Abstract
The tubulin homolog FtsZ forms a polymeric membrane-associated ring structure (Z ring) at midcell that establishes the site of division and provides an essential framework for the localization of a multiprotein molecular machine that promotes division in Escherichia coli. A number of regulatory proteins interact with FtsZ and modulate FtsZ assembly/disassembly processes, ensuring the spatiotemporal integrity of cytokinesis. The Z-associated proteins (ZapA, ZapB, and ZapC) belong to a group of FtsZ-regulatory proteins that exhibit functionally redundant roles in stabilizing FtsZ-ring assembly by binding and bundling polymeric FtsZ at midcell. In this study, we report the identification of ZapD (YacF) as a member of the E. coli midcell division machinery. Genetics and cell biological evidence indicate that ZapD requires FtsZ but not other downstream division proteins for localizing to midcell, where it promotes FtsZ-ring assembly via molecular mechanisms that overlap with ZapA. Biochemical evidence indicates that ZapD directly interacts with FtsZ and promotes bundling of FtsZ protofilaments. Similarly to ZapA, ZapB, and ZapC, ZapD is dispensable for division and therefore belongs to the growing group of FtsZ-associated proteins in E. coli that aid in the overall fitness of the division process.
INTRODUCTION
In Escherichia coli, assembly of the essential tubulin homolog FtsZ into a membrane-associated ring (Z ring) establishes the site of division and provides a structural framework for the localization of the rest of the division machinery (1, 13, 16). The overall concentration of FtsZ is effectively unchanged throughout the cell cycle, and cell division is primarily regulated by the GTP hydrolysis-coupled assembly/disassembly processes of FtsZ at midcell (33, 53). A number of FtsZ-regulatory proteins, some of which colocalize with FtsZ at midcell, promote the FtsZ assembly/disassembly processes and thereby play critical roles in exercising spatiotemporal control of cytokinesis (13, 29). An understanding of the precisely orchestrated molecular interactions between FtsZ and its regulators that ultimately govern the formation of a stable yet dynamic midcell Z-ring at the onset of division remains incomplete.
During the early stages of cytokinesis, the midcell division site in E. coli is characterized by a dynamic Z-ring structure made up of FtsZ and FtsZ-stabilizing proteins FtsA, ZipA, ZapA, ZapB, and ZapC (2, 11, 14, 15, 18, 21, 23, 25, 31, 44, 52). FtsZ stabilizers are involved in the maturation of early FtsZ assemblies by mechanisms that include tethering FtsZ at the membrane, stabilizing FtsZ protofilament associations by cross-linking or bundling separate protofilaments, and/or preventing depolymerization through the inhibition of GTP hydrolysis (11, 14, 17, 24, 25, 41, 42). The stabilized FtsZ ring promotes recruitment of late-assembly divisome components in a largely linear hierarchy to form a constriction-competent multiprotein septal ring (13). FtsA and ZipA not only are essential for tethering FtsZ to the membrane at the initiation of E. coli division but are also required for recruiting downstream divisome proteins, although certain FtsA mutants allow cytokinesis in the absence of the less widely conserved ZipA protein (19, 42, 43). Additionally, members of a growing group of FtsZ-associated proteins, including ZapA, along with its binding protein ZapB, and ZapC, also play important roles in modulating Z-ring assembly early in division (14, 18, 21, 25). These proteins, collectively called Z-associated proteins (Zaps), show no homology at the sequence level but execute partially redundant functions in affecting FtsZ assembly. Zap proteins do not have roles in tethering FtsZ to the membrane or recruiting downstream divisome proteins to the Z-ring and are nonessential for division. Accordingly, the interactions of Zap proteins with FtsZ are characterized by modest phenotypes in single mutants but strong synergistic division phenotypes when expression of two or more Zap proteins is altered (15, 17, 21, 25). The widely conserved FtsZ regulator ZapA dynamically associates with the Z ring and imparts FtsZ-ring stability by cross-linking and bundling adjacent FtsZ protofilaments, as well as by promoting longitudinal interactions between FtsZ monomers (11, 36). ZapB, another nonessential FtsZ regulator, localizes to the divisome via ZapA and enhances the role of ZapA in FtsZ bundling (17). The recently identified cell division factor ZapC binds and promotes FtsZ bundling at midcell (14, 25). While some FtsZ-interacting proteins such as FtsA and ZapA are highly conserved among bacteria, others, such as ZipA, ZapB, and ZapC, are restricted to the Gammaproteobacteria class of bacteria (14, 15, 25, 44).
Here we report the discovery of YacF as a cell division factor belonging to the Zap family of proteins in E. coli that binds the conserved C-terminal tail of FtsZ and bundles FtsZ polymers at midcell. YacF encodes a nonessential FtsZ-associated protein, removal of which leads to synthetic detrimental effects in cells carrying a conditional ftsZ allele that is impaired in its enzymatic activity or in cells lacking ZapA. Overexpression of YacF interferes with normal FtsZ ring assembly, leading to filamentation. In vitro, YacF interacts directly with FtsZ and promotes FtsZ polymer bundling with a reduction in FtsZ GTPase activity. YacF, which we propose to call ZapD, is therefore a member of the group of FtsZ-associated proteins that bind and bundle FtsZ at midcell and likely enhance the overall division efficiency in E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
Bacterial strains and plasmids used in this study are listed in Table 1. Strain construction was by transformation or by generalized transduction using P1 (51).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Sourceb or reference |
|---|---|---|
| Strains | ||
| MG1655 | F− λ−ilvG rfb50 rph1 | Laboratory collection |
| TB28 | MG1655 ΔlacZYA | 6 |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | Laboratory collection |
| JOE309 | MC4100 araD+ | 9 |
| EC307 | MC4100 leu::Tn10 (Tetr) ftsZ84 (Ts) | 28 |
| MDG148 | JOE309 leu::Tn10 (Tetr) ftsA12 (Ts) | 9 |
| PS223 | W3110 zipA1 (Ts) | 42 |
| JW0099 | AG1 pCA24N-zapD | 30 |
| BL21(λDE3) pET11b-FtsZ | F−ompT hsdSB (rB− mB−) dcm gal λ(DE3) pET11b-FtsZ | L. Romberg |
| BL21(λDE3) pLysS | F−ompT hsdSB (rB− mB−) dcm gal λ(DE3) pLysS | Agilent |
| AJ18-79 | JD315 pNG162-zapD | |
| GF73 | TB28 pCA24N | |
| JD160 | BL21(λDE3) pLysS pET28b-His10-Smt3-zapD | |
| JD257 | TB28 ΔzapA::frt | |
| JD305 | TB28 ΔzapA::frt ΔzapC::Kan | |
| JD310 | TB28 ΔzapD::Kan | |
| JD315 | TB28 ΔzapA::frt ΔzapD::Kan | |
| JD317 | TB28 ΔzapC::Kan | |
| JD330 | EC307 ΔzapD::Kan | |
| JD345 | TB28 ΔzapD::frt ΔzapC::Kan | |
| JD360 | TB28 ΔzapA::frt ΔzapD::frt ΔzapC::Kan | |
| JD381 | TB28 pCA24N-zapD | |
| Plasmids | ||
| pCA24N | pT5-lac, lacIq, Cmr | 30 |
| pCP20 | pSC101 ori(Ts) cI857 λPRflp, Ampr, Cmr | 12 |
| pDSW208-ZapD-eYFP | pJC58-zapD | |
| pET28b-His10-Smt3 | pBR322 ori pT7-His10-Smt3, Kanr | S. Shuman |
| pJC58 | pDSW208(eYFP) MCS-eyfp cloning vector | 9 |
| pKD46 | pSC101 ori(Ts) pBAD gam bet exo, Ampr | 12 |
| pNG162 | pSC101, Specr | 20 |
Amp, ampicillin; Cm, chloramphenicol, Kan, kanamycin; Spec, spectinomycin; Tet, tetracycline.
