Abstract
The TonB system of proteins is required for the energy-dependent active transport of iron-bound substrates across the outer membrane of Gram-negative bacteria. We have identified three TonB systems within the human pathogen Vibrio vulnificus. The TonB1 system contains the TonB1, ExbD1, and ExbB1 proteins, whereas both the TtpC2-TonB2 and TtpC3-TonB3 systems contain an additional fourth protein, TtpC. Here we report that TtpC3, although highly related to TtpC2, is inactive in iron transport, whereas TtpC2 is essential for the function of the TtpC2-TonB2 system in V. vulnificus. This protein, together with TonB2, is absolutely required for both the uptake of endogenously produced iron-bound siderophores as well as siderophores produced from other organisms. Through complementation we show that V. vulnificus is capable of using different TtpC2 proteins from other Vibrio species to drive the uptake of multiple siderophores. We have also determined that aerobactin, a common bacterial siderophore involved in virulence of enteric bacteria, can only be brought into the cell using the TtpC2-TonB2 system, indicating an important evolutionary adaptation of TtpC2 and TonB2. Furthermore, in the absence of TonB1, TtpC2 is essential for a fully virulent phenotype as demonstrated using 50% lethal dose (LD50) experiments in mice.
INTRODUCTION
Vibrio vulnificus is an opportunistic marine pathogen that is capable of causing fatal septicemia in both humans and eels (19, 37). Human infections are generally caused by ingestion of contaminated seafood or through open wounds that come in contact with contaminated seawater (33, 49). Primary sepsis is often associated with diseases that are linked to elevated iron levels, including cirrhosis, hemochromatosis, and thalassemia (19).
Iron is an essential element that is required for the survival of almost all living organisms. Iron is highly versatile, ranging in processes from signaling to metabolism, and can act as both an electron donor and acceptor (12). Even though it is the fourth most abundant element in the Earth's crust, it remains a major challenge to obtain by many microorganisms. Iron forms insoluble ferric hydroxides when exposed to aerobic conditions (12). In addition, microbes that colonize mammalian hosts are confronted with high-affinity iron-binding proteins such as transferrin and lactoferrin, which only exacerbate iron limitation inside the host environment (9). Microorganisms have evolved specific iron-sequestering systems that focus around siderophores, which are low-molecular-weight high-affinity iron-chelating molecules, and the cell surface receptors responsible for binding these molecules (1, 11, 24).
In Gram-negative bacteria, the outer membrane (OM) protein receptors are responsible for binding an assortment of minerals and nutrients, including iron-bound siderophores. The inner membrane (IM) is the site of respiration that results in the generation of energy in the forms of the proton motive force (PMF) and adenosine triphosphate (ATP). The PMF energy is transduced to the OM receptors by the TonB system of proteins, thus activating them to a structure that recognizes the iron-siderophore complex (7, 12, 42).
The TonB system has been extensively studied in Escherichia coli, where it has been shown to consist of three integral inner membrane proteins: TonB, ExbB, and ExbD (5, 6, 8). Both ExbB and ExbD have been shown to promote conformational changes in TonB by transferring energy from the PMF, thereby allowing TonB to span the periplasm and make contact with TonB-dependent OM receptors (16, 21, 25, 28–32, 40, 43, 50). All members of the Vibrionaceae have two very similar TonB systems, the TonB1 and TtpC2-TonB2 systems, and in the case of a smaller subset of organisms in this family, a third TonB system, TtpC3-TonB3. Both the TtpC2-TonB2 and TtpC3-TonB3 systems contain an additional protein, TtpC (2, 26, 27, 48, 52).
TtpC is highly conserved in all pathogenic Vibrio species analyzed to date (26, 27, 48). This protein is 45 kDa in size and contains three transmembrane domains with its carboxy terminus anchored in the IM and its amino-terminal region in the periplasm (26, 48; our unpublished results). Cross-linking experiments in V. anguillarum have shown that TtpC (herein named TtpC2 to differentiate it from TtpC3 found in the TtpC3-TonB3 system) is capable of forming complexes with other members of the TtpC-TonB2 system and potentially outer membrane receptor proteins (48). TtpC is an essential component of the TtpC-TonB2 system in both V. cholerae and V. anguillarum (25, 48). Although TtpC can be found in all Vibrio species as well as many aquatic bacteria, little is known about its function within the TtpC-TonB2 system.
In this study, we demonstrate that TtpC2 is essential for the TtpC2-TonB2 system in V. vulnificus and that TtpC3 cannot replace it. In addition, the TtpC2 proteins from other Vibrio species can complement a ΔttpC2 mutation and function as part of the TtpC2-TonB2 system of V. vulnificus, underlying the importance and ubiquity of this protein. We also present data showing the ability of this protein to intervene in the uptake of endogenous and heterologous siderophores. Finally, through virulence assays, we show that TtpC2 is crucial for a fully virulent phenotype in V. vulnificus infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. V. vulnificus was routinely grown in tryptic soy broth with the addition of 10 g/liter of NaCl for a total of 1.5% NaCl (TSBS) or in minimal CM9 media (1× M9 salts is 60 g Na2HPO4, 30 g KH2PO4, 50 g NaCl, 10 g NH4Cl per liter [pH 7.2]), 0.2% Casamino Acids, 0.5% glucose, 10 μM CaCl2, 100 μM MgSO4) (10). E. coli was routinely grown in LB broth. Antibiotics were used at the following final concentrations: 10 μg/ml chloramphenicol (Cm) for V. vulnificus and 30 μg/ml Cm for E. coli when appropriate. Thiosulfate-citrate-bile salts-sucrose agar (TCBS) (Becton, Dickinson) was used as the selective medium for V. vulnificus during conjugations.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristica | Reference or source |
|---|---|---|
| Strains | ||
| V. vulnificus | ||
| CMCP6 | Wild type | J. Rhee |
| AA-14 | ΔvenB | 2 |
| AA-6 | ΔtonB1 | 2 |
| AA-7 | ΔtonB2 | 2 |
| VSRK301 | ΔttpC2 | This study |
| AA-8 | ΔtonB3 | 2 |
| VSRK198 | ΔttpC3 | This study |
| AA-9 | ΔtonB1 ΔtonB2 | 2 |
| VSRK357 | ΔtonB1 ΔttpC2 | This study |
| AA-10 | ΔtonB1 ΔtonB3 | 2 |
| VSRK199 | ΔtonB1 ΔttpC3 | This study |
| AA-11 | ΔtonB2 ΔtonB3 | 2 |
| VSRK275 | ΔttpC2 ΔttpC3 | This study |
| AA-12 | ΔtonB1 ΔtonB2 ΔtonB3 ΔlacZ | 2 |
| VSRK202 | ΔtonB1 ΔtonB2 ΔttpC3 | This study |
| VSRK304 | ΔtonB1 ΔttpC2 ΔttpC3 | This study |
| AA-16 | ΔtonB1 ΔtonB2 ΔtonB3 ΔvenB | 2 |
| VSRK283 | ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB | This study |
| V. parahaemolyticus | ||
| RIMD | Wild type | T. Honda |
| E. coli | ||
| S17-1 λpir | λpir lysogen; thi pro hsdR hsdM+recA RP4-2 Tc∷Mu Km∷Tn7; Tpr Smr | 46 |
| Plasmids | ||
| pCR2.1 | TA cloning vector; Kmr Ampr | Invitrogen |
| pMMB208 | Broad-host-range expression vector; CmrPtac | 36 |
| pDM4 | Suicide vector with oriR6K; CmrsacB | 35 |
| pRK2013 | Helper plasmid; Kmr | 15 |
| ptonB1 | V. vulnificus CMCP6 tonB1 cloned into pMMB208 | 2 |
| ptonB2 | V. vulnificus CMCP6 tonB2 cloned into pMMB208 | 2 |
| pttpC2(Vvul) | V. vulnificus CMCP6 ttpC2 cloned into pMMB208 | This study |
| pttpC2(Vcho) | V. cholerae CA401 ttpC2 cloned into pMMB208 | This study |
| pttpC2(Vpara) | V. parahaemolyticus RIMD ttpC2 cloned into pMMB208 | This study |
| pttpC2(Valg) | V. alginolyticus 12G01 ttpC2 cloned into pMMB208 | This study |
| pttpC2(Vang) | V. anguillarum 775 ttpC2 cloned into pMMB208 | This study |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant; Tpr, trimethoprim resistant.
