΔerbR). The ΔndvB mutant had an intermediate growth rate that was substantially slower than that of the wild-type strain (Fig. 4A). In agreement with previous work, the ΔexaA, ΔpqqC, and ΔerbR mutants did not grow on ethanol (13, 14). The growth phenotypes were specific to the ethanol medium, since each of the strains exhibited wild-type growth rates on arginine minimal medium (Fig. 4B) and LB broth (Fig. 4C). The compromised planktonic growth of the ndvB mutant on ethanol is consistent with the regulatory link between ndvB and the ethanol oxidation genes.
Fig 4.
Inactivation of ndvB compromises planktonic growth on ethanol. The PA14 wild-type strain (WT), the ΔndvB mutant, and three ethanol oxidation gene mutants (ΔexaA, ΔpqqC, and ΔerbR) were grown in M63 containing ethanol (A) or arginine (B) as the sole carbon source or in LB medium (C). Growth was monitored by measuring absorbance at 600 nm. The growth curves are representative of at least two replicate experiments. Statistically significant differences in growth between the wild-type and mutant strains (P < 0.05) were observed for ethanol but not for the other conditions.
Inactivation of ethanol oxidation genes increases the antibiotic sensitivity of biofilms.
The biofilm-specific induction of ethanol oxidation genes was surprising given that the biofilms in these experiments were grown with medium lacking known inducers (i.e., ethanol and other alcohols) (12, 30). This observation suggested that the genes may have a function in biofilms that is unrelated to ethanol oxidation. Since ndvB is important for biofilm-specific antibiotic resistance (21), we tested whether the ethanol oxidation genes contribute to antibiotic resistance. We obtained transposon insertion mutants of 4 ethanol oxidation genes (exaA, exaC, pqqC, and eraR) from the PA14 transposon mutant library (18). We tested their antibiotic resistance phenotypes under both planktonic and biofilm growth conditions by determining the minimum bactericidal concentrations (MBCs) of ciprofloxacin, an antibiotic used to treat P. aeruginosa infections (11). We found that the mutants did not differ substantially from the wild-type strain in growth rate, biofilm formation ability, or planktonic antibiotic resistance (Table 4 and data not shown). However, compared to the wild-type strain, the mutants were more sensitive to ciprofloxacin when growing in biofilms (Table 4). These data suggested that ethanol oxidation genes contribute to the antibiotic resistance of biofilms.
Table 4.
Minimum bactericidal concentrations of ciprofloxacin for PA14 transposon mutantsa
| Strainb | Ciprofloxacin MBC (μg/ml) |
|
|---|---|---|
| MBC-P | MBC-B | |
| PA14 | 3 | 20 |
| PA14 ΔndvB | 3 | 5 |
| exaA::MrT7 | 3 | 10 |
| exaC::MrT7 | 3 | 10 |
| pqqC::MrT7 | 3 | 10 |
| eraR::MrT7 | 3 | 10 |
Minimum bactericidal concentrations were determined for planktonic (MBC-P) and biofilm (MBC-B) cultures grown in M63-arginine.
Transposon mutants were retrieved from the nonredundant library of PA14 transposon insertion mutants (18).
To validate these results, we tested the antibiotic resistance phenotypes of unmarked deletions of exaA, pqqC, and erbR. We determined MBCs for planktonic and biofilm cells (Table 5) and MICs for planktonic cells (Table 6). Tobramycin sensitivity was observed in the deletion mutant biofilms, although ciprofloxacin sensitivity was not observed (Table 5). Consistent with the role of these genes in biofilm-specific antibiotic resistance, mutants with double deletions (i.e., ΔndvB plus deletion of one of the three genes listed above) displayed increased susceptibility to tobramycin in comparison to those with single deletions (Table 5). Furthermore, we performed an MBC assay comparing the resistance of ΔndvB biofilms grown in ethanol minimal medium to that of the ΔndvB biofilms grown in arginine minimal medium. We found that growth in ethanol resulted in a slight but reproducible increase in resistance to tobramycin, suggesting that activation of the ethanol oxidation genes partially restores antibiotic resistance.
Table 5.