The source was this study, unless otherwise noted.
Growth conditions.
E. coli strains were grown and maintained in LB (0.5% NaCl) at 30°C unless otherwise stated. Strains containing plasmid pDSW208-ZapD-eYFP or pNG162-ZapD were used for imaging in this study. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 12.5 μg/ml. Other details, including inducer isopropyl β-d-1-thiogalactopyranoside (IPTG) concentrations, are specified in the figure legends.
Plasmid and strain construction. (i) Plasmid construction.
Plasmid pDSW208-ZapD-eYFP expressing hybrid protein under the control of the IPTG-inducible trc promoter was constructed by amplifying zapD using the SalI yacF 5P and SalI yacF 3P primers from the JW0099 ASKA plasmid clone (30). The PCR product was digested by SalI, cloned into pJC58 to create a C-terminal enhanced yellow fluorescent protein (eYFP) fusion protein, and verified by sequence analysis. To generate a zapD-complementing vector, zapD was PCR amplified using yacF 5P GW and yacF 3P GW primers from the JW0099 ASKA plasmid clone into pDONR221 plasmid (Invitrogen) to create a Gateway entry clone. After sequence verification, the entry clone was recombined into an IPTG-inducible pNG162 destination vector to create the pNG162-zapD expression clone according to the Gateway protocol. A list of the primers used in the study is included in Table S1 in the supplemental material.
(ii) Construction of strains carrying deletions in genes of interest.
Chromosomal deletions in genes of interest were obtained from the Keio Collection (4) for zapD, zapC, zapB, and zapA and P1 transduced into the TB28 strain background to create isogenic strains and verified by PCR for presence of a kanamycin resistance cassette. Where necessary, an antibiotic resistance gene was eliminated with the aid of FLP expression plasmid and verified by PCR for loss of the resistance marker (12).
(iii) Construction of strains for protein purification.
Using primers SUMO-5 YacF BamHI and SUMO-3 YacF HindIII, zapD was amplified by PCR and the product was digested by BamHI and HindIII and cloned into the same sites of pET28b-His10-Smt3 vector. The sequence-verified clone was transformed into BL21λDE3/pLysS to express ZapD.
Microscopy.
Microscopy was performed using a 100× oil immersion objective on a Nikon Eclipse Ti microscope with Chroma filters. Images were captured digitally using a Nikon DigiSight Monochrome charge-coupled-device (CCD) camera and analyzed using Nikon Elements Basic software. Color images were pseudocolored using the same software and assembled using Adobe Photoshop.
(i) Live bacterial cell imaging.
Screening of plasmid ZapD-eYFP fusion protein in various strains was performed individually on agarose pads on glass slides. For temperature-sensitive (Ts) strains, cells grown at the restrictive temperature were visualized by setting the objective heater (TC-500 temperature controller; 20/20 Inc.) on the microscope to 42°C. Descriptions of strain-specific growth and induction conditions are included in the figure legends.
(ii) Indirect immunofluorescence.
For visualizing FtsZ in cells overexpressing ZapD, cells containing pCA24N-ZapD were grown overnight at 30°C. Cells were fixed in methanol-acetone, and the presence of FtsZ was probed with an anti-FtsZ rabbit polyclonal antibody (Genscript) at 1:10,000 and a Texas-Red conjugated secondary antibody as described previously (28). Samples were also harvested for Western blot analyses prior to fixation.
(iii) TEM.
To visualize the morphology of the FtsZ polymers under conditions identical to those used for FtsZ sedimentation studies (described below), aliquots of experimental and control reaction mixtures were removed, negatively stained with 2% uranyl acetate, and examined by conventional transmission electron microscopy (TEM). Images were collected on a JEOL 2100F TEM operated at 200 kV and recorded on a 2k-by-2k CCD camera at 2.5 μm underfocus and at a nominal magnification of ×30,000 to ×50,000.
Protein-interaction platform (PIP) assays using Saccharomyces cerevisiae.
Full-length ftsZ, ftsZ1–372, ftsZ1–314, ftsZ289–383, ftsZ364–383, ftsZ374–383, and zapD were PCR amplified with a stop codon into pDONR223 (Invitrogen) plasmids to create Gateway entry clones. After sequence verification, yeast expression plasmids were created using site-specific recombination (Gateway) to generate FtsZ and ZapD fusion proteins that are expressed under the control of a regulatable GAL1 promoter. A pAG413-egfp-zapD expression plasmid was transformed into Saccharomyces cerevisiae strain S288C MATa, and pAG416-μNS-ftsZ or pAG416-μNS-ftsZ deletions or pAG416-μNS-zapD expression plasmids were transformed into strain S288C MATα. The strains that conditionally express the fluorophore fusion plasmid were mated with those that conditionally express the μNS fusion plasmid, and diploids were selected. Visualizations of live yeast expressing both the flurophore and μNS fusions were conducted using 96-well glass-bottomed plates as described previously (48).
Protein purification.