Sequence identity.
The percentage of identity was found through the align function of the BLASTp interface (3). V. vulnificus strain CMCP6 (accession number PRJNA62909) was used for the BLAST analysis against V. parahaemolyticus strain RIMD (accession number PRJNA57969), V. alginolyticus strain 12G01 (accession number PRJNA54241), V. cholerae strain CA401 (accession number AF047974.1), V. anguillarum strain 775 (accession number PRJNA68057), and E. coli K-12 (accession number PRJNA57779).
Construction and complementation of V. vulnificus mutants.
In-frame deletions of the entire coding sequences of genes were generated using splicing by overlap extension (SOE) PCR (22, 45). Upstream and downstream regions (approximately 700 bp) flanking each gene were amplified with specific primers. The two 700-bp fragments with overlapping primer ends were spliced using SOE PCR with the two outside primers. The amplified fragment was cloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA), digested with appropriate restriction enzymes, and subcloned into the suicide vector pDM4, which was previously digested with the same restriction enzymes. E. coli S17-1 λpir transformed with the pDM4 (35) derivatives was conjugated with V. vulnificus, and exconjugants were selected on TCBS agar with 2 μg/ml Cm. For the excision of the suicide vector, clones were incubated in the absence of Cm and plated on TSAS plates (TSBS with 1.5% agar) containing 15% sucrose. Those colonies that grew on these plates were screened for Cm sensitivity, and deletions within the genes of interest were confirmed by PCR.
Complementation of deleted genes was achieved by amplifying each gene of interest by PCR with primers containing restriction sites. The fragments were cloned into the pCR2.1 vector (Invitrogen, Carlsbad, CA), sequenced, and then subcloned into pMMB208 (36) under the control of the Ptac promoter. This promoter is under the control of the lacI gene harbored in the vector. The constructs were transformed into V. vulnificus strains by triparental conjugation with the plasmid helper pRK2013 (15). In order to induce transcription of the cloned genes, 1 mM isopropyl-d-thiogalactopyranoside (IPTG) was added to the solid and/or broth medium.
CAS assays.
Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.02 into 25 ml of CM9 media and incubated at 37°C to an OD600 of 0.5 (∼3 h). Cells were normalized to an OD600 of 0.5 in 1 ml of CM9 media. Cells were then centrifuged, and 0.5 ml of supernatant was used in the chrome azurol S (CAS) assay. A ratio of 1:1 of cell supernatant to CAS solution (600 μM hexadecyltrimethylammonium bromide [HDTMA], 150 μM FeCl3, 150 μM CAS) was mixed and placed into 0.2-cm cuvettes, and the OD630 was read using a spectrophotometer at 20 min after mixing. The assay was normalized to the wild type (WT), and the final values were inverted to show a positive change for ease of reading.
Bioassays.
Bacteria were grown overnight in TSBS with the appropriate antibiotics and diluted 1/400 into TSAS containing 100 μg/ml ethylenediamine-di-(o-hydroxyphenylacetic) acid (EDDA) or CM9 minimal media containing 30 μg/ml of EDDA. In these experiments, bacterial cells were included in the agar and, upon solidification, plates were spot inoculated with different iron sources. After incubation of the plates at 37°C, halos of bacterial growth surrounding the locations of the spots indicated positive results. In these experiments, ferric ammonium citrate (FAC), which does not require active transport, was included to confirm that the strain inoculated in the agar was viable. The purified compounds spotted on top of the bioassay plates to determine the functionality of the different TonB cluster of genes in V. vulnificus were as follows: 1 mg/ml iron-free vibriobactin (EMC Microcollections, Germany), 1 mg/ml iron-free anguibactin (our laboratory), 1 mg/ml iron-free aerobactin, 1 mg/ml iron-free enterobactin, 1 mg/ml iron-free ferrioxamine (all three from EMC Microcollections, Germany), and 500 μg/ml FAC (Sigma). Each bioassay plate was spotted with 2 μl of each indicated iron source. To test vibrioferrin, WT V. parahaemolyticus was streaked onto the plate. Vulnibactin and the hydroxamate siderophore, both from V. vulnificus, were tested by streaking either WT or the ΔvenB strain onto the bioassay plate. Halo growth around these streaks indicates that the strain within the plate can use the siderophore being produced by the streaked strain.
Virulence experiments.
Overnight cultures were diluted to an OD600 of 0.02 into 25 ml of TSBS and incubated at 37°C to an OD600 of 0.5. Cells were pelleted and washed twice in phosphate-buffered saline (PBS). Cells were diluted to an OD600 of 1.0 and serially diluted in PBS. Five 4- to 6-week-old CD1 mice (Charles River Laboratories) per dilution were injected intraperitoneally with 0.1 ml of the strain of interest. Three serial dilutions for each strain were evaluated. Mortality was monitored for 48 h postinfection, and 50% lethal dose (LD50) calculations were determined by the Reed-Muench method (44).
Radioactive 55Fe uptake assays.