Minimum bactericidal concentrations for PA14 deletion mutants
| Strain | MBC (μg/ml) |
|||
|---|---|---|---|---|
| Tobramycin | Ciprofloxacin | |||
| MBC-P | MBC-B | MBC-P | MBC-B | |
| PA14 | 16 | 200 | 2 | 20 |
| PA14 ΔndvB | 16 | 50 | 2 | 10 |
| PA14 ΔexaA | 8 | 50 | 1 | 20 |
| PA14 ΔpqqC | 16 | 100 | 2 | 20 |
| PA14 ΔerbR | 16 | 50 | 1 | 20 |
| PA14 ΔndvB ΔexaA | 8 | 25 | 1 | 10 |
| PA14 ΔndvB ΔpqqC | 16 | 25 | 1 | 10 |
| PA14 ΔndvB ΔerbR | 16 | 25 | 0.5 | 20 |
Table 6.
MICs for planktonic cultures of PA14 deletion mutants
| Strain | MIC (μg/ml) |
|
|---|---|---|
| Tobramycin | Ciprofloxacin | |
| PA14 | 2 | 1 |
| PA14 ΔndvB | 2 | 1 |
| PA14 ΔexaA | 2 | 1 |
| PA14 ΔpqqC | 2 | 1 |
| PA14 ΔerbR | 2 | 1 |
To confirm the importance of these genes in antibiotic resistance, we tested whether induction of ndvB, erbR, exaA, and pqqC in the wild-type background increases planktonic antibiotic resistance. The genes were cloned into pJB866, a vector carrying the Pm promoter, which can be induced by m-toluic acid (35, 36). We determined the MICs of PA14 carrying each vector under inducing conditions (2 mM m-toluic acid). Compared to the vector control, the vectors with cloned genes increased the tobramycin MIC of PA14 by 2-fold for ndvB, exaA, and pqqC, with no MIC change for erbR. The vectors also increased the gentamicin MIC by 2-fold (for erbR) or 4-fold (for exaA and pqqC). The ndvB vector increased the ciprofloxacin MIC of PA14 by 2-fold.
We also expressed erbR, ndvB, exaA, and pqqC in the respective deletion mutant strains. Expression of erbR in an ΔerbR strain resulted in an 8-fold increase in resistance to ciprofloxacin in an MIC assay. Expression of ndvB in an ΔndvB mutant strain resulted in a 2-fold increase in resistance to ciprofloxacin. However, expression of exaA or pqqC had no effect on their respective deletion mutant strains. Thus, increased expression of several ethanol oxidation genes resulted in increased resistance to tobramycin, gentamicin, and ciprofloxacin, further indicating that these genes contribute to antibiotic resistance.
Expression of erbR and exaA is restored in ΔndvB biofilms after incubation with wild-type periplasmic extract.
The ndvB-dependent induction of the ethanol oxidation genes in biofilms is likely an ethanol-independent mechanism, since the gene expression experiments were carried out in medium lacking ethanol. In Rhizobium species, NdvB-derived cyclic glucans are important for symbiotic interactions with plants, possibly by mediating signaling between bacteria and plants (2). We reasoned that a signaling mechanism involving glucans might be responsible for the gene expression changes we observed in P. aeruginosa biofilms. Since both NdvB-derived glucans and the ethanol dehydrogenase localize to the periplasm (14, 21), it seemed possible that a periplasmic signaling molecule present in wild-type biofilms might be absent in ΔndvB biofilms. Thus, adding the signal in trans to ΔndvB biofilms might restore expression of ethanol oxidation genes to wild-type levels.
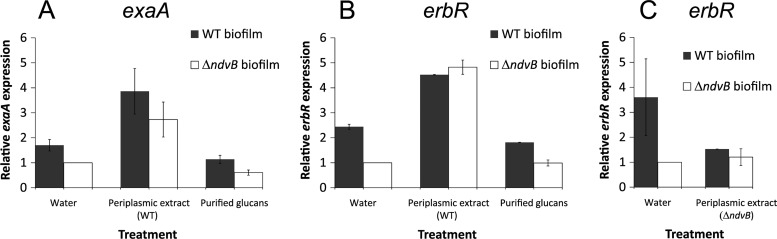
We isolated periplasmic extracts from late-stationary-phase planktonic cultures of the wild-type strain, a condition that results in ndvB gene expression and glucan production (21). To determine whether a component of the extract might affect transcription of ethanol oxidation genes, we added wild-type periplasmic extract to preformed wild-type and ΔndvB biofilms and incubated for 8 h. We assayed erbR and exaA expression and found that the wild-type periplasmic extract restored gene expression in the ΔndvB biofilms to wild-type levels and also increased expression in the wild-type biofilms (Fig. 5A and B). In a control experiment, periplasmic extract isolated from ΔndvB cultures did not restore wild-type expression of erbR (Fig. 5C).
Fig 5.