One liter of bacterial cultures was grown, and the bacteria were induced to express FtsZ. FtsZ was purified, and concentrations were determined by a bicinchoninic acid (BCA) assay as described previously (46). ZapD was purified by growing 0.5-liter bacterial cultures with the fusion plasmid at 37°C to an optical density at 600 nm (OD600) of ∼0.5, at which point ZapD expression was induced with 0.85 mM IPTG followed by growth for 4 h at 30°C. Cells were harvested by centrifugation, and pellets were resuspended in ice-cold lysis buffer (50 mM NaPO4, 0.3 M NaCl, 10 mM imidazole, pH 8.0) and stored at −80°C. After a couple of freeze-thaw cycles, cells were sonicated and centrifuged at 50,000 × g in a Beckman Type 60 Ti rotor, supernatants were loaded into Ni2+-nitrilotriacetic acid-agarose (NiNTA) columns (Qiagen) and washed, and the ZapD fusion protein was eluted in buffer containing 50 mM NaPO4, 0.3 M NaCl, and 250 mM imidazole (pH 8.0). The eluate was digested with the Smt3-specific protease Ulp1 during overnight dialysis in buffer (50 mM Tris-Cl, 150 mM NaCl, 1 mM dithiothreitol [DTT], 10% glycerol, pH 8.0) at 4°C (37). The released His10-Smt3 was removed by passing the dialysate over an NiNTA resin column. The flowthrough containing tag-free ZapD was collected and separated on a Sephadex 75 (GE) size exclusion column. Peak fractions were pooled and concentrated using an Amicon Ultra-15 filter with buffer exchange (50 mM MOPS [morpholinepropanesulfonic acid; pH 6.5], 50 mM KCl, 10 mM MgCl2, 0.2 mM DTT, 2% glycerol) and stored at −80°C. Aliquots of the protein in the storage buffer were separated on a 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel to obtain estimates of purification, and concentrations were determined by A280 measurements.
Endogenous ZapD levels.
Wild-type strain was grown to an OD600 of 0.7 to 0.8 at 37°C, at which point various dilutions were plated on LB agar to obtain CFU counts per milliliter. Whole-cell proteins from defined volumes of these cultures were harvested, serially diluted, separated on a 12.5% SDS-PAGE gel, and transferred onto nitrocellulose blots. Purified tag-free ZapD dilutions of known concentrations were run in parallel. Blots were probed with a peptide-derived anti-ZapD rabbit polyclonal antibody (Genscript), and intensities were measured by ImageJ (NIH) analyses.
FtsZ sedimentation assay.
Purified FtsZ (5 μM) was added to purified ZapD (1 or 5 μM) in FtsZ polymerization buffer (50 mM MOPS [pH 6.5], 50 mM KCl, 2.5 mM MgCl2), and GDP or GTP (1 mM) was added last as described previously (39). Reaction mixtures (100 μl) were processed at room temperature and spun down using a TLA100.2 rotor at 80,000 rpm for 12 min. At that point, 80 μl of supernatant was carefully removed and 4× loading dye was added to reach a final 1× concentration. The rest of the supernatant was discarded, and the pellets were resuspended in the original reaction volume buffer plus 1× loading dye (final concentration). The supernatants (10 μl) and pellets (10 μl) were resolved in a 12.5% SDS-PAGE gel.
FtsZ GTPase assay.
The GTPase activity of FtsZ was measured using an EnzCheck Free Phosphate Assay kit (Molecular Probes) essentially under the FtsZ sedimentation assay conditions described above. Purified FtsZ (5 μM) with and without purified ZapD (1 or 5 μM) was added to reaction mixtures in polymerization buffer (50 mM MOPS [pH 6.5], 50 mM KCl, 2.5 mM MgCl2) containing 1 mM GTP, and absorbance at 360 nm was monitored every 10 s for 30 min at room temperature. Standard curves with known amounts of phosphate were calculated to determine the amount of inorganic phosphate released.
Analysis of proteins.
Western blot analysis was performed using standard protocols unless otherwise mentioned. Proteins were prepared from cell pellets as described previously (27). Densities of protein bands were determined by ImageJ analysis (NIH).
RESULTS
Identification of ZapD (YacF) as a putative divisome component.
YacF, a previously uncharacterized protein, was identified as localizing to midcell in an FtsZ-dependent manner in a previously described protein localization screen conducted in E. coli (27). YacF (∼28.3 kDa) is predicted to be a nonessential cytoplasmic protein (DUF1342 superfamily), and orthologs are distributed in 191 species of proteobacteria (3). Sequence analysis of YacF does not reveal any substantial clues to its specific functions; neither does the structure of a close homolog (42% sequence identity, 66% homology) from Vibrio parahemolyticus (PDB:2OEZ) available from a structural genomic initiative (5). The yacF gene is located at the 2.4-min region of the E. coli chromosome adjacent to the cell wall synthesis and cell division mur-ftsZ gene cluster and is predicted to be cotranscribed with the upstream coaE encoding the essential dephospho-coenzyme A (dephospho-CoA) kinase and perhaps the downstream yacG encoding an endogenous DNA gyrase inhibitor, although yacG may have an independent promoter (34, 49, 55). Both coaE and yacF are also located in one polycistronic unit in closely related species. As we demonstrate below, the yacF gene product is a Z-associated protein, and we propose to rename the gene zapD.
ZapD-eYFP localizes to midcell in a manner dependent on FtsZ but not on FtsA, ZipA, ZapA, or ZapC.
To confirm results obtained from a protein localization microscopy screen, we monitored the localization pattern of a ZapD-eYFP fusion in an ftsZ84 (Ts) strain; cells carrying ftsZ84 form filaments with no visible constrictions at the restrictive temperature because of their inability to assemble an FtsZ ring at the cell division site (45). At the restrictive temperature, in the absence of FtsZ localization, ZapD-eYFP failed to localize to potential division sites and was diffusely distributed, indicating that FtsZ is required for the localization of ZapD to midcell (Fig. 1A and B).
Fig 1.