Overnight cultures grown in TSBS with Cm were subcultured to an OD600 of 0.02 into 20 ml of CM9 with 1 mM IPTG plus Cm and grown at 37°C to an OD600 of 0.3. Cells were pelleted and washed once with M9 salts plus 100 μM nitrilotriacetic acid (NTA). Cells were then diluted to an OD600 of 0.3 in a total of 8 ml and placed into 50-ml conical tubes. Cells were shaken at 37°C for 2 h to deplete iron. To label the siderophore, 55FeCl3 (2 mCi; Perkin-Elmer, Life Sciences Inc., Boston, MA) was diluted 1:10 in 0.5 M HCl and 1 μl was added to 48 μl H2O along with 1 μl of 5.3 mM desferrioxamine G (EMC Microcollections, Germany). The labeling was performed at 37°C for 15 min. Ten microliters of 1 M potassium cyanide (KCN) was added to the appropriate cultures 10 min before the addition of [55Fe]ferrioxamine to act as a negative control. At time zero, 1.87 μl (18.7 pmol [55Fe]ferrioxamine) was added to 8 ml of culture and allowed to continue shaking at 37°C. One milliliter of cells was removed at the appropriate times and passed over a 0.45-μm surfactant-free hydroanalysis mixed cellulose ester Triton-free (HATF) filter (Millipore, Billerica, MA). To measure [55Fe]ferrioxamine internalization rather than binding, the ferrioxamine receptor was quenched by washing twice with 10 ml of 0.1 M LiCl. Filters were dried, and the amount of 55Fe was determined by scintillation counting. Two biological replicates were done for each strain.
RESULTS
V. vulnificus contains two TtpC proteins associated with the TtpC2-TonB2 and TtpC3-TonB3 systems.
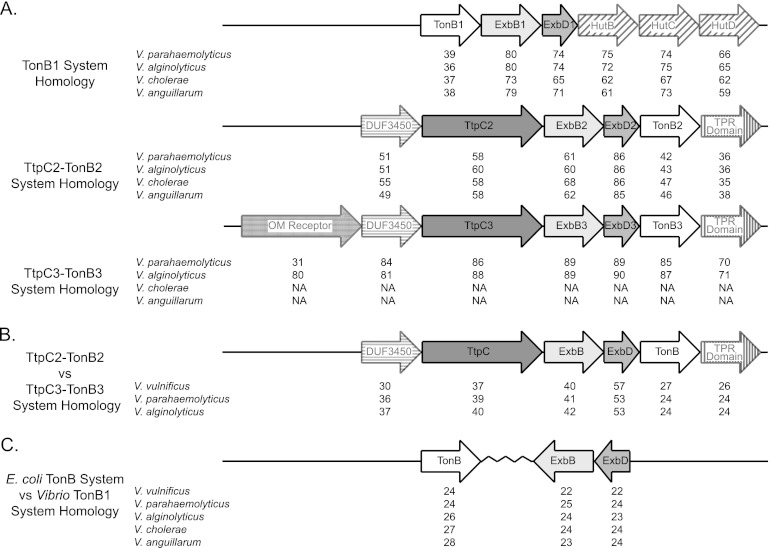
The bacterium V. vulnificus contains three TonB systems (2) (Fig. 1A). The TonB1 system consists of three genes, tonB1, exbB1, and exbD1, and was first elucidated in V. cholerae (17, 20). The TonB1 system is present in all members of the Vibrionaceae examined. The TonB1 system of V. vulnificus, as well as every other member of the Vibrionaceae, is associated with genes needed for heme uptake (Fig. 1A). Unlike E. coli, the TonB1 system of the Vibrionaceae contains all three genes in one operon (Fig. 1A and C). In this study, we show the homology of the V. vulnificus TonB1 system of proteins, TonB1, ExbB1, and ExbD1, with that of other Vibrio TonB1 systems (Fig. 1A). The TonB1 protein of V. vulnificus is on average ∼40% identical to TonB1 proteins from other members of the Vibrionaceae; however, the percentages of identity of both the ExbB1 and ExbD1 proteins are much higher (around 75%). This higher level of identity is also seen in the heme utilization proteins, HutB, HutC, and HutD (Fig. 1A). Comparing the different TonB systems of assorted Vibrio species to the E. coli TonB system, we found that the TonB1 system had the highest degree of identity with an average of only ∼25% for any given protein (Fig. 1C and data not shown).
Fig 1.
The TonB systems of V. vulnificus and their homology to other bacterial species. (A) The three TonB systems of V. vulnificus are depicted. The percentages of identity of the V. vulnificus proteins to the V. parahaemolyticus, V. alginolyticus, V. cholerae, and V. anguillarum proteins are shown. The percentage of identity is listed below each protein being compared. NA represents the lack of that system in the bacterium. (B) The TtpC2-TonB2 system of V. vulnificus, V. parahaemolyticus, and V. alginolyticus is compared to the TtpC3-TonB3 system from the same species. The percentage of identity is listed below each protein being compared. (C) The TonB system of E. coli is compared against the TonB1 systems of V. vulnificus, V. parahaemolyticus, V. alginolyticus, V. cholerae, and V. anguillarum. The percentage of identity is listed below each protein being compared.
All of the vibrios examined thus far contain a second TonB system termed the TtpC2-TonB2 system. This is unlike E. coli that contains only a single TonB system. The TtpC2-TonB2 system has shown uptake specificity to certain iron-bound substrates in both V. anguillarum and V. cholerae (26, 48). Unlike the TonB1 system, the TtpC2-TonB2 system consists of four proteins: TonB2, ExbD2, ExbB2, and TtpC2 (25, 26, 50; unpublished data) (Fig. 1A). In addition, the Vibrionaceae have two genes surrounding the TtpC2-TonB2 system. These include a hypothetical protein with a domain of unknown function (DUF3450) and a protein that contains a tetratricopeptide repeat (TPR) domain that can be involved in protein-protein interactions (Fig. 1A). The role, if any, that these two additional genes play in the TtpC2-TonB2 system is currently being investigated. The percentages of identity of the V. vulnificus TtpC2-TonB2 system and these two additional proteins are compared to four other Vibrio species in Fig. 1A. Similar to TonB1, the TonB2 protein of V. vulnificus has about 45% identity with the other TonB2 proteins in the TtpC2-TonB2 system. ExbD2 shares the largest amount of identity between the members of the Vibrionaceae evaluated here, with the identity close to 85%. Both TtpC2 and ExbB2 of V. vulnificus share about 60% identity to the corresponding proteins from other Vibrio species.
A few Vibrio species, including V. vulnificus, V. parahaemolyticus, and V. alginolyticus, contain a third TonB system, TtpC3-TonB3 (2, 26, 27). This system also contains the fourth essential protein TtpC. In this study, we show the TtpC3-TonB3 system and its organization (Fig. 1A). The highest percentage of identity between Vibrio species was seen with the TtpC3-TonB3 system, close to ∼85% (Fig. 1A). Overall, this system appears very similar to the TtpC2-TonB2 system in its general grouping and orientation, with the exception of one gene. Unlike the TtpC2-TonB2 system, the TtpC3-TonB3 system contains an OM receptor protein upstream of DUF3450. When the OM receptor of V. vulnificus is compared to that of V. alginolyticus, the percentage of identity is very similar to other proteins in this operon. On the other hand, the OM receptor protein of V. parahaemolyticus shows a very low percentage of identity (only 31% compared to V. vulnificus).
