Abstract
The halophilic archaeon Haloferax volcanii utilizes fructose as a sole carbon and energy source. Genes and enzymes involved in fructose uptake and degradation were identified by transcriptional analyses, deletion mutant experiments, and enzyme characterization. During growth on fructose, the gene cluster HVO_1495 to HVO_1499, encoding homologs of the five bacterial phosphotransferase system (PTS) components enzyme IIB (EIIB), enzyme I (EI), histidine protein (HPr), EIIA, and EIIC, was highly upregulated as a cotranscript. The in-frame deletion of HVO_1499, designated ptfC (ptf stands for phosphotransferase system for fructose) and encoding the putative fructose-specific membrane component EIIC, resulted in a loss of growth on fructose, which could be recovered by complementation in trans. Transcripts of HVO_1500 (pfkB) and HVO_1494 (fba), encoding putative fructose-1-phosphate kinase (1-PFK) and fructose-1,6-bisphosphate aldolase (FBA), respectively, as well as 1-PFK and FBA activities were specifically upregulated in fructose-grown cells. pfkB and fba knockout mutants did not grow on fructose, whereas growth on glucose was not inhibited, indicating the functional involvement of both enzymes in fructose catabolism. Recombinant 1-PFK and FBA obtained after homologous overexpression were characterized as having kinetic properties indicative of functional 1-PFK and a class II type FBA. From these data, we conclude that fructose uptake in H. volcanii involves a fructose-specific PTS generating fructose-1-phosphate, which is further converted via fructose-1,6-bisphosphate to triose phosphates by 1-PFK and FBA. This is the first report of the functional involvement of a bacterial-like PTS and of class II FBA in the sugar metabolism of archaea.
INTRODUCTION
Various halophilic archaea, including Haloarcula marismortui and Haloferax volcanii, have been reported to utilize fructose as carbon and energy sources (29, 34, 46). The pathway of fructose degradation has been studied so far mainly in the Haloarcula species H. vallismortis and H. marismortui (6, 7, 29). On the basis of enzyme analyses, a modified version of the Embden-Meyerhof (EM) pathway has been proposed, involving fructose phosphorylation via ketohexokinase to fructose-1-phosphate, which is further phosphorylated to fructose-1,6-bisphosphate (FBP) by fructose-1-phosphate kinase (1-PFK). FBP is subsequently cleaved by FBP aldolase (FBA) to dihydroxyacetone phosphate and glyceraldehyde-3-phosphate, which are degraded to pyruvate following classical enzymes of the EM pathway. In vivo evidence for the operation of an EM pathway in fructose degradation was demonstrated in H. marismortui by labeling experiments with [13C]fructose using growing cultures (29). With the same labeling techniques, glucose degradation in H. marismortui was shown to be degraded in vivo via an Entner-Doudoroff (ED) type pathway (29), which is in accordance with the proposed semiphosphorylated ED pathway for glucose degradation in haloarchaea (43).
Although several enzymes of the proposed modified EM pathway in haloarchaea, ketohexokinase, 1-PFK, and class I and class II type FBA, have been purified (21, 31, 36, 37), the genes encoding these enzymes have not been identified so far, and their functional involvement in fructose catabolism has not been demonstrated. Recently, the fructose-specific upregulation of a gene (HVO_1500) encoding putative 1-PFK has been reported for H. volcanii, and the role of a transcription regulator (GlpR) in fructose catabolism has been analyzed (39). So far, fructose transport in haloarchaea and other archaeal species has not been studied in detail. For H. volcanii a secondary Na+/fructose symport system was proposed on the basis of fructose uptake experiments in cell suspensions (46).
In contrast to that in archaea, fructose uptake in the bacterial domain is well studied and usually involves the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS), which phosphorylates fructose during transport to fructose-1-phosphate. In general, phosphotransferase systems are composed of five components, two cytoplasmic proteins, protein kinase enzyme I (EI) and histidine protein (HPr), and the substrate-specific enzyme II (EII). EII consists of two soluble components (EIIA and EIIB) and a transmembrane component, EIIC, that carry out both the transport and concomitant phosphorylation of the substrate across the membrane. The transfer of the phosphoryl group from PEP to sugar proceeds via the transient phosphorylation of EI, HPr, EIIA, and EIIB (for reviews, see references 9, 19, and 33).
So far, PTS-like sugar uptake systems have not been reported in the archaeal domain. In those archaea analyzed, e.g., in the hyperthermophilic Sulfolobus species and Thermococcales, ABC transporters appear to be the predominant sugar transport systems (1, 32).
However, the recent genome sequence analysis of H. volcanii revealed the presence of a gene cluster (HVO_1495 to HVO_1499) encoding homologs of five components of a complete bacterial PTS (EIIB, EI, HPr, EIIA, and EIIC) (26). This putative PTS might be involved in fructose transport in H. volcanii, since genes encoding putative 1-PFK (HVO_1500) and FBA (HVO_1494), i.e., key enzymes of the proposed modified EM pathway, were located adjacent to the PTS gene cluster (Fig. 1A).
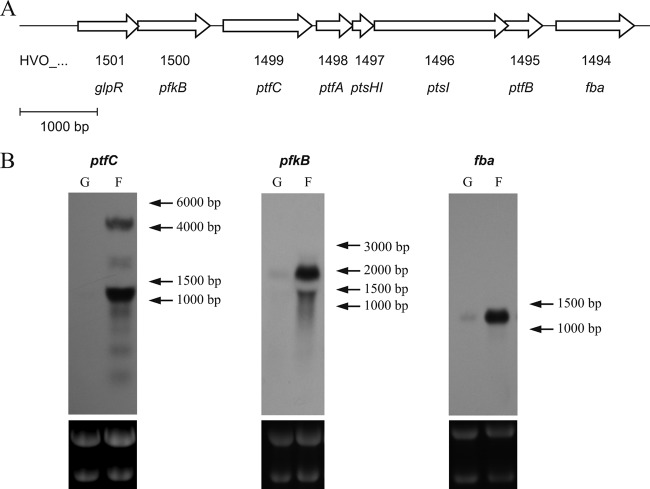
Fig 1.
Genomic arrangement and Northern blot analyses of genes involved in fructose metabolism in H. volcanii. (A) Schematic representation of fructose-specific PTS cluster and neighboring genes. Open reading frames are displayed as open arrows. glpR (HVO_1501) encodes a transcription regulator (39); pfkB (HVO_1500) encodes putative 1-phosphofructokinase (1-PFK) of the PfkB family; the HVO_1499 to HVO_1495 cluster (ptfC, ptfA, ptsH1, ptsI, and ptfB) encodes all five components of the putative fructose-specific PTS (EIIC, EIIA, HPr, EI, and EIIB); fba encodes a putative class II fructose-1,6-bisphosphate aldolase (FBA). The bar indicates a length of 1,000 bp. (B) Northern blot analyses of genes involved in fructose metabolism of H. volcanii. Total RNA was isolated from glucose (G)- or fructose (F)-grown cells and analyzed with specific probes for ptfC (HVO_1499), pfkB (HVO_1500), and fba (HVO_1494). To control equal loading, the 23S and 16S rRNAs were visualized by ethidium bromide staining (lower panels).
In this paper, the functional involvement of this putative PTS and of 1-PFK and FBA in fructose degradation was analyzed in H. volcanii, one of the model organisms of haloarchaea to study, e.g., metabolic pathways and regulation (45). In H. volcanii, protocols to generate in-frame deletion mutants and systems for the homologous overexpression of halophilic proteins are available (2, 4, 12). Transcriptional analyses of putative genes encoding EIIC, 1-PFK, and FBA were performed, and deletion mutants of these genes were prepared and analyzed in growth experiments. Finally, 1-PFK and FBA were characterized after homologous overexpression.