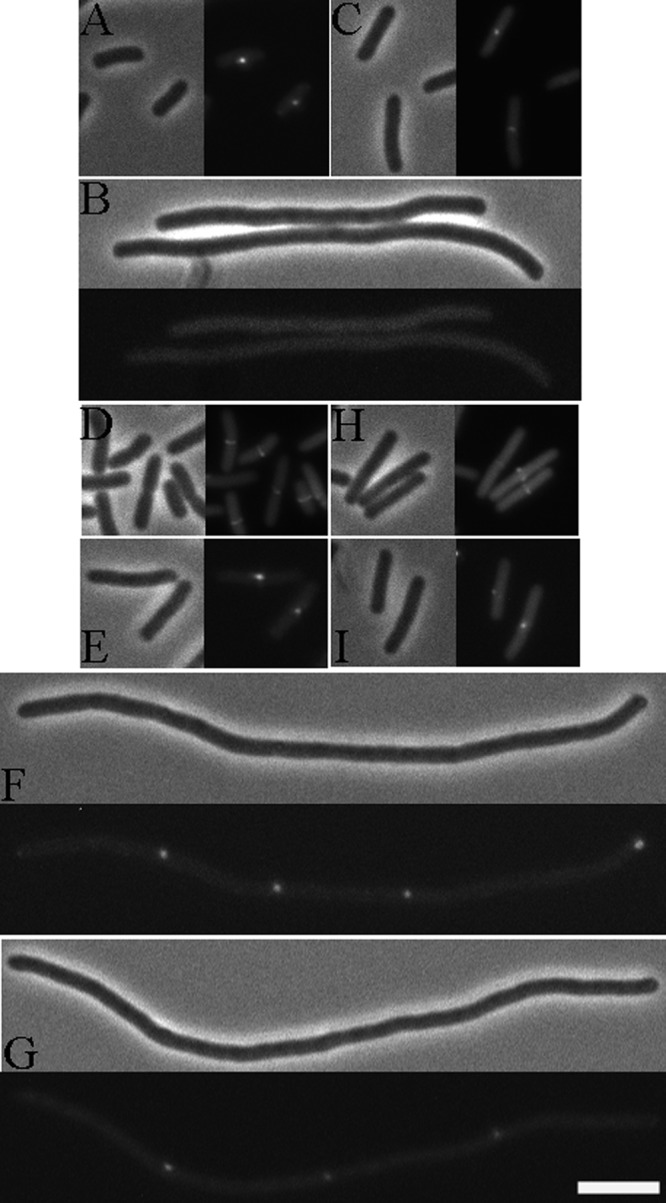
ZapD-eYFP localizes to midcell in an FtsZ-dependent manner. (A) Phase and fluorescent images of ZapD-eYFP localizing YFP to midcell at the permissive temperature (30°C) in ftsZ84 (Ts) cells. (B) Diffuse localization of ZapD-eYFP at the restrictive temperature (42°C) in ftsZ84 (Ts) cells. Strain EC307 carrying plasmid pDSW208-ZapD-eYFP was grown in LB at 30°C with 20 μM IPTG until an OD600 of ∼1.0 was reached, at which point cells were immobilized on agarose pads and visualized. For visualization at the restrictive temperature, the plasmid-bearing conditional strain was grown at 30°C with 20 μM IPTG until an OD600 of ∼0.2 was reached, at which point cells were diluted at 1:5 into fresh LB with IPTG, transferred to 42°C, and grown for an additional 1 h; then, filaments were immobilized on agarose pads for microscopy. (C) Phase and fluorescent images of ZapD-eYFP localizing to midcell in zapD cells. Strain JD310 carrying the plasmid pDSW208-ZapD-eYFP was grown in LB at 30°C with 20 μM IPTG until an OD600 of ∼1.0 was reached, at which point cells were immobilized on agarose pads and visualized. (D to G) Phase and fluorescent images of ZapD-eYFP localizing to midcell in ftsA12 and zipA1 cells, respectively, at permissive (30°C; D and E) and at restrictive (42°C; F and G) temperatures. Strains MDG148 and PS223, each carrying plasmid pDSW208-ZapD-eYFP, were grown in LB at the permissive temperature with 20 μM IPTG until an OD600 ∼1.0 of was reached, and cells were visualized on agarose pads. For growth at the restrictive temperature, cells were grown at 30°C in LB with 20 μM IPTG until an OD600 of ∼0.2 was reached, at which point cells were diluted at 1:5 in fresh LB with IPTG, transferred to 42°C, and grown for another hour, at which point cells were immobilized on agarose pads and visualized. (H and I) Phase and fluorescent images of ZapD-eYFP fusion localizing to midcell in a ZapA- and ZapC-independent manner. Strain JD257 and strain JD317 bearing plasmid pDSW208-ZapD-eYFP were each grown at 30°C with 20 μM IPTG until an OD600 of ∼1.0 was reached, at which point cells were visualized on agarose pads. Bar, 5 μm. Note that localization of ZapD-eYFP as foci instead of bands in certain strain backgrounds appears to be a property of the particular fusion protein used in this study.
To elucidate the divisome components that are required for the recruitment of ZapD to midcell, we visualized ZapD-eYFP localization in cells lacking early septal ring components, namely, FtsA, ZipA, ZapA, and ZapC. ZapD-eYFP localized to potential division sites in FtsA- or ZipA-depleted filaments at the restrictive temperature (Fig. 1D to G) and to midcell in cells lacking ZapA, ZapC, or ZapD (Fig. 1H, I, and C), indicating that FtsA, ZipA, ZapA, and ZapC are dispensable for the localization of ZapD to midcell.
ZapD interacts with the conserved C-terminal tail of FtsZ.
Since ZapD and FtsZ colocalize at midcell, we examined whether ZapD interacts with FtsZ by the use of the yeast PIP assay as described previously (14). Briefly, the PIP assay relies on the reoviral scaffolding protein μNS, which forms large focal inclusions in yeast. When a query protein is fused to μNS and a likely interaction partner fused to a fluorophore, interactions can be identified by screening for yeast that form fluorescent foci. In yeast expressing both μNS-FtsZ and green fluorescent protein-ZapD (GFP-ZapD), we found that ZapD interacts with FtsZ, as evidenced by the formation of fluorescent focal platforms (Fig. 2B). The interaction between FtsZ and ZapD was also detected when FtsZ was fused to the fluorophore and ZapD to μNS (Fig. 2D). While GFP-ZapD alone was diffusely localized (Fig. 2A), yeast coexpressing GFP-ZapD and μNS-ZapD displayed fluorescent focal platforms (Fig. 2C). In control reactions, both FtsZ and ZapD formed characteristic focal platforms when fused to μNS whereas GFP-FtsZ expressed alone was diffusely localized (see Fig. S1 in the supplemental material). These results suggest a direct interaction between FtsZ and ZapD and that ZapD likely forms homo-oligomers in the cell.