To characterize the TtpC2-TonB2 and TtpC3-TonB3 systems, each was compared against each other in three different Vibrio species (Fig. 1B). In V. vulnificus, these two systems appear to be very different from each other with identity ranging from 27% to 57%. This amount of identity was also true in both V. parahaemolyticus and V. alginolyticus. ExbD3 remained consistently higher in identity, as was seen in other comparisons. It should be noted that the level of TtpC2 to TtpC3 identity was low (around 40%).
The V. vulnificus TtpC2 protein is essential for the TtpC2-TonB2 system in mediating transport of endogenous siderophores.
Bioassays were used to investigate the essentiality of ttpC2 within the TtpC2-TonB2 system for the uptake of the endogenous siderophores, vulnibactin and the uncharacterized hydroxamate compound (47), in V. vulnificus. We began by testing an assortment of mutants that were deleted in different TonB systems (Table 2). This large assortment of mutants allowed us to test not only the role played by TtpC2 in the TtpC2-TonB2 system but also to test the ability of each of the three TonB systems to use these two siderophores.
Table 2.
TtpC2 is an essential component of the TtpC2-TonB2 system in mediating the uptake of endogenous siderophores in V. vulnificus
| V. vulnificus strain or genotype | Growth on indicated iron sourcea |
||
|---|---|---|---|
| FAC | Vulnibactin | Hydroxamate | |
| WT | + | + | + |
| ΔtonB1 | + | + | + |
| ΔtonB2 | + | + | + |
| ΔttpC2 | + | + | + |
| ΔtonB3 | + | + | + |
| ΔtonB1 ΔtonB2 | + | − | − |
| ΔtonB1 ΔttpC2 | + | − | − |
| ΔtonB1 ΔtonB3 | + | + | + |
| ΔtonB2 ΔtonB3 | + | + | + |
| ΔtonB1 ΔtonB2 ΔtonB3 | + | − | − |
| ΔtonB1 ΔttpC2 ΔttpC3/pMMB208 | + | − | − |
| ΔtonB1 ΔttpC2 ΔttpC3/pttpC2(Vvul) | + | + | + |
Growth was determined by the presence of a halo (+) or lack thereof (−) around the iron source indicated. FAC, which does not require active transport, was added as a positive control to confirm that the strain imbedded within the plate was viable. Two microliters of FAC was spotted (500 μg/ml). Wild-type V. vulnificus was streaked onto the plate, and the halo of growth was monitored around the streak. A V. vulnificus ΔvenB mutant strain that is deficient in the production of vulnibactin was streaked onto the plate, and the halo of growth was monitored around the streak.
We previously demonstrated that both the TonB1 and TtpC2-TonB2 systems were capable of mediating vulnibactin uptake (2), but it was unknown which TonB system was responsible for mediating the uptake of the hydroxamate siderophore. Additionally, we previously investigated only tonB2 within the TtpC2-TonB2 system and not ttpC2. As shown in Table 2, single mutant strains that were deleted for either tonB1 or ttpC2 were capable of using both native siderophores. When the ΔtonB1 ΔttpC2 double mutant was tested, both native siderophores could no longer be acquired by V. vulnificus, indicating that both siderophores could use either the TonB1 system or the TtpC2-TonB2 system. In addition, these data demonstrate that TtpC3 cannot be used in place of TtpC2 since TtpC3 is still produced by these strains. We also created mutants that were deleted for tonB2 instead of ttpC2 and tested their ability to use the two endogenous siderophores. As shown in Table 2, all of these mutations showed the same results as when ttpC2 was deleted. Once again, a ΔtonB1 ΔtonB2 double mutant was not capable of using these siderophores as iron sources. Both siderophores were also unable to be used when all three TonB systems were deleted. We tested the ΔtonB1 ΔttpC2 ΔttpC3 triple deletion strain with a complementing plasmid expressing TtpC2. In this complemented strain, both native siderophores could once again be used as iron sources (Table 2).
We further analyzed these mutants through the use of chrome azurol S (CAS) assays. This colorimetric method can be used to detect the secretion of iron-bound siderophores. The endogenous siderophores of V. vulnificus are secreted in iron-depleted conditions. If the cells contain a functional TonB system capable of mediating the uptake of these iron-bound siderophores, they will be brought back into the cell. By testing different TonB mutants, one can assess if that particular TonB system can be used to power the uptake of each of these siderophores. Vibrio vulnificus will continue to produce and secrete siderophores until it has arrived at the optimal iron level. If a TonB system is nonfunctional and these siderophores cannot be brought back into the cell, an accumulation of siderophores builds up in the medium. By harvesting the supernatant from these cultures and mixing it with the CAS reagents, one can determine through a color change if a siderophore is unable to be brought back into the cell.
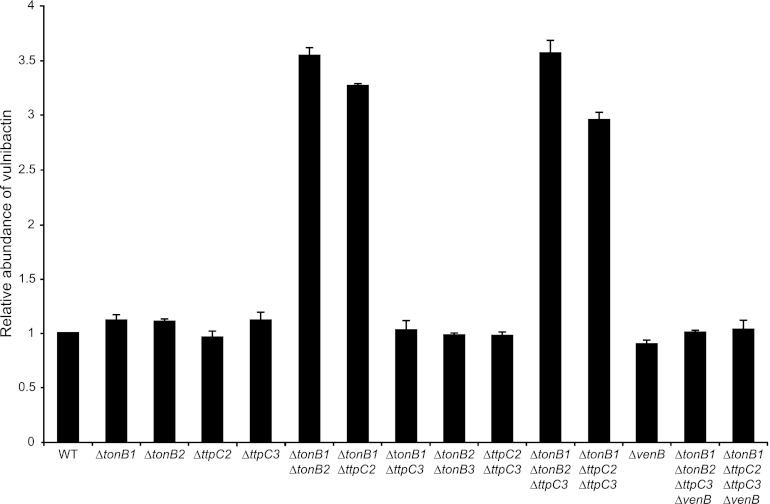
The results from these experiments are shown in Fig. 2. Both the TonB1 and TtpC2-TonB2 systems are used for the uptake of endogenous siderophores, confirming our bioassay results. Single and/or double mutations that allow either the TonB1 system or the TtpC2-TonB2 system to still function show the same amount of siderophore production as WT. Only when genes in both the TonB1 and TtpC2-TonB2 systems are deleted do we see an increase in relative production of siderophores. It is important to point out that the same result is seen independently of whether tonB2 or ttpC2 is deleted, once again confirming the essentiality of TtpC2 in the TtpC2-TonB2 system. By analyzing a ΔvenB mutant that does not produce vulnibactin, as well as this mutation in conjunction with other TonB system deletions, we have determined that vulnibactin and not the hydroxamate siderophore is the primary siderophore secreted under conditions of iron limitation (Fig. 2).
Fig 2.