The data indicate that fructose uptake in H. volcanii involves a functional PTS, forming fructose-1-phosphate that is further phosphorylated to FBP by 1-PFK. The cleavage of FBP to glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP) is catalyzed by a class II type FBA. This is the first report of the functional involvement of a bacterial type PTS and of class II FBA in sugar uptake and metabolism in archaea.
MATERIALS AND METHODS
Growth of Haloferax volcanii and preparation of cell extracts.
H. volcanii H26, containing a uracil auxotrophy (ΔpyrE2), was used for the construction of in-frame deletion mutants by using the so-called pop-in/pop-out strategy (4, 12, 23). Wild-type and mutant strains were grown aerobically at 42°C in a modified complex medium (20) using 1% Casamino Acids instead of yeast extract and tryptone. The medium was supplemented with trace elements (20). For pop-out selection, the medium was supplemented with 5-fluoroorotic acid (50 μg/ml) and uracil (30 μg/ml). Instead of using complex medium, a synthetic medium (20) containing 100 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.2) and 25 mM glucose as the sole carbon source was used for the pop-out selection of HVO_1494.
Growth experiments for the phenotypic analysis of mutants and complementation experiments were carried out in minimal medium, with slight modifications, as described previously (49). This medium was prepared without histidine and molybdate and was supplemented with 0.8 μg/ml thiamine and 0.1 μg/ml biotine. As carbon sources, d-glucose, d-fructose, d-xylose (each 25 mM), 40 mM acetate, and 1% Casamino Acids were used. If necessary, uracil (ΔpyrE2) was added to the medium. The organisms were incubated aerobically at 42°C, and growth was monitored by measuring the absorbance at 600 nm during the time period. The determination of fructose in the supernatant of fructose-grown H. volcanii was carried out as described previously (29).
H. volcanii H1209 was used for the homologous overexpression of proteins (3). H1209 was transformed with pTA963 carrying HVO_1494 (coding for FBA) or HVO_1500 (coding for 1-PFK) under the control of the inducible tryptophan promoter. Overexpression was performed in complex medium containing the respective compounds according to Allers et al. (3).
Enzymatic activity was measured in crude extracts of cells grown to log phase on fructose or glucose in minimal medium (optical density at 600 nm [OD600], 1.2). Cells were centrifuged at 10,000 × g for 5 min at 4°C, and pellets were resuspended in 50 mM Tris-HCl, pH 8.0, and 2 M KCl. After the disruption of cells by sonication and centrifugation for 30 min at 10,000 × g at 4°C, the supernatant was used for the measurement of enzyme activities. The protein concentration was determined by the Bradford method with bovine serum albumin as the standard (13).
RNA isolation, reverse transcriptase PCR (RT-PCR), and Northern blotting.
Total RNA was isolated by an improved method of Chomczynski and Sacchi (16) using 1-bromo-3-chloropropane for phase separation (15). Two to 5 ml of mid-exponential-phase H. volcanii cultures (OD600, 1.2), grown on either glucose or fructose, was centrifuged at 10,000 × g for 2 min, followed by resuspension in 1 ml Tri reagent (Sigma-Aldrich) with further treatment as specified by the manufacturer.
For RT-PCR, the RNA was first treated with DNase I as described by the producer (Fermentas). DNA-free total RNA was transcribed into cDNA with the GoScript reverse transcription system (Promega) as described by the manufacturer. Each reaction mixture contained 400 ng of DNA-free RNA as the template, 0.025 μg/μl random hexamers (Fermentas), and 2 mM MgCl2. As a control, reverse transcriptase was excluded. The ensuing PCRs were performed with Taq DNA polymerase (Peqlab) and 4 ng of cDNA as the template. The PCR was carried out with an annealing temperature of 64°C for 25 cycles and a 1-min synthesis time. As a constitutive expression control, ribosomal protein L10 gene (ribL) was used (14). Primer sequences for RT-PCR are described in Table S1 in the supplemental material.
For Northern blotting, the total RNA (15 μg per lane) was denatured for 10 min at 65°C and separated by electrophoresis (100 V, 60 min) on a 1.2% formaldehyde agarose gel in 1× MOPS buffer (20 mM MOPS, pH 7.0, 5 mM sodium acetate, 2 mM EDTA, and 0.75% formaldehyde). A RiboRuler High Range RNA ladder (Fermentas) was used for molecular size determination. After electrophoresis, the RNA was transferred to a positively charged nylon membrane (Roche Diagnostics) by vacuum blotting using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0). RNA was fixed to the membrane by incubation at 120°C for 30 min. Probes for the specific detection of transcript were generated by PCR using digoxigenin (DIG)-11-dUTP (Roche Diagnostics) and Taq DNA polymerase (Peqlab) according to the protocols of the suppliers. Primer sequences for amplification are summarized in Table S1 in the supplemental material. After denaturing at 100°C for 5 min, each probe was incubated with the respective membrane in DIG Easy Hyb (Roche Diagnostics) at 50°C for 16 h. The subsequent chemiluminescent detection of the labeled DNA fragments was carried out as described by the manufacturer (Roche Diagnostics).
Generation of in-frame deletion mutants of H. volcanii.
Knockout mutants of HVO_1499 (ptfC), HVO_1500 (pfkB), and HVO_1494 (fba) were constructed by the pop-in/pop-out strategy. In each case, two PCR fragments of about 600 bp, containing the up- and downstream sequences of the corresponding gene, were amplified using Pwo DNA polymerase (Peqlab). Both fragments were fused by PCR, generating an in-frame deletion fragment that was ligated into pTA131 (4) followed by transformation in E. coli XL1-Blue MRF′. After plasmid preparation from Escherichia coli and sequencing, each plasmid was transformed in H. volcanii H26 (17, 18). Growth in uracil-free medium was used to select for clones that had integrated the vector into their genome via homologous recombination at the indicated locus (pop-in). Clones that experienced a second homologous recombination (pop-out) were selected in medium containing uracil and 5-fluoroorotic acid. Successful deletion was verified by Southern blot analysis. Therefore, DNA was isolated with the Wizard DNA isolation kit (Promega) and digested with restriction endonucleases (see Table S2 in the supplemental material). The further procedure was performed as described previously (28). The primers that were used for the generation of deletion constructs are listed in Table S2 in the supplemental material.
Cloning, complementation, and homologous overexpression.
HVO_1499 (ptfC), HVO_1500 (pfkB), and HVO_1494 (fba) were amplified by PCR using Pwo polymerase (Peqlab) and genomic DNA of H. volcanii H26, followed by restriction enzyme cleavage and ligation in pTA963 according to the cloning strategy of Allers et al. (3). For primer sequences and restriction enzymes used, see Table S1 in the supplemental material. The ligated plasmids were transformed and multiplied in E. coli XL1-Blue MRF′, followed by purification and sequencing.
For complementation experiments, the deletion strains ΔptfC, ΔpfkB, and Δfba were transformed with the plasmids containing the respective wild-type genes under the control of the tryptophan promoter. As a negative control, each mutant was transformed with pTA963 without an insert. Growth experiments with the complemented and control strains were performed in minimal medium with fructose, glucose, xylose, acetate, or Casamino Acids. Up to 200 μM tryptophan was added to fully complement the deletion mutants.