Fig 2.
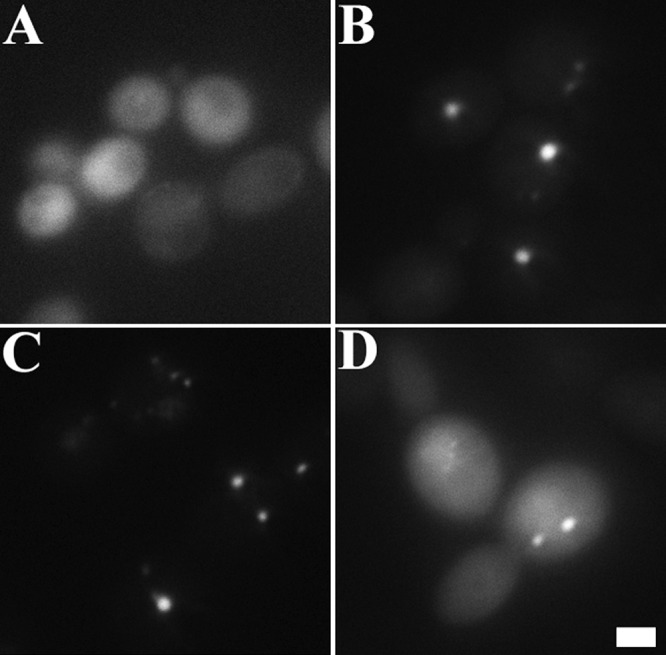
ZapD interacts with FtsZ and with itself in a protein interaction platform yeast assay. (A) Yeast cells expressing GFP-ZapD showed diffusely localized fluorescence at 3 to 4 h postinduction of fusion protein; (B) coexpression of both GFP-ZapD and μNS-FtsZ fusion proteins displayed fluorescent foci; (C) coexpression of GFP-ZapD and μNS-ZapD fusions also displayed focal interactions; (D) yeast cells coexpressing FtsZ fused to GFP and ZapD fused to μNS also displayed fluorescent focal platforms. Bar, 2 μm.
To narrow down the region of FtsZ that interacts with ZapD, we next tested the ability of ZapD to interact with various deletions of FtsZ fused to μNS in the yeast PIP assay. The deletion constructs used were FtsZ1–372 (lacking the conserved C-terminal tail), FtsZ1–314 (lacking the conserved tail and the variable linker domain), FtsZ289–383 (lacking the N-terminal and part of the conserved C-terminal globular domains), and FtsZ364–383 and FtsZ374–383 (carrying various lengths of the conserved C-terminal tail). Because the 17-amino-acid C-terminal tail of FtsZ has been shown to be essential in its interactions with other FtsZ stabilizers such as FtsA and ZipA (24, 26, 32), we hypothesized that this domain might be important for ZapD binding. Indeed, we find that, analogous to the interactions of FtsA and ZipA, ZapD requires the conserved C-terminal FtsZ tail for its interaction with FtsZ (Table 2).
Table 2.
The conserved C-terminal tail of FtsZ is required for ZapD interaction
| μNS fusiona | Interaction with GFP-ZapDb |
|---|---|
| FtsZ1–383 | + |
| FtsZ1–372 | − |
| FtsZ1–314 | − |
| FtsZ289–383 | + |
| FtsZ364–383 | + |
| FtsZ374–383 | + |
Full-length FtsZ and various FtsZ deletions fused to the μNS reoviral scaffolding protein were coexpressed with ZapD fused to GFP in yeast cells and visualized 3 to 4 h postinduction.
+ indicates yeast cells displaying fluorescent foci representing interactions between ZapD and the relevant FtsZ construct. − indicates diffuse localization of GFP-ZapD, likely representing no interaction between ZapD and the relevant FtsZ construct.
Overexpression of ZapD leads to filamentation.
The localization and protein-protein interaction data determined with ZapD suggested that ZapD may be a cell division factor that modulates FtsZ assembly. Therefore, we explored the in vivo effects of overexpression of ZapD in wild-type cells. Uninduced expression of ZapD from the leaky T5/lac promoter of pCA24N led to significant filamentation, with highly aberrant FtsZ assemblies (Fig. 3C and D). Medial FtsZ rings were observed in wild-type cells grown under the same conditions and bearing the pCA24N vector alone (Fig. 3A and B). The cellular FtsZ levels remained essentially unchanged upon overexpression of ZapD (see Fig. S2 in the supplemental material). These data suggest that overexpression of ZapD interferes with cell division, perhaps through modulating FtsZ polymer assembly.
Fig 3.
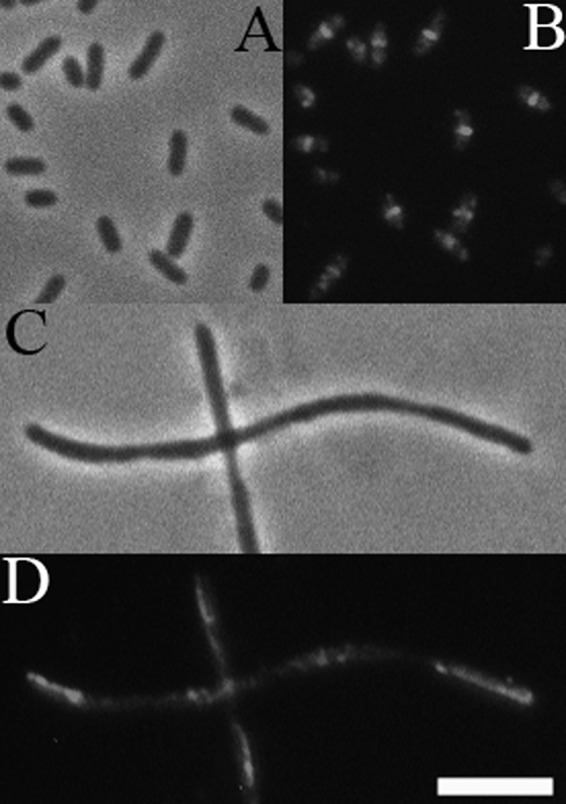
Overexpression of ZapD interferes with FtsZ ring assembly. (A and B) Phase and fluorescent images of pCA24N vector alone in wild-type cells. Medial FtsZ rings are detected by indirect immunofluorescence with an anti-FtsZ antibody in GF73 cells. (C and D) Leaky expression from pCA24N-zapD construct leads to formation of long nonseptate filaments with aberrant FtsZ assemblies in wild-type cells. Loose helices and long rod-shaped patches of FtsZ are seen in JD381 cells overexpressing ZapD. Bar, 5 μm.