TtpC2 within the TtpC2-TonB2 system is essential for vulnibactin uptake. Liquid chrome azurol S (CAS) assays were used to determine the relative production of V. vulnificus native siderophore vulnibactin. Supernatant from log phase cultures grown in minimal media were mixed with CAS solution at a 1:1 ratio. The OD630 was determined after 20 min of incubation at room temperature. Samples were normalized to WT.
The V. vulnificus TtpC2 protein is essential for the TtpC2-TonB2 system in mediating transport of exogenous siderophores.
In order to better survive in the various environments and hosts that V. vulnificus inhabits, it has gained the ability to use many different iron sources, including the use of exogenous siderophores, i.e., siderophores produced by other bacterial and fungal species (2, 4). In order to assess which siderophores V. vulnificus can use, and specifically which TonB system is responsible for their uptake, we performed bioassays. Table 3 shows which siderophores can be used by V. vulnificus and, specifically, which TonB system is utilized. Vibriobactin, a siderophore produced by V. cholerae (18), was able to be used by V. vulnificus through both the TonB1 and TtpC2-TonB2 systems. Single mutants either in the TonB1, TtpC2-TonB2, or TtpC3-TonB3 system were still capable of using vibriobactin. Only when a double mutant in tonB1 with either tonB2 or ttpC2 was used could vibriobactin no longer be taken into the cell. Triple mutants in all three TonB systems, as well as quadruple mutants that were deleted for vulnibactin production, were also unable to use vibriobactin. This indicates that vulnibactin production and transport do not affect the outcome of vibriobactin uptake. Complementation of a quadruple mutant with tonB1, ttpC2, or tonB2 restored the uptake of exogenous vibriobactin (Table 3). Similar results were seen for the siderophores vibrioferrin, enterobactin, and ferrioxamine produced by V. parahaemolyticus, E. coli, and Streptomyces species (14, 41, 52, 54). This is the first reported case of V. vulnificus utilizing both vibriobactin and enterobactin. Previous studies have shown that ferrioxamine can be utilized by V. vulnificus (4); however, our results are the first report indicating which TonB system is used for its uptake. We also show that aerobactin can be taken up by V. vulnificus (Table 3). Unlike the other siderophores tested, ttpC2 or tonB2 single mutants resulted in the loss of aerobactin uptake, indicating that aerobactin can only be transported using the energy provided by the TtpC2-TonB2 system and not by the TonB1 or TtpC3-TonB3 system.
Table 3.
TtpC2 is an essential component of the TtpC2-TonB2 system in mediating the uptake of exogenous siderophores in V. vulnificus
| V. vulnificus strain or genotype | Growth on indicated iron sourcea |
||||||
|---|---|---|---|---|---|---|---|
| FAC | Vibriobactin | Vibrioferrin | Ferrioxamine | Aerobactin | Enterobactin | Anguibactin | |
| WT | + | + | + | + | + | + | − |
| ΔtonB1 | + | + | + | + | + | + | − |
| ΔtonB2 | + | + | + | + | − | + | − |
| ΔttpC2 | + | + | + | + | − | + | − |
| ΔtonB3 | + | + | + | + | + | + | − |
| ΔttpC3 | + | + | + | + | + | + | − |
| ΔtonB1 ΔtonB2 | + | − | − | − | − | − | − |
| ΔtonB1 ΔttpC2 | + | − | − | − | − | − | − |
| ΔtonB1 ΔtonB3 | + | + | + | + | + | + | − |
| ΔtonB1 ΔttpC3 | + | + | + | + | + | + | − |
| ΔtonB2 ΔtonB3 | + | + | + | + | − | + | − |
| ΔtonB1 ΔtonB2 ΔtonB3 | + | − | − | − | − | − | − |
| ΔtonB1 ΔttpC2 ΔttpC3 | + | − | − | − | − | − | − |
| ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB/pMMB208 | + | − | − | − | − | − | − |
| ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB/ptonB1 | + | + | + | + | − | + | − |
| ΔtonB1 ΔtonB2 ΔttpC3 ΔvenB/ptonB2 | + | + | + | + | + | + | − |
| ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB/pttpC2(Vvul) | + | + | + | + | + | + | − |
Growth was determined by the presence of a halo (+) or lack thereof (−) around the iron source indicated. Two microliters of each iron source was spotted on the surface of the plates in the following concentrations: FAC, 500 μg/ml; vibriobactin, 1.0 mg/ml; anguibactin, 1.0 mg/ml; aerobactin, 1.0 mg/ml; enterobactin, 1.0 mg/ml; and ferrioxamine, 1.0 mg/ml. Wild-type V. parahaemolyticus producing vibrioferrin was streaked onto the plate, and the halo of growth was monitored around the streak.
Although bioassays are a highly sensitive method for detecting different iron sources, they are only capable of giving an endpoint reading. To determine if the assorted V. vulnificus mutant strains were capable of operating in iron transport, the kinetics of iron uptake using radioactive [55Fe]ferrioxamine was evaluated. Vulnibactin is a catecholate siderophore, whereas ferrioxamine possesses hydroxamate groups for binding the iron atom. In our study, we have been unable to perform radioactive 55Fe transport assays with catecholate siderophores, i.e., enterobactin and vibriobactin in members of the Vibrionaceae. For this reason we used [55Fe]ferrioxamine in our uptake studies.
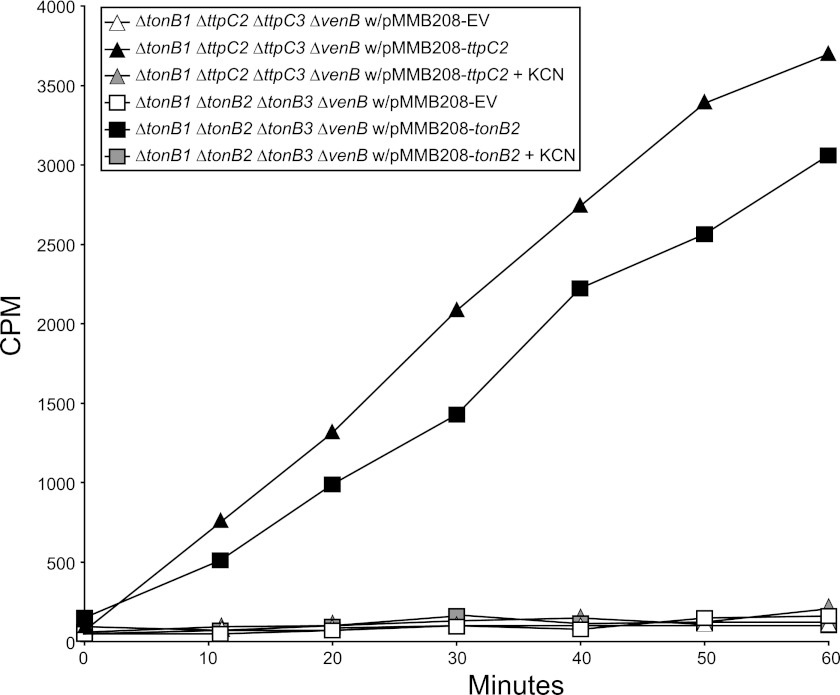
The V. vulnificus ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB and ΔtonB1 ΔtonB2 ΔtonB3 ΔvenB mutants were each evaluated with and without complementing plasmids expressing TtpC2 and TonB2, respectively. Cultures were grown in minimal media containing nitrilotriacetate (NTA) to remove any excess iron, and then [55Fe]ferrioxamine was added. Samples of the cells were removed at 10-min intervals, and the amount of iron taken into the cell was determined by scintillation counting (Fig. 3). When either of these strains was complemented with an empty vector, we saw no uptake of [55Fe]ferrioxamine (Fig. 3, open symbols). When these strains were complemented with the missing component of the TtpC2-TonB2 system, both strains were capable of bringing [55Fe]ferrioxamine into the cell that was mediated by the TtpC2-TonB2 system (Fig. 3, black symbols). Additionally, a duplicate set of complemented strains was treated with potassium cyanide (KCN) to stop cellular respiration and cease the production of the PMF. When KCN was added, all [55Fe]ferrioxamine uptake was prevented (Fig. 3, gray symbols), indicating a necessity to have the PMF generated. These results demonstrate that TtpC2 is essential in the TtpC2-TonB2 system in the mediation of the uptake of [55Fe]ferrioxamine and that the generation of the PMF is necessary.