Expression of erbR and exaA in biofilms treated with periplasmic extract or purified glucans. Preformed wild-type and ΔndvB biofilms were treated for 8 h with water (negative control), wild-type periplasmic extract, ΔndvB periplasmic extract, or purified glucans. Expression of exaA (A) and erbR (B and C) was measured by qPCR and is given relative to that of the untreated ΔndvB biofilms. Each panel shows the results of two or three biological replicates tested in triplicate qPCRs.
These data suggest that a periplasmic molecule is capable of inducing erbR and exaA gene expression in the ΔndvB mutant. We suspected that the signal might be the NdvB-derived glucans. Therefore, we purified glucans from PA14 periplasmic extracts by gel filtration (21). However, treatment of the ΔndvB biofilms with purified glucans did not induce expression of erbR or exaA (Fig. 5A and B). Thus, the induction of ethanol oxidation genes appears to be modulated by an unidentified periplasmic molecule that is dependent on the presence of ndvB.
DISCUSSION
The increased antibiotic resistance of biofilms continues to confound treatment of bacterial infections (4, 15). We investigate ndvB, a gene which contributes to the increased antibiotic resistance of P. aeruginosa biofilms. ndvB encodes a glucosyltransferase required for the formation of cyclic periplasmic glucans (21, 27). We have previously shown that ndvB-derived glucans can interact with antibiotics and, thus, sequester these compounds away from their cellular targets (21). In this study, we explore other functions of P. aeruginosa glucans by examining how global gene expression is affected by their presence or absence. DNA microarray and qPCR analyses revealed that several genes required for ethanol oxidation are responsive to the presence of ndvB in the genome. These genes have greater expression levels in biofilms than in planktonic cells, and their inactivation increased the antibiotic sensitivity of bacteria growing in biofilms. Additional genes were also identified as ndvB responsive in this study, but their role in antibiotic resistance is unknown.
The P. aeruginosa ethanol oxidation system has been studied only in planktonic cultures, and its function in biofilms is unexplored. We found that eight ethanol oxidation genes are induced in P. aeruginosa biofilms. The cause of the induction is unclear, since these genes are not expressed in planktonic cultures in the absence of known inducers (12, 30). One explanation is that ethanol is produced in wild-type, but not ΔndvB, biofilms at levels sufficient to activate the ethanol oxidation genes. The most likely route of ethanol production in P. aeruginosa is as an anaerobic fermentation product. Steep oxygen gradients exist in laboratory biofilms exposed to air, and it is possible that a subpopulation of bacteria grows anaerobically (33). Nevertheless, ethanol production has not been demonstrated in P. aeruginosa cultures. In anaerobic conditions, P. aeruginosa can convert pyruvate to lactate, acetate, and succinate, a process which contributes to long-term anaerobic survival (9). It was suggested that ethanol can also be produced as a pyruvate fermentation by-product; however, ethanol production was not detected (9). These observations and the absence of pyruvate in the growth media of our experiments argue against significant ethanol production in the biofilms.
If ethanol production in P. aeruginosa biofilms is not responsible for the gene expression changes, then what is responsible? Expression of the ethanol oxidation genes is regulated by three sensor kinases, three response regulators, and additional regulators that have been postulated (22). This complex regulatory scheme implies that several physiological signals, the natures of which are unknown, are integrated. Intriguingly, inactivation of several enzymes involved in central metabolism inhibits the induction of ethanol oxidation genes, suggesting that some internal metabolites are involved in the induction (12, 22). Our results indicate that an ndvB-dependent signaling function that can induce expression of the ethanol oxidation genes is found in the P. aeruginosa periplasm (Fig. 5), though NdvB-derived glucans themselves do not appear to be the signal. It is possible that a signaling function associates with the glucans but was lost during the glucan purification procedure.
An interesting finding of this study was the antibiotic sensitivity found in several mutants of ethanol oxidation genes. The sensitivity was observed only when the bacteria grew as biofilms. Furthermore, double mutants (ΔndvB plus deletion of exaA, pqqC, or erbR) were more sensitive than the corresponding single mutants. Together, these results suggest that ndvB promotes antibiotic resistance in two ways: drug sequestration by cyclic glucans (21) and activation of ethanol oxidation genes. The mechanism by which the ethanol oxidation genes promote antibiotic resistance is unclear but may be related to the redox activity of the ethanol dehydrogenase enzyme. Several studies have demonstrated that PQQ, the cofactor of the dehydrogenase, can protect bacteria from oxidative damage (24, 31). Since bactericidal antibiotics are thought to kill bacteria in part by increasing oxidative stress (17), perhaps PQQ and the ethanol dehydrogenase alleviate antibiotic-induced oxidative stress. Future investigations based on these findings may reveal the nature of this protective mechanism and illuminate the role of cyclic glucans and ethanol oxidation genes in P. aeruginosa biofilms.