The homologous overexpression for protein characterization of 1-PFK and fructose-1,6-bisphosphate aldolase (FBA) was performed in H. volcanii H1209 transformed with the respective plasmid according to Allers et al. (3). Expression was induced by 3 mM tryptophan (3) followed by further growth for 16 h at 42°C. Cells were harvested by centrifugation at 15,000 × g for 20 min at 10°C. Each pellet was resuspended in 0.1 M Tris-HCl, pH 8.0, 2 M KCl, 5 mM imidazole and disrupted in a French pressure cell at 14,000 lb/in2. The cell lysate was centrifuged at 100,000 × g at 4°C, and the supernatant was applied to a 1 ml nickel-nitrilotriacetic acid (Ni-NTA) Superflow cartridge (Qiagen) that was equilibrated in the same buffer. After a washing step, protein was eluted by stepwise increasing the imidazole concentration up to 500 mM. Eluted protein (1 ml) was applied to a HighLoad 16/60 Superdex 200 preparation-grade column (GE Healthcare) that was equilibrated with 50 mM Tris-HCl, pH 8.0, containing 2 M KCl. Protein was eluted by an isocratic flow at 1 ml/min. The protein concentration was determined by the Bradford method (13) with bovine serum albumin as the standard, and the purity of the protein was analyzed by SDS-PAGE.
Characterization of 1-PFK and FBA.
Enzyme activities were measured spectrophotometrically in a total volume of 200 μl at 42°C and at 340 nm. We ensured that the auxiliary enzymes in the coupled assays were not rate limiting. One unit (U) is defined as the conversion of 1 μmol substrate per min.
1-PFK was measured by coupling the ATP-dependent formation of fructose-1,6-bisphosphate (FBP) to NADH oxidation via pyruvate kinase (PK) and lactate dehydrogenase (LDH) as described previously (25). In detail, the standard assay mixture contained 100 mM Tris-HCl, pH 7.5, 7 mM MgCl2, 0.3 mM NADH, 1 mM phosphoenolpyruvate, 2 mM ATP, 0.5 M KCl, 0.1 to 20 mM fructose-1-phosphate or fructose-6-phosphate, 1.65 U LDH, 2 U PK, and purified protein (0.05 to 0.6 μg). The apparent Km for ATP was measured at concentrations from 0.05 to 2 mM in the presence of 2 mM fructose-1-phosphate (F1P). The pH dependence of the enzyme was measured between pH 5.5 and 9 with standard assay conditions containing bis-Tris (pH 5.5 to 7.5) and Tris-HCl (pH 7.5 to 9), each at 20 mM. The effect of potassium on 1-PFK was carried out in a standard assay with KCl concentrations of up to 3 M.
FBA activity was determined by coupling the FBP-dependent formation of dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) to the oxidation of NADH via triosephosphate isomerase (TIM) and glycerol-3-phosphate dehydrogenase (GDH) as described previously (24). The standard assay mixture contained 100 mM Tris-HCl, pH 7.5, 0.3 mM NADH, 1 M KCl, 0.75 mM MnCl2, 1 mM cysteine, 0.1 to 20 mM FBP, 4.7 U GDH, 14 U TIM, and purified protein (0.01 to 0.4 μg). The effect of pH was determined at pH values ranging from 5.5 to 9 using the same buffers as those described for 1-PFK. The influence of potassium on enzymatic activity was measured from 0 to 3 M. To analyze the dependence on divalent cations, 2 mM EDTA was added to the standard assay. The influence of different divalent cations on the activity of FBA was determined in a discontinuous assay. The assay contained 100 mM Tris-HCl, pH 7.5, 1 M KCl, 0.75 mM ions (Fe2+, Mn2+, or Zn2+), 1 mM cysteine, 7.5 mM FBP, and purified protein. During incubation (0 to 100 s), aliquots were taken and the reaction was stopped by the addition of 80 mM EDTA. The amount of GAP formed was determined by coupling the glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-dependent conversion of GAP to 1-arseno-3-phosphoglycerate to the reduction of NAD+ as described previously (10). The assay contained 100 mM Tris-HCl, pH 7.5, 3 mM NAD+, 20 mM potassium arsenate, 14 U TIM, and 8 U GAPDH. The reverse rate, e.g., the formation of FBP from GAP and DHAP, was monitored in a stopped assay. The reaction mixture contained 100 mM Tris-HCl, pH 7.5, 1 M KCl, 0.75 mM MnCl2, 1 mM cysteine, 1 mM GAP, 5 mM DHAP, and purified protein. During incubation (0 to 180 s), aliquots were taken and the reactions were stopped with 80 mM EDTA. The amount of GAP was determined as described above, except that TIM was omitted.
RESULTS
Recent genome analyses of H. volcanii indicate the presence of a cluster of five chromosomal genes, HVO_1495 to HVO_1499 (ptfB, ptsI, ptsH1, ptfA, and ptfC; ptf stands for phosphotransferase system for fructose), encoding homologs of the five bacterial PTS components EIIB, EI, HPr, EIIA, and EIIC (26). Directly upstream and downstream of this cluster, respectively, genes coding for putative fructose-1-phosphate kinase (1-PFK) (HVO_1500 [pfkB]) and fructose-1,6-bisphosphate aldolase (FBA) (HVO_1494 [fba]) were annotated (Fig. 1A). To prove the functional involvement of these putative genes in fructose catabolism, the fructose-specific regulation of transcripts and of enzyme activities were analyzed, followed by growth studies of in-frame deletion mutants. Further, putative 1-PFK and FBA were characterized after homologous overexpression.
The PTS cluster HVO_1495 to HVO_1499 is involved in fructose transport.
H. volcanii was grown on minimal media containing fructose and, for comparative analyses, glucose as carbon and energy sources. The cells grew on fructose (10 mM) with doubling times of about 4 h to an OD600 of 1.5, and during growth fructose was completely consumed (see Fig. S1 in the supplemental material).
Total RNA was prepared from log-phase cells grown on fructose or glucose. The transcriptional regulation of the putative PTS was analyzed by RT-PCR and Northern blotting with a probe specific for HVO_1499 (ptfC), which encodes the putative fructose-specific transmembrane component EIIC. RT-PCR analysis indicates the presence of ptfC transcripts in fructose-grown cells and their absence from glucose-grown cells, indicating the fructose-specific induction of ptfC (see Fig. S2 in the supplemental material). Northern blotting with a ptfC probe revealed a 4,000-bp transcript in fructose-grown cells which was not detectable in glucose-grown cells (Fig. 1B). The transcript size of about 4,000 bp corresponded well to the sum of base pairs calculated for the five genes of the PTS cluster HVO_1495 to HVO_1499 (4,079 bp), indicating that the complete PTS cluster in H. volcanii was expressed as a cotranscript specifically upregulated by fructose. An additional strong transcript signal of about 1,100 bp, corresponding to the size of ptfC, was detected in fructose-grown cells, also suggesting the monocistronic transcription of ptfC.
To prove the functional involvement of the PTS in fructose metabolism, ptfC was deleted in frame using the recently developed pop-in/pop-out method (4, 12, 23). Successful deletion was confirmed by Southern blot analysis (see Fig. S3A in the supplemental material). The ΔptfC mutant did not grow on fructose, whereas growth on glucose was unaffected (Fig. 2A; also see Fig. S4A in the supplemental material). The ability to grow on fructose was recovered by the complementation of the mutant in trans (Fig. 2A). These results indicate that ptfC, encoding the fructose-specific membrane component EIIC, and the entire PTS operon in H. volcanii are functionally involved in fructose uptake as part of fructose catabolism.
Fig 2.