Fig 3.
TtpC2 is essential for a functional TtpC2-TonB2 system in the uptake of radioactive [55Fe]ferrioxamine. Ferrioxamine uptake assays were performed as described in Materials and Methods. The strains with the following genotypes were used: ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB with pMMB208 (empty vector), pMMB208-ttpC2, and pMMB208-ttpC2 + KCN; ΔtonB1 ΔtonB2 ΔtonB3 ΔvenB with pMMB208 (empty vector), pMMB208-tonB2, and pMMB208-tonB2 + KCN. Samples were removed at the times indicated, and the amount of [55Fe]ferrioxamine brought into the cells was determined by scintillation counting. This is a representative experiment that was done in triplicate.
Complementation of a V. vulnificus ΔttpC2 mutant can be accomplished by using other TtpC2 proteins from members of the Vibrionaceae.
We have previously demonstrated that the TonB2 protein of V. anguillarum can be substituted in a ΔtonB2 strain of V. cholerae to restore the function of the TtpC2-TonB2 system (48). To initiate experiments to determine the role of TtpC2 in the TtpC2-TonB2 system, TtpC2 proteins from other Vibrio species were substituted into a V. vulnificus ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB mutant. Complementing plasmids were moved into this strain, and bioassays were utilized to determine which siderophores V. vulnificus was able to use. The results of this study are shown in Table 4. The complementing TtpC2 proteins from V. cholerae, V. parahaemolyticus, V. alginolyticus, and V. anguillarum yielded the same results as when complementing with V. vulnificus TtpC2, demonstrating that siderophores previously utilized by the V. vulnificus TtpC2-TonB2 system can still be used when TtpC2 proteins from other Vibrio species are substituted. Both anguibactin and ferrichrome were still unable to be utilized by V. vulnificus, indicating that V. vulnificus most likely does not contain an OM receptor for these siderophores.
Table 4.
Complementation of ttpC2 from other Vibrio species restores the TtpC2-TonB2-mediated uptake of various iron sourcesa
| Iron sourceb | TtpC from other Vibrio species containing the indicated complementing plasmid |
|||||
|---|---|---|---|---|---|---|
| pMMB208 | pttpC2(Vvul) | pttpC2(Vcho) | pttpC2(Vpar) | pttpC2(Valg) | pttpC2(Vang) | |
| FAC | + | + | + | + | + | + |
| Vulnibactin | − | + | + | + | + | + |
| Hydroxamate | − | + | + | + | + | + |
| Vibriobactin | − | + | + | + | + | + |
| Vibrioferrin | − | + | + | + | + | + |
| Aerobactin | − | + | + | + | + | + |
| Anguibactin | − | − | − | − | − | − |
| Enterobactinc | − | + | + | + | + | + |
| Ferrichrome | − | − | − | − | − | − |
| Ferrioxamine | − | + | + | + | + | + |
The embedded strain VSRK283 (ΔtonB1 ΔttpC2 ΔttpC3 ΔvenB) contained the complementing plasmid pMMB208 expressing TtpC2 from the Vibrio species containing the plasmids listed at the top of the table.
Growth was determined by the presence of a halo (+) or lack thereof (−) around the iron source indicated. Two microliters of each iron source was spotted on the surface of the plates in the following concentrations: FAC, 500 μg/ml; vibriobactin, 1.0 mg/ml; aerobactin, 1.0 mg/ml; anguibactin, 1.0 mg/ml; enterobactin, 1.0 mg/ml; ferrichrome, 1.0 mg/ml; and ferrioxamine, 1.0 mg/ml. Wild-type V. vulnificus and V. parahaemolyticus producing vulnibactin and vibrioferrin, respectively, were streaked onto the plate, and the halo of growth was monitored around the streak. A V. vulnificus ΔvenB mutant was streaked onto the plate to test for growth around the hydroxamate siderophore.
Growth around enterobactin was only seen when the imbedded strain being tested was grown in a minimal medium CM9 plate and not in a rich medium TSAS plate.
TtpC2 is required for full virulence of V. vulnificus.
To determine if TtpC2 is essential in the TtpC2-TonB2 system for virulence, LD50 studies were conducted in the mouse model. We have previously demonstrated that both the TonB1 and TtpC2-TonB2 systems are essential for full virulence in V. vulnificus. In our previous study, we deleted both tonB1 and tonB2 and saw an increase in LD50 values (2). In this study, we specifically determined if TtpC2 and TtpC3 were needed for virulence. When only the TonB1 system (ΔttpC2 ΔttpC3) or the TtpC2-TonB2 system (ΔtonB1 ΔttpC3) was functional, we found that the LD50 values were approximately the same as WT (Table 5). When both the TonB1 and TtpC2-TonB2 systems were nonfunctional (ΔtonB1 ΔttpC2 ΔttpC3), we saw a one-log increase in the LD50 values. From these experiments, we conclude that not only are both the TonB1 and TtpC2-TonB2 systems needed for full virulence but that ttpC2 within the TtpC2-TonB2 system is essential for this trait.
Table 5.
Virulence of V. vulnificus strains in the mouse model using LD50
| V. vulnificus strain or genotype | LD50a |
|---|---|
| Wild type | 1.47 × 105 |
| ΔtonB1 ΔttpC2 ΔttpC3 | 4.22 × 106 |
| ΔtonB1 ΔttpC3 | 4.22 × 105 |
| ΔttpC2 ΔttpC3 | 3.16 × 105 |
Five animals per dilution were inoculated by intraperitoneal injection with the indicated strains, and mortality was recorded at 48 h. LD50s were calculated as described in Materials and Methods.