Loss of ZapD enhances the division defects of the ftsZ84 conditional allele.
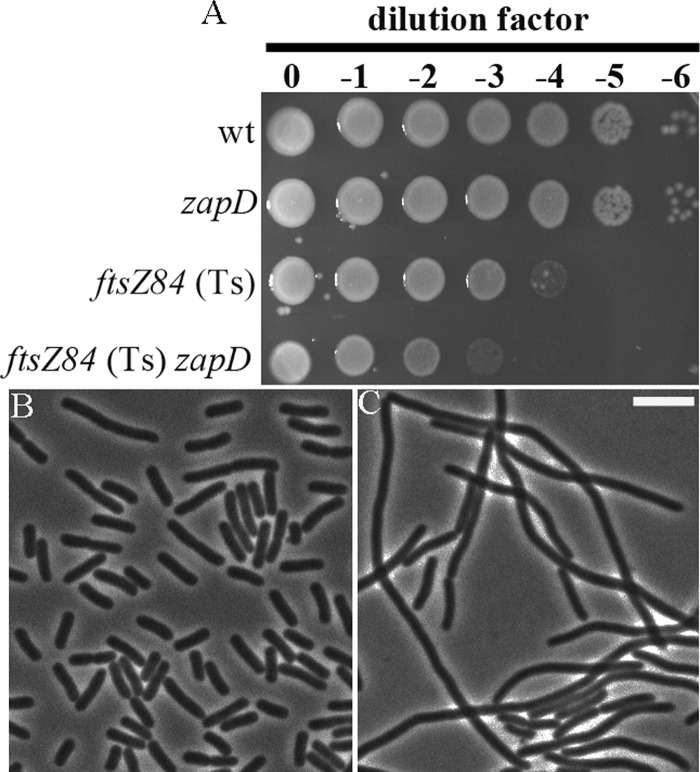
While overexpression of ZapD interferes with cell division, cells lacking zapD are viable, with no discernible division phenotypes. In light of ZapD's dependence on FtsZ but not on the other early assembly division proteins for localization to the divisome, we examined whether ZapD has a biological role in the early stages of cell division. We determined the synergistic effects of removal of zapD in cells carrying the ftsZ84 (Ts) allele. Cells with the conditional ftsZ84 (Ts) allele are filamentous at the restrictive temperature at 42°C because of the inability of FtsZ to localize to division sites. We observed that ftsZ84 (Ts) zapD cells exhibited a growth defect in LB no-salt agar, a more stringent condition for controlling ftsZ84 levels, as ftsZ expression increases at higher salt concentrations (Fig. 4A) (54). The synthetic sick phenotype of ftsZ84 (Ts) zapD was further supported by visualization of extensively filamentous cells at the permissive temperature (30°C) in comparison to ftsZ84 (Ts) cells alone (Fig. 4B). The observed phenotypic changes in ftsZ84 (Ts) zapD cells were not a result of altered FtsZ levels due to the lack of ZapD (see Fig. S2 in the supplemental material).
Fig 4.
Synthetic sick phenotype of ftsZ84 (Ts) cells in the absence of zapD. (A) Overnight cultures of wild-type (wt; TB28), zapD (JD310), ftsZ84 (Ts) (EC307), and ftsZ84 (Ts) zapD (JD330) cells grown in LB at 30°C were normalized to an OD600 of 2.0 and serially diluted, and 6-μl aliquots were spotted on LB no-salt agar plates and incubated overnight at 30°C. The relative plating efficiencies were determined by comparison with control strains. (B and C) Phase images of ftsZ84 (Ts) (EC307) and ftsZ84 (Ts) zapD (JD330) cells. Overnight cultures grown in LB at 30°C were subcultured at 1:100 to an OD600 of 0.6 to 0.8. Samples were immobilized on agarose pads and visualized by microscopy. Bar, 5 μm.
ZapD shares an overlapping function with ZapA.
Further evidence for a role for ZapD in promoting FtsZ assembly comes from synergistic studies of ZapD and a known FtsZ cross-linking and bundling protein, ZapA. Cells lacking both ZapA and ZapD were longer on average than mutants with a zapA or zapD mutation alone, with significant variations in cell lengths seen across the population (Table 3). The elongation phenotype of the double mutant could be rescued by providing ZapD in trans, showing that the phenotype was due to lack of ZapD and not to polar downstream effects on yacG expression (Table 3). The complementing vector alone did not rescue the zapA zapD phenotype in trans (data not shown). These results indicate that ZapD shares a functional role with ZapA in FtsZ bundling during the early stages of division. Introduction of a zapB allele did not affect the cell lengths significantly (data not shown). Interestingly, cells lacking zapC and zapD did not display a significant increase in cell length beyond that of cells lacking zapC alone (Table 3). However, removal of both zapC and zapD in the absence of ZapA resulted in very poor viability, increased filamentation, and a wider distribution of cell lengths across the cell population, indicating an overlapping function for ZapD in enhancing Z-ring stability (Table 3).
Table 3.
Synergism of zapD with known Z-associated (Zap) proteins
| Genotypea | Cell length (μm ± SD) | No. of cells measuredb |
|---|---|---|
| Wild type | 3.4 ± 1.1 | 604 |
| zapA | 4.0 ± 1.9 | 605 |
| zapC | 3.9 ± 1.6 | 601 |
| zapD | 3.2 ± 0.9 | 601 |
| zapA zapD | 5.2 ± 3.7 | 602 |
| zapA zapD pNG162-zapDc | 4.2 ± 2.1 | 640 |
| zapC zapD | 4.2 ± 1.3 | 618 |
| zapA zapC | 6.2 ± 5.2 | 545 |
| zapA zapC zapD | 7.0 ± 6.3 | 625 |
The wild-type strain was TB28, and isogenic single (zapA = JD257; zapC = JD317; zapD = JD310)-, double (zapA zapC = JD305; zapA zapD = JD315; zapC zapD = JD345)-, and triple (zapA zapC zapD = JD360)-mutant strains were constructed by P1 transduction.
Data represent total numbers of cells measured for length using ObjectJ (NIH). Cells were grown in LB broth at 30°C to an OD600 of 0.6 to 0.8, at which point samples were immobilized on agarose pads and visualized by phase microscopy.