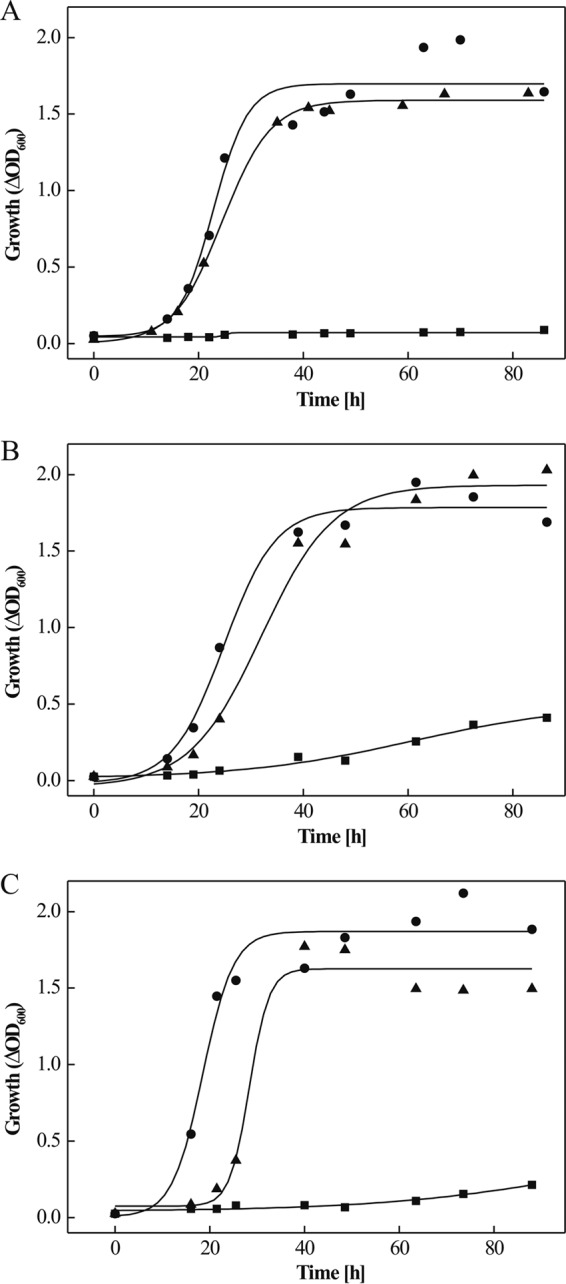
Growth analyses of in-frame deletion mutants of genes involved in fructose degradation in H. volcanii. (A to C) Growth of H. volcanii ΔptfC (A), ΔpfkB (B), and Δfba (C) strains on 25 mM fructose (■) compared to the wild type (•) and complementation strains with functional genes (▲). Precultures for growth experiments were grown in complex medium containing 1% Casamino Acids (A and B) or in synthetic medium containing 25 mM glucose (C). Growth was measured by determining the optical density at 600 nm (OD600).
1-PFK, encoded by pfkB, is involved in fructose catabolism. (i) Transcription analysis and mutant characterization.
pfkB (HVO_1500), encoding putative 1-PFK, is located upstream of the PTS operon (Fig. 1A). With Northern blot analysis using a pfkB-specific probe, a 1,800-bp transcript was detected with RNA from fructose-grown cells, which was present at a significantly lower level in glucose-grown cells, indicating the high upregulation of pfkB by fructose (Fig. 1B). The transcript length of about 1,800 bp indicates the cotranscription of pfkB and HVO_1501, encoding the transcriptional regulator GlpR (39). The cotranscription of pfkB and glpR and upregulation by fructose has already been shown previously by RT-PCR measurements (39). pfkB was deleted in frame, and successful deletion was confirmed by Southern blot analyses (see Fig. S3B in the supplemental material). The ΔpfkB mutant did not grow on fructose, but growth on glucose was not affected (Fig. 2B; also see Fig. S4B in the supplemental material). A fructose-positive growth phenotype was fully recovered by complementation in trans (Fig. 2B). These data indicate that pfkB is essential for fructose catabolism.
(ii) Characterization of 1-PFK.
Cell extracts of H. volcanii grown on fructose showed about 3-fold higher 1-PFK activity (0.1 U/mg) than glucose-grown cells (0.031 U/mg), supporting a specific role in fructose metabolism. To prove that pfkB encodes a functional 1-PFK, the gene was homologously expressed in H. volcanii H1209 using the vector pTA963 (2). The recombinant enzyme was purified as a His tag fusion protein by Ni-NTA affinity chromatography, followed by gel filtration on Superdex. The apparent molecular mass of native enzyme, determined by gel filtration, was about 70 kDa. Denaturing SDS-PAGE (Fig. 3A) revealed only one subunit of about 38 kDa, indicating a dimeric form of native 1-PFK.
Fig 3.
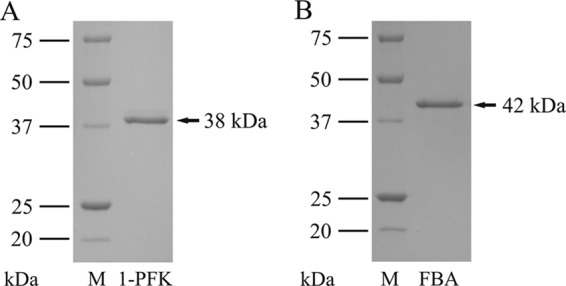
SDS-PAGE of purified, His-tagged 1-phosphofructokinase (1-PFK) (A) and fructose-1,6-bisphosphate aldolase (FBA) (B) of H. volcanii. M, molecular mass marker.
Enzyme activity was tested with fructose-1-phosphate (F1P) and fructose-6-phosphate (F6P) as substrates. With both substrates, the enzyme followed Michaelis-Menten kinetics with apparent Km and Vmax values for F1P of 0.31 mM and 308 U/mg and F6P of 1.12 mM and 30.7 U/mg, respectively. The catalytic efficiency (kcat/Km) of the enzyme for F1P (1.2 × 106 s−1 M−1) was about 36-fold higher than that for F6P (0.034 × 106 s−1 M−1), indicating that F1P is the physiological substrate defining the enzyme as F1P-specific kinase (1-PFK). The apparent Km and Vmax for the cosubstrate ATP was 0.08 mM and 200 U/mg, respectively. 1-PFK showed the highest activity at about 0.5 M KCl, and the pH optimum was between 7.5 and 9.
FBA encoded by fba is a bacterial-like class II aldolase involved in fructose catabolism. (i) Transcription analysis and mutant characterization.
HVO_1494 (fba), located downstream of the PTS operon, encodes a putative fructose-1,6-bisphosphate aldolase (FBA) that likely is involved in cleaving fructose-1,6-bisphosphate, the product of 1-PFK. Northern blot analysis revealed highly increased transcript levels of fba in fructose-grown cells compared to those in glucose-grown cells, indicating fructose-specific upregulation (Fig. 1B). Also, FBA activity in cell extracts was 8-fold upregulated in fructose-grown cells (0.39 U/mg) compared to that in glucose-grown cells (0.052 U/mg). To prove the functional involvement of fba in fructose degradation, it was deleted in frame. The Δfba mutant (see Fig. S3C in the supplemental material) lost the ability to grow on fructose, but growth on glucose was not inhibited and was even slightly stimulated (Fig. 2C; also see Fig. S4C in the supplemental material). A fructose-positive growth phenotype was recovered by complementation with fba in trans (Fig. 2C). The data indicate that fba is essential for growth on fructose.
(ii) Characterization of FBA.
fba was overexpressed in H. volcanii, and the recombinant enzyme was purified as a His tag fusion protein by Ni-NTA chromatography, followed by gel filtration on Superdex. The apparent molecular mass of the native enzyme was 80 kDa and that of the subunit about 42 kDa (Fig. 3B), indicating that FBA is a homodimeric protein. FBA catalyzed the cleavage of fructose-1,6-bisphosphate to dihydroxyacetone phosphate and GAP with apparent Km and Vmax values of 0.24 mM and 55 U/mg, respectively. The aldolase also catalyzed the aldol condensation reaction, i.e., FBP formation from GAP and DHAP, at a rate of about 20 U/mg. The enzyme showed the highest activities at 1 M KCl and 1 mM cysteine. The pH optimum was 7.5. The enzyme required divalent cation for activity. The highest activity was obtained with MnCl2 (100%, 50 U/mg), which could be partially replaced by FeSO4 (40% residual activity) rather than by ZnCl2 (<3% residual activity). The addition of EDTA (2 mM) to the enzyme resulted in complete inhibition of activity, which could be reversed by the addition of Mn2+ (3 mM) (data not shown). The metal dependence of FBA activity defines the enzyme as class II aldolase.