DISCUSSION
V. vulnificus can cause severe septic infections in individuals with abnormally large amounts of iron in their blood (19, 23). A high concentration of iron has been shown to be a key virulence factor in many bacterial infections, including V. vulnificus (12, 53). With the production of two siderophores, vulnibactin and a hydroxamate-type compound (39, 47), V. vulnificus is poised to be a serious human pathogen (47). One of the methods used by bacteria to internalize iron is through the use of siderophores whose complexes with ferric iron can be internalized by first interacting with an OM receptor protein. Since there is no energy in the OM, the TonB systems are responsible for harnessing the energy generated by the PMF in the IM and transferring it to the cognate OM receptors, thereby activating their ability to bind iron-carrying siderophores (7, 12, 43). Unlike the single well-studied TonB system in E. coli, V. vulnificus contains three TonB systems (2, 27).
Initial in silico work led us to discover that V. vulnificus contains two ttpC genes, one associated with the TtpC2-TonB2 system and another with what we have termed the TtpC3-TonB3 system (2, 27). In both V. cholerae and V. anguillarum, TtpC2 has been shown to be essential for the function of the TtpC2-TonB2 system (26, 48). In this study, we have demonstrated that TtpC2 is an essential component of the TtpC2-TonB2 system in V. vulnificus, whereas TtpC3 is nonfunctional for this system. Although the TtpC2-TonB2 and TtpC3-TonB3 systems have the same four genes and similar surrounding DNA, the two systems are remarkable different. The TtpC2-TonB2 system is present in all members of the Vibrionaceae, whereas the TtpC3-TonB3 system is only present in a small subset of Vibrio species as well as a limited number of other bacteria (26, 27). Here we show that on average the TtpC2-TonB2 system shares around 50% homology with TtpC2-TonB2 systems in other vibrios. In contrast, the TtpC3-TonB3 system is much more highly conserved with an average identity close to 90%. When we compared the TtpC2-TonB2 system to the TtpC3-TonB3 system, we found that their amino acid sequence identity was extremely low (only about 30%), indicating that they are extremely different at the amino acid level, although they appear similar when comparing gene length, grouping, and orientation.
In this study, the essentiality of TtpC for iron transport in V. vulnificus was determined using nutritional bioassays, CAS assays, and [55Fe]ferrioxamine uptake assays. Our results indicate that either the TonB1 system or the TtpC2-TonB2 system can be used to energize the endogenous siderophore vulnibactin for iron transport in V. vulnificus. In addition, we show that TtpC3 cannot complement a ΔttpC2 mutation to create a functional TtpC2-TonB2 system. Our previous studies have shown that the TtpC3-TonB3 system of V. vulnificus cannot be used for the uptake of vulnibactin under the same conditions utilized in this study and that the ttpC3-tonB3 operon is only expressed in serum (2). Thus, TtpC3 is very different from TtpC2 and cannot substitute when TtpC2 is lost. Our results also show that strains deficient in TtpC2 or TonB2 but complemented with either TtpC2 or TonB2 had the same kinetics for the uptake of [55Fe]ferrioxamine, indicating that both TtpC2 and TonB2 are essential components of the TtpC2-TonB2 system. Furthermore, we have replaced V. vulnificus TtpC2 with TtpC proteins from other Vibrio species and showed that they can function in the uptake of both endogenous as well as exogenous siderophores.
In addition to vulnibactin, V. vulnificus produces a recently identified hydroxamate-type siderophore (47). This hydroxamate siderophore was tested on bioassay plates to determine which TonB system is used for its uptake. Similar to vulnibactin, the hydroxamate siderophore can be taken into the cell using either the TonB1 system or the TtpC2-TonB2 system. This is the first report indicating which TonB systems can be utilized for growth when the hydroxamate siderophore is used. Another important conclusion from these experiments is that vulnibactin is the dominant siderophore produced and/or has a much higher affinity for iron than the hydroxamate siderophore under the conditions we tested. The hydroxamate siderophore may be the dominant siderophore produced under different environmental and growth conditions. Changes in temperature, salt concentrations, or even growth in hosts (eels or humans) might increase the production of this siderophore.
Our results in this study using exogenously produced siderophores have not only confirmed the essentiality of ttpC2 in the TtpC2-TonB2 system but have also uncovered siderophores that were previously unknown to be used by V. vulnificus. It has previously been shown that V. vulnificus can grow using the hydroxamate siderophores ferrioxamine and aerobactin (4, 51). It was unknown, however, which TonB system was used for their uptake. Here we have shown that both the TonB1 and TtpC2-TonB2 systems can be utilized for ferrioxamine uptake. In addition, we have shown that aerobactin can only be used by V. vulnificus through the TtpC2-TonB2 system. This specificity for the TtpC2-TonB2 system is also found in both V. cholerae and V. anguillarum for the transport of enterobactin and anguibactin, respectively (48). Anguibactin has been shown to be the most important virulence factor for V. anguillarum, further solidifying the importance of the TtpC2-TonB2 system (10). In a human host, the ability to internalize aerobactin could become crucial in a V. vulnificus infection since this siderophore is produced by both pathogenic and nonpathogenic E. coli strains in the intestine and could result in a single or combined bacterial infection (13, 38). In this study, we have also demonstrated for the first time that both vibrioferrin and enterobactin, produced by V. parahaemolyticus and E. coli, respectively, can be used by V. vulnificus. A previous report had claimed that vibrioferrin was unable to be utilized by V. vulnificus (4). In our study, this is not the case; in fact, vibrioferrin can be used by either the TonB1 system or the TtpC2-TonB2 system. Surprisingly, enterobactin was only utilized by V. vulnificus when bioassays were conducted in minimal media with minimal EDDA compared to TSAS media with excess EDDA. In V. vulnificus, enterobactin can be used by either the TonB1 system or the TtpC2-TonB2 system. In contrast, enterobactin can only be brought into cells using the TtpC2-TonB2 system in both V. cholerae and V. anguillarum.
We still do not know whether TtpC2 or TonB2 makes contact with the OM receptor. Our previous study in V. anguillarum using heteronuclear single quantum correlation (HSQC) approaches, showed that TonB2 cannot interact with the amino terminal end of the FatA receptor for anguibactin, suggesting that it does not interact with this receptor (34). It is possible that both TtpC2 as well as TonB2 are needed in order to make contact with the OM receptor. Experiments designed to address this possibility are under way.
Previous studies have shown that the TonB systems are important in virulence for V. vulnificus (2). Here we have shown that either the TonB1 system or the TtpC2-TonB2 system is vital for a fully virulent phenotype in a mouse model. A log difference is seen when both the TonB1 and TtpC2-TonB2 systems are deleted compared to WT. LD50 values are comparable when testing the WT strain against strains that have only one of the two TonB systems still functional. In conclusion, both the TtpC2 and TonB2 proteins are essential for iron transport of many siderophores by V. vulnificus and both the TonB1 and the TtpC2-TonB2 systems are essential for the pathogenesis of the disease caused by this human pathogen.