Strain AJ18-79 was subcultured at a dilution of 1:100 in LB with IPTG added to reach a 50 μM final concentration at 30°C and grown to an OD600 of 0.6 to 0.8, at which point cells were visualized by phase microscopy.
Purified ZapD is dimeric in solution.
The genetics and cell biological data are consistent with a role for ZapD in promoting FtsZ assembly in E. coli. To test this possibility in vitro, we purified both proteins to conduct sedimentation, electron microscopy, and GTPase assays. To avoid tag-related artifacts in determining FtsZ-ZapD interactions, we purified tag-free ZapD to ≥95% purity (see Fig. S3A in the supplemental material). Gel filtration data show that a majority of purified ZapD eluted at ∼44 ± 6 kDa (see Fig. S3B in the supplemental material). Based on the predicted molecular mass of ZapD, these data suggest that purified ZapD is likely to be mostly dimeric in solution, with some higher-order oligomers also being present.
ZapD interacts directly with FtsZ and promotes FtsZ assembly in vitro.
In order to determine whether ZapD and FtsZ interact directly, we conducted an FtsZ sedimentation assay. To obtain approximate cellular levels of ZapD for comparison with levels of FtsZ, we determined the intracellular concentrations of ZapD by the use of quantitative Western blotting with an anti-ZapD polyclonal antibody. Approximately 500 to 800 molecules of ZapD were calculated to be present per cell during the logarithmic phase, indicating that ZapD is a low-abundance protein and that its cellular concentration is approximately one-fifth of previously published FtsZ cellular concentrations (35, 47).
Purified FtsZ (5 μM) and ZapD (1 or 5 μM) were incubated in the presence of GDP or GTP in polymerization reactions. In control reactions in the absence of ZapD and in the presence of GDP, FtsZ was mostly present in the supernatant (Fig. 5). Upon addition of GTP, polymeric FtsZ was present in the pellet (Fig. 5). However, in the presence of GTP and either 1 or 5 μM ZapD, ∼5.5- or 13-fold-more FtsZ was present in the pellet, respectively (Fig. 5A and B) and ZapD copelleted with FtsZ (Fig. 5A). In the absence of FtsZ, ZapD (5 μM) alone was present, mostly in the supernatant, with either GTP or GDP (Fig. 5). These sedimentation results are consistent with the yeast protein-protein interaction data and indicate that ZapD directly binds and pellets FtsZ.
Fig 5.
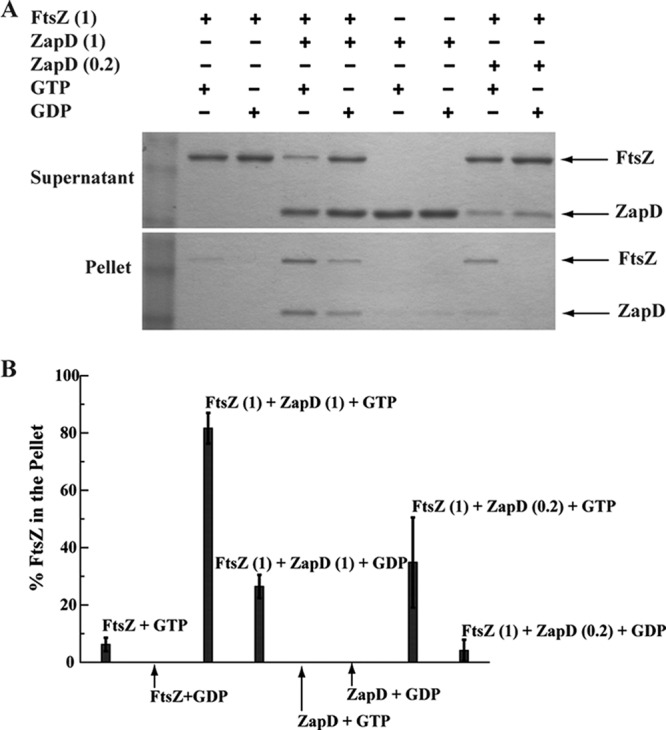
ZapD interacts directly with FtsZ in vitro and pellets FtsZ. (A) Sedimentation reaction mixtures of FtsZ and ZapD in buffer containing MOPS (pH 6.5), 50 mM KCl, and 2.5 mM MgCl2 with 1 mM GTP or GDP were spun, and equivalent aliquots of supernatant and pellet fractions were separated on an SDS-PAGE gel and visualized by staining. Where present, FtsZ was at 5 μM and ZapD at 1 or 5 μM. (B) Amounts of FtsZ present in pellets in reaction mixtures with and without ZapD in the presence of GDP or GTP are shown as averages of the results of 3 independent experiments. The percentage of FtsZ in each pellet was quantified based on band intensities of total FtsZ present in the pellet and supernatant fractions.
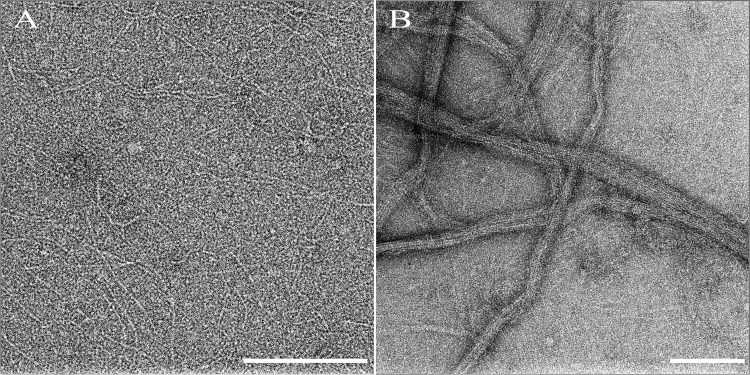
To ascertain the morphology of the polymeric state of FtsZ in the sedimentation assays, reactions were further visualized by transmission electron microscopy. Characteristic protofilaments of FtsZ were visualized upon addition of GTP (Fig. 6A), and no polymers were visualized in the presence of GDP (data not shown). Addition of substoichiometric amounts of ZapD to FtsZ in the presence of GTP did not significantly alter the morphology of the FtsZ protofilaments (see Fig. S4 in the supplemental material). However, upon addition of equimolar ZapD to FtsZ in reaction mixtures containing GTP, extensive bundling of FtsZ protofilaments was seen by electron microscopy (Fig. 6B). Taken together, these data indicate that ZapD promotes FtsZ bundling through specific stabilizing interactions.