FBA has a gluconeogenetic function during growth on acetate, d-xylose, and Casamino Acids.
To analyze a potential role of reversible FBA of H. volcanii in FBP formation in the course of gluconeogenesis, the growth of the Δfba mutant with acetate, d-xylose, or Casamino Acids was analyzed. As shown in Fig. S5 in the supplemental material, the Δfba mutant did not grow on either of these substrates. The growth-positive phenotype on each substrate could be restored with fba by complementation in trans (see Fig. S5). Since the catabolic pathways of acetate, xylose, and Casamino Acids in H. volcanii do not contain FBP as an intermediate, the requirement of FBA for growth on these substrates indicates an anabolic function catalyzing fructose-1,6-bisphosphate formation in the course of gluconeogenesis.
DISCUSSION
In the present communication, the genes and enzymes involved in fructose uptake and its degradation to triosephosphates were identified on the basis of genome organization, transcriptional analyses, and gene deletion studies of candidate genes and enzyme characterization. The data indicate that fructose uptake is mediated by a bacterial-like PEP-dependent phosphotransferase system (PTS), generating fructose-1-phosphate, which is further phosphorylated to fructose-1,6-bisphosphate (FBP) by fructose-1-phosphate kinase (1-PFK). FBP is cleaved to triosephosphates by a bacterial like class II fructose-1,6-bisphosphate aldolase (FBA) (Fig. 4).
Fig 4.
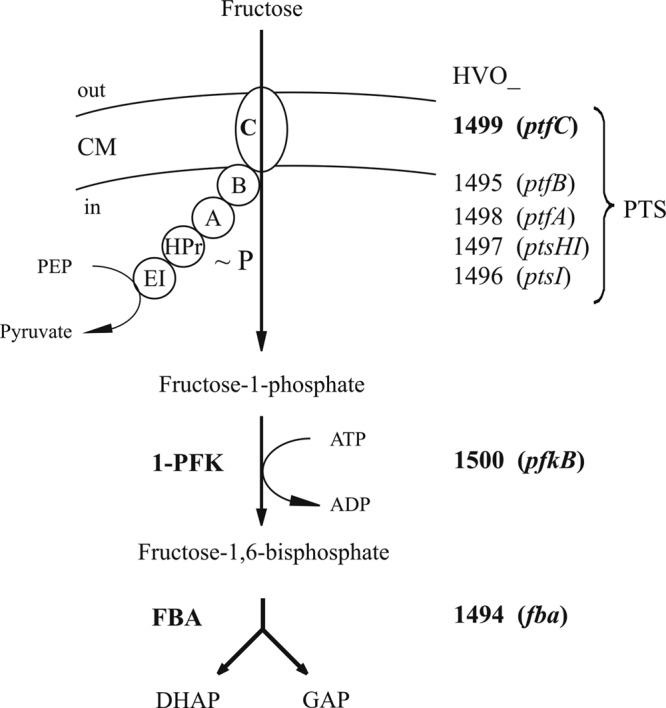
Proposed pathway of fructose uptake and degradation to triose phosphates in the haloarchaeon H. volcanii. Fructose uptake by PTS is shown schematically. It involves the putative cytoplasmic components EI, HPr, EIIA, and EIIB (encoded by HVO_1496 [ptsI], HVO_1497 [ptsH1], HVO_1498 [ptfA], and HVO_1495 [ptfB], respectively) and the transmembrane component EIIC (encoded by HVO_1499 [ptfC]), forming fructose-1-phosphate. Note that the Haloferax PTS, in contrast to bacterial counterparts, consists of five separate components. ∼P indicates the phosphoryl group which is transferred from PEP to fructose; the transient phosphorylation of PTS components is not shown. Fructose-1-phosphate is phosphorylated by 1-PFK (encoded by HVO_1500 [pfkB]) to fructose-1,6-bisphosphate, which is cleaved to dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) by FBA (encoded by HVO_1494 [fba]). Proteins and genes that were proven to be functionally involved in fructose uptake and degradation are marked in boldface. CM, cytoplasmic membrane.
A bacterial-like PTS cluster in H. volcanii is involved in fructose catabolism.
H. volcanii contains a gene cluster, HVO_1495 to HVO_1499 (ptfB, ptsI, ptsH1, ptfA, and ptfC), encoding all five homologs, EIIB, EI, HPr, EIIA, and EIIC, of a bacterial-like fructose-specific PTS. Northern blot analyses indicate that this PTS cluster forms an operon which was highly induced by fructose, indicating a functional role in fructose catabolism. This was proven with a deletion mutant of ptfC, encoding the fructose-specific membrane component EIIC. The ΔptfC mutant did not grow on fructose, indicating that EIIC was essential for fructose uptake. In a previous report, fructose uptake in H. volcanii was analyzed in cell suspensions, and the authors suggested the presence of an Na+-linked fructose symport system (46). However, the fructose-negative growth phenotype of the H. volcanii ΔptfC mutant excludes an effective fructose uptake system operating besides the PTS.
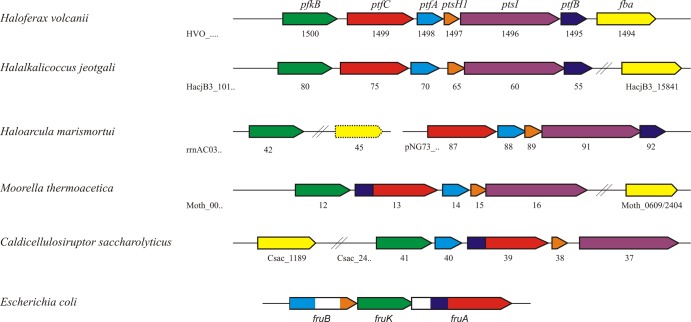
This is the first report on the functional involvement of a bacterial-like PTS in sugar metabolism in the archaeal domain. Recent sequenced haloarchaeal genomes indicate the presence of homologs of the complete H. volcanii PTS cluster, e.g., in Haloarcula marismortui, Haloarcula hispanica, Halalkalicoccus jeotgali, and Haloterrigena turkmenica. The five PTS components in these haloarchaea show high degrees of sequence identity, e.g., about 70% for ptfC, and an identical organization pattern, as shown for Halalkalicoccus jeotgali and Haloarcula marismortui (Fig. 5), suggesting a role of these phosphotransferase systems in fructose uptake. Besides the PTS operon described here, H. volcanii contains a second, incomplete PTS (HVO_2101 to HVO_2104) missing the EI component, the function of which is not known (26). Further, a single HPr component, ptsH2 (HVO_1543), is present in H. volcanii, and it has been shown to be transcriptionally linked to genes involved in glycerol metabolism (26, 38).
Fig 5.
Genomic organization of fructose-specific PTS, 1-PFK, and FBA in H. volcanii compared to selected haloarchaea and bacteria. Genes are indicated as colored arrows: 1-phosphofructokinase (1-PFK; HVO_1500 [pfkB]) in green, EIIC (HVO_1499 [ptfC]) in red, EIIA (HVO_1498[ptfA]) in blue, HPr (HVO_1497 [ptsH1]) in orange, EI (HVO_1496 [ptsI]) in purple, EIIB (HVO_1495 [ptfB]) in dark blue, and fructose-1,6-bisphosphate aldolase (FBA; HVO_1494 [fba]) in yellow. EIIC and EIIB homologs in M. thermoacetica (Moth_0013) and C. saccharolyticus (Csac2439) are fused. fruB and fruA of E. coli represent fusions of EIIA-HPr and EIIB-EIIC, respectively; white areas in these genes show regions with domains that probably are nonfunctional. rrnAC0345 codes for a class I FBA in Haloarcula marismortui (yellow with dashed line).