ACKNOWLEDGMENTS
This work was supported by grant AI19018 from the National Institutes of Health to J.H.C. Ryan J. Kustusch was supported by a postdoctoral fellowship training grant and Carole J. Kuehl by a graduate student training grant, both from NIH training grant 5T32-AI 007471-13 in Molecular Microbiology and Immunology.
Footnotes
Published ahead of print 13 April 2012
REFERENCES
- 1. Actis LA, Tolmasky ME, Crosa JH. 1999. Bacterial plasmids: replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Front. Biosci. 4:D43–D62 [DOI] [PubMed] [Google Scholar]
- 2. Alice AF, Naka H, Crosa JH. 2008. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect. Immun. 76:4019–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aso H, Miyoshi S, Nakao H, Okamoto K, Yamamoto S. 2002. Induction of an outer membrane protein of 78 kDa in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. FEMS Microbiol. Lett. 212:65–70 [DOI] [PubMed] [Google Scholar]
- 5. Bassford PJ, Jr, Bradbeer C, Kadner RJ, Schnaitman CA. 1976. Transport of vitamin B12 in tonB mutants of Escherichia coli. J. Bacteriol. 128:242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradbeer C, Woodrow ML. 1976. Transport of vitamin B12 in Escherichia coli: energy dependence. J. Bacteriol. 128:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295–307 [DOI] [PubMed] [Google Scholar]
- 8. Braun V, et al. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bullen JJ, Rogers HJ, Griffiths E. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1–35 [DOI] [PubMed] [Google Scholar]
- 10. Crosa JH. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature 284:566–568 [DOI] [PubMed] [Google Scholar]
- 11. Crosa JH. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crosa JH, Mey AR, Payne SM. 2004. Iron transport in bacteria. ASM Press, Washington, DC [Google Scholar]
- 13. Der Vartanian M, et al. 1992. Role of aerobactin in systemic spread of an opportunistic strain of Escherichia coli from the intestinal tract of gnotobiotic lambs. Infect. Immun. 60:2800–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ernst JF, Bennett RL, Rothfield LI. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Germon P, Ray MC, Vianney A, Lazzaroni JC. 2001. Energy-dependent conformational change in the TolA protein of Escherichia coli involves its N-terminal domain, TolQ, and TolR. J. Bacteriol. 183:4110–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldberg MB, et al. 1992. Characterization of a Vibrio cholerae virulence factor homologous to the family of TonB-dependent proteins. Mol. Microbiol. 6:2407–2418 [DOI] [PubMed] [Google Scholar]
- 18. Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383–385 [PubMed] [Google Scholar]
- 19. Gulig PA, Bourdage KL, Starks AM. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118–131 [PubMed] [Google Scholar]
- 20. Henderson DP, Payne SM. 1994. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Higgs PI, Myers PS, Postle K. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Higuchi R, Krummel B, Saiki RK. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hor LI, Chang YK, Chang CC, Lei HY, Ou JT. 2000. Mechanism of high susceptibility of iron-overloaded mouse to Vibrio vulnificus infection. Microbiol. Immunol. 44:871–878 [DOI] [PubMed] [Google Scholar]
- 24. Köster WL, Actis LA, Waldbeser LS, Tolmasky ME, Crosa JH. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829–23833 [PubMed] [Google Scholar]
- 25. Kuehl CJ, Crosa JH. 2009. Molecular and genetic characterization of the TonB2-cluster TtpC protein in pathogenic vibrios. Biometals 22:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuehl CJ, Crosa JH. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol. 5:1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kustusch RJ, Kuehl CJ, Crosa JH. 2011. Power plays: iron transport and energy transduction in pathogenic vibrios. Biometals 24:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsen RA, et al. 2007. His(20) provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsen RA, Letain TE, Postle K. 2003. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol. Microbiol. 49:211–218 [DOI] [PubMed] [Google Scholar]
- 30. Larsen RA, Postle K. 2001. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276:8111–8117 [DOI] [PubMed] [Google Scholar]
- 31. Larsen RA, Thomas MG, Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809–1824 [DOI] [PubMed] [Google Scholar]
- 32. Larsen RA, Wood GE, Postle K. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943–953 [DOI] [PubMed] [Google Scholar]
- 33. Linkous DA, Oliver JD. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207–214 [DOI] [PubMed] [Google Scholar]
- 34. López CS, Peacock RS, Crosa JH, Vogel HJ. 2009. Molecular characterization of the TonB2 protein from the fish pathogen Vibrio anguillarum. Biochem. J. 418:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morales VM, Bäckman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47 [DOI] [PubMed] [Google Scholar]
- 37. Morris JGJ. 1988. Vibrio vulnificus—a new monster of the deep? Ann. Intern. Med. 109:261–263 [DOI] [PubMed] [Google Scholar]
- 38. Nowrouzian F, Wold AE, Adlerberth I. 2001. P fimbriae and aerobactin as intestinal colonization factors for Escherichia coli in Pakistani infants. Epidemiol. Infect. 126:19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okujo N, et al. 1994. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 7:109–116 [DOI] [PubMed] [Google Scholar]
- 40. Ollis AA, Manning M, Held KG, Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pollack JR, Neilands JB. 1970. Enterobactin, an iron transport compound from Salmonella typhimurium, Biochem. Biophys. Res. Commun. 38:989. [DOI] [PubMed] [Google Scholar]
- 42. Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol. 422:245–269 [DOI] [PubMed] [Google Scholar]
- 43. Postle K, Larsen RA. 2007. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20:453–465 [DOI] [PubMed] [Google Scholar]
- 44. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hygiene 27:493–497 [Google Scholar]
- 45. Senanayake SD, Brian DA. 1995. Precise large deletions by the PCR-based overlap extension method. Mol. Biotechnol. 4:13–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 47. Simpson LM, Oliver JD. 1983. Siderophore production by Vibrio vulnificus. Infect. Immun. 41:644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stork M, Otto BR, Crosa JH. 2007. A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 189:1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strom MS, Paranjpye RN. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177–188 [DOI] [PubMed] [Google Scholar]
- 50. Swayne C, Postle K. 2011. Taking the Escherichia coli TonB transmembrane domain “offline”? Nonprotonatable Asn substitutes fully for TonB His20. J. Bacteriol. 193:3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanabe T, et al. 2005. A novel aerobactin utilization cluster in Vibrio vulnificus with a gene involved in the transcription regulation of the iutA homologue. Microbiol. Immunol. 49:823–834 [DOI] [PubMed] [Google Scholar]
- 52. Wang Q, Liu Q, Cao X, Yang M, Zhang Y. 2008. Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: their roles in iron utilization and virulence. Arch. Microbiol. 190:595–603 [DOI] [PubMed] [Google Scholar]
- 53. Wright AC, Morris JGJ. 1991. The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect. Immun. 59:192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamamoto S, Okujo N, Yoshida T, Matsuura S, Shinoda S. 1994. Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem. 115:868–874 [DOI] [PubMed] [Google Scholar]