ACKNOWLEDGMENTS
This work was supported by grants from Cystic Fibrosis Canada and the National Sciences and Engineering Research Council.
We thank Jon Labriola for technical assistance.
We dedicate this article to the late Eva Markvoort, a courageous woman who continues to inspire us.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid 38:35–51 [DOI] [PubMed] [Google Scholar]
- 2. Breedveld MW, Miller KJ. 1994. Cyclic β-glucans of members of the family Rhizobiaceae. Microbiol. Rev. 58:145–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CLSI 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard—eighth edition. CLSI document M07-A8Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 5. Development Core Team R 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 6. Dunlap J, Minami E, Bhagwat AA, Keister DL, Stacey G. 1996. Nodule development induced by mutants of Bradyrhizobium japonicum defective in cyclic β-glucan synthesis. Mol. Plant Microbe Interact. 9:546–555 [DOI] [PubMed] [Google Scholar]
- 7. Dylan T, et al. 1986. Rhizobium meliloti genes required for nodule development are related to chromosomal virulence genes in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 83:4403–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dylan T, Nagpal P, Helinski DR, Ditta GS. 1990. Symbiotic pseudorevertants of Rhizobium meliloti ndv mutants. J. Bacteriol. 172:1409–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eschbach M, et al. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186:4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gentleman RC, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 12. Gliese N, Khodaverdi V, Gorisch H. 2010. The PQQ biosynthetic operons and their transcriptional regulation in Pseudomonas aeruginosa. Arch. Microbiol. 192:1–14 [DOI] [PubMed] [Google Scholar]
- 13. Gliese N, Khodaverdi V, Schobert M, Gorisch H. 2004. AgmR controls transcription of a regulon with several operons essential for ethanol oxidation in Pseudomonas aeruginosa ATCC 17933. Microbiology 150:1851–1857 [DOI] [PubMed] [Google Scholar]
- 14. Gorisch H. 2003. The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1647:98–102 [DOI] [PubMed] [Google Scholar]
- 15. Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108 [DOI] [PubMed] [Google Scholar]
- 16. Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 17. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 18. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loewus FA. 1952. Improvement in anthrone method for determination of carbohydrates. Anal. Chem. 24:219 [Google Scholar]
- 20. Mah T-F, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 21. Mah T-F, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 22. Mern DS, Ha SW, Khodaverdi V, Gliese N, Gorisch H. 2010. A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: indication of four levels of sensor kinases and response regulators. Microbiology 156:1505–1516 [DOI] [PubMed] [Google Scholar]
- 23. Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1:Unit 1B.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Misra HS, et al. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578:26–30 [DOI] [PubMed] [Google Scholar]
- 25. Pardee AB, Jacob F, Monod J. 1959. Genetic control and cytoplasmic expression of inducibility in the synthesis of β-galactosidase by E. coli. J. Mol. Biol. 1:165–178 [Google Scholar]
- 26. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 27. Sadovskaya I, et al. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated β-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904 [DOI] [PubMed] [Google Scholar]
- 28. Sauer K. 2003. The genomics and proteomics of biofilm formation. Genome Biol. 4:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schobert M, Gorisch H. 1999. Cytochrome c550 is an essential component of the quinoprotein ethanol oxidation system in Pseudomonas aeruginosa: cloning and sequencing of the genes encoding cytochrome c550 and an adjacent acetaldehyde dehydrogenase. Microbiology 145(Part 2):471–481 [DOI] [PubMed] [Google Scholar]
- 30. Schobert M, Gorisch H. 2001. A soluble two-component regulatory system controls expression of quinoprotein ethanol dehydrogenase (QEDH) but not expression of cytochrome c(550) of the ethanol-oxidation system in Pseudomonas aeruginosa. Microbiology 147:363–372 [DOI] [PubMed] [Google Scholar]
- 31. Shrivastava M, Rajpurohit YS, Misra HS, D'Souza SF. 2010. Survival of phosphate-solubilizing bacteria against DNA damaging agents. Can. J. Microbiol. 56:822–830 [DOI] [PubMed] [Google Scholar]
- 32. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 33. Walters MC, III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Whiteley M, et al. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864 [DOI] [PubMed] [Google Scholar]
- 35. Zhang L, Hinz AJ, Nadeau JP, Mah T-F. 2011. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J. Bacteriol. 193:5510–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L, Mah T-F. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190:4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]