However, phosphotransferase systems have not been found in any other archaeal genomes (8, 9). Since phosphotransferase systems are also absent from eukarya but are frequently present in the bacterial domain, the haloarchaeal fructose-specific PTS likely originated from bacteria via lateral gene transfer. Based on the sequence comparison of the transmembrane EIIC proteins, the bacterial phosphotransferase systems can be divided into 7 subfamilies, including the glucose family, specific for glucose and glucosides, and the fructose family, specific for fructose and mannitol (9). The EIIC component of H. volcanii contains a PTS EIIC type 2 domain (PS51104), which attributes the haloarchaeal PTS to the fructose/mannitol PTS family. More refined sequence analysis according to TIGRFAMs (http://www.jcvi.org/cgi-bin/tigrfams/index.cgi) classifies the H. volcanii EIIC as a member of the fructose-specific EIIC type 2 subfamily (TIGR01427). Members of this subfamily phosphorylate fructose specifically at the 1 position. Within the bacterial domain, the haloarchaeal EIIC showed the highest degree of sequence identities (44 to 48%), predominantly in EIIC homologs of members related to Clostridia, including Moorella thermoacetica and Caldicellulosiruptor saccharolyticus.
However, the genomic organization of the haloarchaeal PTS, which is composed of separate genes encoding separate single protein components, differs significantly from most bacterial counterparts. Usually bacterial fructose-specific phosphotransferase systems contain fusions of genes encoding fused proteins. For example, in E. coli, fruA and fruB encode fusion proteins of EIIB/EIIC and of HPr/EIIA, respectively (Fig. 5). For a recent review of the genome organization of various bacterial PTS clusters and of phylogenetic analyses of the individual PTS components, see references 9 and 19. In contrast, the fructose PTS in haloarchaea appear not to contain any fusion protein. The highest similarity of genome organization of haloarchaeal PTS was found in the bacteria Moorella thermoacetica and Caldicellulosiruptor saccharolyticus (Fig. 5). With the exception of fused EIIB/EIIC components, these bacteria show separate PTS genes. Due to high similarity in both the sequence and organization of the PTS genes, the haloarchaeal PTS might be acquired via horizontal gene transfer from Clostridia-related Gram positives, e.g., from Moorella and Caldicellulosiruptor species. A recent phylogenetic analysis of putative EI and HPr of Haloarcula marismortui PTS also suggested the bacterial origin of these components (19).
1-PFK of H. volcanii.
HVO_1500, located directly upstream of the PTS cluster, was identified as gene pfkB, encoding functional 1-PFK in H. volcanii. pfkB was transcriptionally induced, and the ΔpfkB strain did not grow on fructose. Recombinant 1-PFK, obtained after homologous overexpression, was characterized and showed a higher catalytic efficiency for F1P as the substrate than for F6P, defining the enzyme as functional 1-PFK. Homologs of 1-PFKs with high sequence identity (about 55%) were found in other halophilic archaea, e.g., in Haloarcula marismortui, Halorhabdus utahensis, Haloarcula hispanica, Halalkalicoccus jeotgali, and Haloterrigena turkmenica, suggesting a catalytic function similar to that in H. volcanii. In accordance with genes of H. volcanii, the pfkB genes of Halalkalicoccus jeotgali (Fig. 5) and Haloterrigena turkmenica are located next to the homologous PTS cluster.
The H. volcanii 1-PFK, characterized as a 70-kDa homodimer, differs from 1-PFK of Haloarcula vallismortis, which was reported to be a 70-kDa monomeric enzyme (36). The gene encoding the characterized 1-PFK in Haloarcula vallismortis has not been identified. 1-PFK of H. volcanii belongs to the pfkB family of sugar kinases, showing considerable sequence identity to bacterial 1-PFK homologs, e.g., 31% to 1-PFK of E. coli, which is encoded by the fruK gene (30).
The 1-PFKs from Clostridium pasteurianum and Enterobacter aerogenes, which have been implicated in fructose degradation in connection with fructose-specific PTS, have been characterized (40, 48). The A. aerogenes enzyme showed a molecular mass of about 70 kDa; the C. pasteurianum enzyme was reported to be a 63-kDa heterodimer (40, 48) with kinetic constants for F1P and ATP and high Vmax values of about 300 U/mg, which are very similar to data reported for the H. volcanii enzyme.
FBA from H. volcanii is a bacterial-like class II enzyme.
HVO_1494, located directly downstream of the PTS cluster, was identified as the gene fba, encoding functional FBA in H. volcanii. fba was highly induced by fructose, and the Δfba strain showed a fructose-negative growth phenotype, indicating a functional role in fructose degradation via the proposed modified Embden-Meyerhof pathway. The Δfba strain grew well on glucose, in accordance with glucose degradation in H. volcanii, via a semiphosphorylated Entner-Doudoroff pathway, which does not require catabolic FBA activity (43). The enhanced growth of the Δfba strain on glucose compared to that of the wild type cannot be explained and remains to be studied.
FBA from H. volcanii was characterized as an 80-kDa homodimeric class II type FBA which requires divalent cations for activity and was completely inactivated by EDTA. The highest activity of haloarchaeal FBA was found with Mn2+, which could partially be replaced by Fe2+ rather than by Zn2+. A class II aldolase from Haloferax mediterranei has been reported to be an 80-kDa homodimer showing highest activity with Fe2+, whereas Mn2+ and Zn2+ did not have a stimulating effect (21). Usually bacterial class II FBAs prefer Zn2+ ions (11).
Sequence alignment of the haloarchaeal FBA and phylogenetic affiliation.
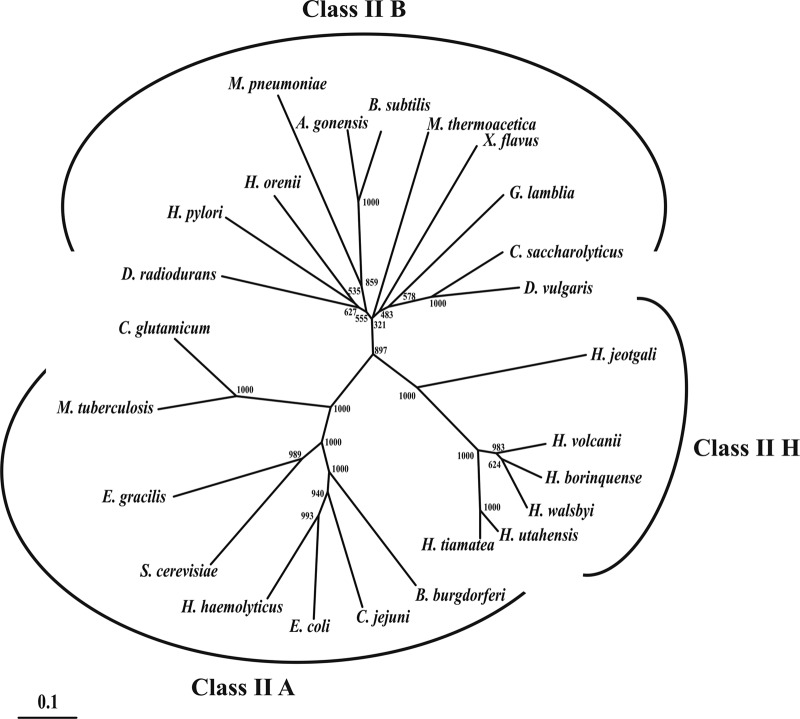
Homologs of H. volcanii class II FBA with sequence identity of up to 90% were found in other haloarchaea, e.g., Halorhabdus, Halogeometricum, Haloquadratum, and Halalkalicoccus species, suggesting a similar catabolic FBA function in fructose catabolism in these organisms. Class II FBA homologs with significant sequence identity (>15%) were not found in any other archaeal genomes; however, a large number of homologs with identities of up to 30% were detected in the bacterial domain. Class II type FBA of bacteria and lower eukarya, e.g., Giardia, Saccharomyces, and Euglena, are divided into two subgroups, A and B, on the basis of sequence similarity and physiological function (27, 35). Class II A members are usually specific for fructose-1,6-bisphosphate as the substrate and are involved in glycolysis and gluconeogenesis. Class II B family include members with a more heterogeneous aldol substrate spectrum that is involved in the degradation of various sugars (in addition to glucose), sugar alcohols, and amino sugars (27, 35). A multiple-sequence alignment of class II FBA from H. volcanii, the homolog from Halogeometricum boringuense, and from bacterial class II FBA subgroups A and B are given in Fig. S6 in the supplemental material. The Haloferax FBA contained, with few deviations, the consensus patterns 1 and 2 of the bacterial class II FBA and also several conserved amino acids, including three histidines involved in metal binding and aspartate, arginine, and glutamate, which have been proposed to be involved in catalysis, as concluded from the crystal structure of the E. coli class II FBA (22). The phylogenetic relationship of FBA of H. volcanii and other haloarchaeal homologs with selected FBAs from bacteria and lower eukarya is shown in Fig. 6. Supported by high bootstrap values, the haloarchaeal FBAs form a distinct cluster, suggesting that haloarchaeal class II FBA forms a separate class II FBA subfamily, designated FBA class II H, (FBA haloarchaea) (Fig. 6). One can speculate that the haloarchaeal class II FBAs originated from bacterial class II FBAs via lateral gene transfer. The separate clustering of the class II H FBAs might be explained by the adaptation of these enzymes to halophilic conditions.
Fig 6.
Phylogenetic relationship of class II fructose-1,6-bisphosphate aldolases from haloarchaea (class II H), bacteria, and lower eukaryotes (class II A/B). The numbers at the nodes are bootstrap values according to neighbor joining (the neighbor-joining algorithm of ClustalX was used to generate the values). The tree is based upon a multiple-sequence alignment that was generated with ClustalX using the gonnet matrix (47). The GenBank accession numbers for the aldolases from the different species are shown in parentheses: class II B, Anoxybacillus gonensis (ABL75360), Bacillus subtilis (AEP88659), Caldicellulosiruptor saccharolyticus (YP_001179983), Deinococcus radiodurans R1 (NP_295312), Desulfovibrio vulgaris (YP_002434443), Giardia lamblia (3GB6_A), Halothermothrix orenii (YP_002509546), Helicobacter pylori (CBI66947), Moorella thermoacetica (YP_429482), Mycoplasma pneumoniae (BAL21593), and Xanthobacter flavus (AAA96742); class II A, Borrelia burgdorferi (AAB91507), Campylobacter jejuni (ZP_06371648), Corynebacterium glutamicum (EHE82675), Escherichia coli (BAE76989), Euglena gracilis (CAA61912), Haemophilus haemolyticus (EGT83318), Mycobacterium tuberculosis (P67475), and Saccharomyces cerevisiae (GAA24665); and class II H, Halalkalicoccus jeotgali (YP_003738319), Haloferax volcanii (YP_003535543), Halogeometricum borinquense (ADQ67306), Halorhabdus tiamatea (ZP_08561145), Halorhabdus utahensis (YP_003129569), and Haloquadratum walsbyi (YP_658423). The scale bar corresponds to 0.1 substitutions per site.
This is the first report of a functional class II aldolase in the sugar catabolism of archaea. So far only archaeal class I type FBP aldolases, which form a Schiff base in the catalytic mechanism and are phylogenetically unrelated to class II FBAs, have been described for the sugar catabolism of archaea. Archaeal class I aldolases were found in the hyperthermophiles Pyrococcus furiosus and Thermoproteus tenax (42), which both degrade glucose or glucose polymers to 100 and 80%, respectively, via modified Embden Meyerhof pathways involving FBP as an intermediate and its cleavage via FBA (see reference 43 and references therein). Putative homologs of class I FBAs were also found in a few haloarchaea, including Haloarcula marismortui (rrnAC0345). Since Haloarcula marismortui does not contain a class II FBA, the class I FBA (Fig. 5) in this organism might be the functional FBA involved in fructose degradation; this remains to be shown. In a previous study, the presence of class I and class II FBAs in halophilic archaea were also concluded from activity measurements in cell extracts either in the presence or absence of EDTA and borohydride, which allow discrimination between ion-dependent (class II) and Schiff base-forming (class I) enzymes, respectively (5).
Recently, a novel bifunctional fructose-1,6-bisphosphate aldolase/phosphatase, catalyzing the unidirectional synthesis of fructose-6-phosphate from GAP and DHAP without releasing fructose-1,6-bisphosphate FBA, was described in most archaeal genomes except for haloarchaea. This bifunctional enzyme belongs to the class I aldolase family and is exclusively involved in gluconeogenesis (41).
Gluconeogenetic function of class II FBA in H. volcanii.
Since H. volcanii does not contain a bifunctional gluconeogenic class I FBA, the reversible class II FBA might have an anabolic function in addition to its role in catabolism. FBA was found to catalyze the reverse reaction, i.e., aldol synthesis from GAP and DHAP. As shown in this study, fba mutants did not grow on either acetate and d-xylose or on Casamino Acids as carbon and energy sources (see Fig. S5 in the supplemental material), although the degradation pathways of these substrates in H. volcanii do not involve FBP as an intermediate and thus do not require cleavage by FBA (28). Thus, the requirement for a functional FBA during growth on these substrates indicates a gluconeogenetic role of FBA in catalyzing the formation of FBP, from which pentose phosphates and other cell constituents are formed. The finding that an fba knockout did not inhibit growth on glucose indicates that anabolic pathways of, e.g., pentose phosphate formation, starting from glucose, do not involve FBP formation via class II FBA in H. volcanii. Soderberg proposed for Halobacterium a route for the generation of ribose-5-phosphate from glucose via gluconate, gluconate-6-phosphate, and ribulose-5-phosphate catalyzed by glucose dehydrogenase, an unknown kinase, 6-phosphogluconate dehydrogenase, and ribose-5-phosphate isomerase (44). Since homologs of most of these genes are also present in H. volcanii, a similar route of pentose-phosphate formation might be operative in this organism. Such a pathway, which does not involve an FBA, could explain why the Δfba mutant in H. volcanii is able to grow on glucose.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thorsten Allers (University of Nottingham, United Kingdom) for providing the plasmid pTA963 and the expression strain H. volcanii H1209.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SCHO 316/11-1).
Footnotes
Published ahead of print 6 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Albers SV, Koning SM, Konings WN, Driessen AJ. 2004. Insights into ABC transport in archaea. J. Bioenerg. Biomembr. 36:5–15 [DOI] [PubMed] [Google Scholar]
- 2. Allers T. 2010. Overexpression and purification of halophilic proteins in Haloferax volcanii. Bioeng. Bugs 1:288–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76:1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allers T, Ngo HP, Mevarech M, Lloyd RG. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altekar W, Dhar NM. 1988. Archaebacterial class I and class II aldolases from extreme halophiles. Orig. Life Evol. Biosph. 18:59–64 [DOI] [PubMed] [Google Scholar]
- 6. Altekar W, Rangaswamy V. 1992. Degradation of endogenous fructose during catabolism of sucrose and mannitol in halophilic archaebacteria. Arch. Microbiol. 158:356–363 [Google Scholar]
- 7. Altekar W, Rangaswamy V. 1990. Indication of a modified EMP pathway for fructose breakdown in a halophilic archaebacterium. FEMS Microbiol. Lett. 69:139–143 [Google Scholar]
- 8. Anderson I, et al. 2011. Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS One 6:e20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barabote RD, Saier MH., Jr 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69:608–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bardey V, et al. 2005. Characterization of the molecular mechanisms involved in the differential production of erythrose-4-phosphate dehydrogenase, 3-phosphoglycerate kinase and class II fructose-1,6-bisphosphate aldolase in Escherichia coli. Mol. Microbiol. 57:1265–1287 [DOI] [PubMed] [Google Scholar]
- 11. Berry A, Marshall KE. 1993. Identification of zinc-binding ligands in the class II fructose-1,6-bisphosphate aldolase of Escherichia coli. FEBS Lett. 318:11–16 [DOI] [PubMed] [Google Scholar]
- 12. Bitan-Banin G, Ortenberg R, Mevarech M. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 14. Brenneis M, Hering O, Lange C, Soppa J. 2007. Experimental characterization of cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 3:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chomczynski P, Mackey K. 1995. Substitution of chloroform by bromo-chloropropane in the single-step method of RNA isolation. Anal. Biochem. 225:163–164 [DOI] [PubMed] [Google Scholar]
- 16. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 17. Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148–152 [DOI] [PubMed] [Google Scholar]
- 18. Cline SW, Schalkwyk LC, Doolittle WF. 1989. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171:4987–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Comas I, Gonzalez-Candelas F, Zuniga M. 2008. Unraveling the evolutionary history of the phosphoryl-transfer chain of the phosphoenolpyruvate:phosphotransferase system through phylogenetic analyses and genome context. BMC Evol. Biol. 8:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dambeck M, Soppa J. 2008. Characterization of a Haloferax volcanii member of the enolase superfamily: deletion mutant construction, expression analysis, and transcriptome comparison. Arch. Microbiol. 190:341–353 [DOI] [PubMed] [Google Scholar]
- 21. D'Souza SE, Altekar W. 1998. A class II fructose-1,6-bisphosphate aldolase from a halophilic archaebacterium Haloferax mediterranei. J. Gen. Appl. Microbiol. 44:235–241 [DOI] [PubMed] [Google Scholar]
- 22. Hall DR, et al. 1999. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J. Mol. Biol. 287:383–394 [DOI] [PubMed] [Google Scholar]
- 23. Hammelmann M, Soppa J. 2008. Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J. Microbiol. Methods 75:201–204 [DOI] [PubMed] [Google Scholar]
- 24. Hansen T, Schönheit P. 2001. Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a non-allosteric enzyme related to the phosphofructokinase-B sugar kinase family, from the hyperthermophilic crenarchaeote Aeropyrum pernix. Arch. Microbiol. 177:62–69 [DOI] [PubMed] [Google Scholar]
- 25. Hansen T, Schönheit P. 2000. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch. Microbiol. 173:103–109 [DOI] [PubMed] [Google Scholar]
- 26. Hartman AL, et al. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5:e9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henze K, Morrison HG, Sogin ML, Muller M. 1998. Sequence and phylogenetic position of a class II aldolase gene in the amitochondriate protist, Giardia lamblia. Gene 222:163–168 [DOI] [PubMed] [Google Scholar]
- 28. Johnsen U, et al. 2009. D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J. Biol. Chem. 284:27290–27303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnsen U, Selig M, Xavier KB, Santos H, Schönheit P. 2001. Different glycolytic pathways for glucose and fructose in the halophilic archaeon Halococcus saccharolyticus. Arch. Microbiol. 175:52–61 (Erratum, 180:503, 2003.) [DOI] [PubMed] [Google Scholar]
- 30. Kornberg HL. 2001. Routes for fructose utilization by Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:355–359 [PubMed] [Google Scholar]
- 31. Krishnan G, Altekar W. 1991. An unusual class I (Schiff base) fructose-1,6-bisphosphate aldolase from the halophilic archaebacterium Haloarcula vallismortis. Eur. J. Biochem. 195:343–350 [DOI] [PubMed] [Google Scholar]
- 32. Lee SJ, Böhm A, Krug M, Boos W. 2007. The ABC of binding-protein-dependent transport in Archaea. Trends Microbiol. 15:389–397 [DOI] [PubMed] [Google Scholar]
- 33. Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87 [DOI] [PubMed] [Google Scholar]
- 34. Oren A, Ginzburg M, Ginzburg BZ, Hochstein LI, Volcani BE. 1990. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium form the Dead Sea. Int. J. Syst. Bacteriol. 40:209–210 [DOI] [PubMed] [Google Scholar]
- 35. Plaumann M, Pelzer-Reith B, Martin WF, Schnarrenberger C. 1997. Multiple recruitment of class-I aldolase to chloroplasts and eubacterial origin of eukaryotic class-II aldolases revealed by cDNAs from Euglena gracilis. Curr. Genet. 31:430–438 [DOI] [PubMed] [Google Scholar]
- 36. Rangaswamy V, Altekar W. 1994. Characterization of 1-phosphofructokinase from halophilic archaebacterium Haloarcula vallismortis. Biochim. Biophys. Acta 1201:106–112 [DOI] [PubMed] [Google Scholar]
- 37. Rangaswamy V, Altekar W. 1994. Ketohexokinase (ATP:D-fructose 1-phosphotransferase) from a halophilic archaebacterium, Haloarcula vallismortis: purification and properties. J. Bacteriol. 176:5505–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rawls KS, Martin JH, Maupin-Furlow JA. 2011. Activity and transcriptional regulation of bacterial protein-like glycerol-3-phosphate dehydrogenase of the haloarchaea in Haloferax volcanii. J. Bacteriol. 193:4469–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rawls KS, Yacovone SK, Maupin-Furlow JA. 2010. GlpR represses fructose and glucose metabolic enzymes at the level of transcription in the haloarchaeon Haloferax volcanii. J. Bacteriol. 192:6251–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sapico V, Anderson RL. 1969. D-fructose 1-phosphate kinase and D-fructose 6-phosphate kinase from Aerobacter aerogenes. A comparative study of regulatory properties. J. Biol. Chem. 244:6280–6288 [PubMed] [Google Scholar]
- 41. Say RF, Fuchs G. 2010. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464:1077–1081 [DOI] [PubMed] [Google Scholar]
- 42. Siebers B, et al. 2001. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710–28718 [DOI] [PubMed] [Google Scholar]
- 43. Siebers B, Schönheit P. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol. 8:695–705 [DOI] [PubMed] [Google Scholar]
- 44. Soderberg T. 2005. Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1:347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soppa J. 2006. From genomes to function: haloarchaea as model organisms. Microbiology 152:585–590 [DOI] [PubMed] [Google Scholar]
- 46. Takano J-I, Kaidoh K, Kamo N. 1995. Fructose transport by Haloferax volcanii. Can. J. Microbiol. 41:241–246 [Google Scholar]
- 47. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Hugo H, Gottschalk G. 1974. Purification and properties of 1-phosphofructokinase from Clostridium pasteurianum. Eur. J. Biochem. 48:455–463 [DOI] [PubMed] [Google Scholar]
- 49. Zaigler A, Schuster SC, Soppa J. 2003. Construction and usage of a onefold-coverage shotgun DNA microarray to characterize the metabolism of the archaeon Haloferax volcanii. Mol. Microbiol. 48:1089–1105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.