Fig 6.
ZapD promotes FtsZ-polymer bundling. (A) Negative stained TEM images of FtsZ (5 μM) in buffer containing MOPS (pH 6.5), 50 mM KCl, and 2.5 mM MgCl2 in the presence of 1 mM GTP show characteristic single protofilaments. (B) Upon addition of ZapD (5 μM), large bundled FtsZ protofilaments were visualized. Bar, 200 nm.
The intrinsic GTPase activity of FtsZ is reduced due to ZapD-mediated FtsZ bundling.
Bundling of FtsZ leads to a decrease in the rate of GTP hydrolysis compared to the rate seen with FtsZ protofilaments alone (38, 40). To examine whether ZapD-mediated FtsZ bundling led to changes in the intrinsic GTPase activity of FtsZ polymers, we measured the GTP hydrolysis rate of FtsZ in the presence and absence of ZapD by the use of a spectrophotometric method for quantifying free inorganic phosphate (22). The GTP hydrolysis rate of 5 μM FtsZ under the conditions tested was ∼2 GTP per FtsZ per min (Fig. 7). We suspect that the slightly lower FtsZ GTP hydrolysis rate determined in this study compared to those reported by others (7) can in part be attributed to differences in the temperatures at which the assays were conducted. Addition of 1 μM ZapD to the reaction did not alter the FtsZ GTPase rates significantly (data not shown). However, addition of 5 μM ZapD to the reaction led to a reduction in the GTP hydrolysis rate of FtsZ of ∼60% (Fig. 7), consistent with the ability of ZapD to promote lateral bundling of FtsZ. ZapD alone did not exhibit measurable GTPase activity (Fig. 7). These data indicate that ZapD-mediated FtsZ bundling lowers the GTP hydrolysis rate of FtsZ.
Fig 7.
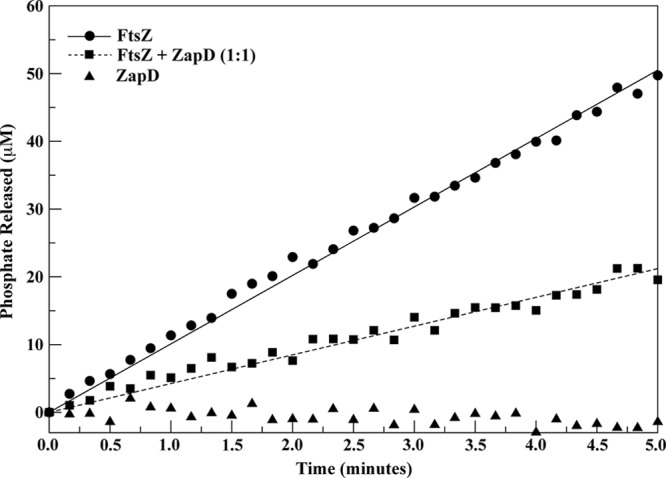
ZapD reduces FtsZ GTPase activity. The GTPase activity of FtsZ was measured using a spectrophotometric method to quantify the amount of released phosphate. Experimental data are represented as the GTPase activity of 5 μM FtsZ in buffer containing MOPS (pH 6.5), 50 mM KCl, and 2.5 mM MgCl2 in the absence of ZapD (filled circles) and in the presence of 5 μM ZapD (filled squares) and as the GTPase activity of 5 μM ZapD in the absence of FtsZ (filled triangles). Data were collected at room temperature every 10 s for a total of 30 min. The fully linear part of the curve was determined as the part with a constant first derivative in time. All curves were found to be essentially linear until approximately 6 min. The data for the first 30 s were discarded for stabilization, and data points from 0.5 to 5.5 min were used in a linear fit to determine the GTPase activity. The straight lines represent linear fits to the experimental data. In all reaction mixtures, GTP was at 1 mM and FtsZ was added last. Standard curves with known concentrations of phosphate were used to convert the A360 readings to phosphate production data. In this representative curve, the GTPase activity of FtsZ alone is 2.02 ± 0.02 GTP/FtsZ/min and FtsZ in the presence of ZapD at a 1:1 ratio is 0.85 ± 0.01 GTP/FtsZ/min.
DISCUSSION
ZapD is a cell division protein that enhances FtsZ assembly in E. coli.
This study provides evidence that ZapD is a bona fide component of the midcell division apparatus in E. coli. ZapD requires FtsZ to be recruited to midcell, where it directly binds and bundles FtsZ polymers. Overexpression of ZapD leads to aberrant FtsZ assemblies and formation of long nonseptate filaments. Although the removal of ZapD does not present any discernible phenotype in terms of cell division or viability in wild-type cells, in cells containing ftsZ84, a heat-sensitive allele with weak GTPase and polymerization activities, removal of ZapD causes division and growth defects similar to those observed in ftsZ84 (Ts) zapA cells (18). These results, coupled with division defects seen in zapA zapD double mutants and zapA zapC zapD triple mutants, indicate that ZapD-mediated FtsZ assembly plays a positive role in maintaining the integrity of the FtsZ ring in the cell.
The ZapD docking site on FtsZ overlaps the FtsZ interaction site of other FtsZ-modulatory proteins, namely, FtsA, ZipA, MinC, and ClpX (8, 26, 32, 50). However, some of the FtsZ residues implicated as being important for FtsZ-FtsA and FtsZ-ZipA interactions are dispensable for ZapD binding. This result indicates that FtsA and ZipA, which have other critical roles in addition to promoting FtsZ-ring stability, and ZapD, whose sole apparent function appears to be in enhancing FtsZ assembly, have different modes of binding the C-terminal tail of FtsZ (26, 32).
Protein-protein interaction data and the properties of purified ZapD in solution (as well as the presence of two molecules in the asymmetric unit for the putative structural homolog from V. parahemolyticus) suggest self-interaction among ZapD molecules. The simplest interpretation of oligomerization of ZapD is that two molecules of ZapD interact with adjacent FtsZ protofilaments, enhancing their lateral association at midcell. However, at this point we cannot exclude the possibility that ZapD dimers promote Z-ring stability by also enhancing the longitudinal interactions in an individual FtsZ protofilament.
Z-associated proteins may promote FtsZ bundling via distinct molecular mechanisms.
Precisely how the Z-associated proteins, which share no primary sequence homology, perform the same overall function of promoting FtsZ bundling is not completely understood. Both ZapC and ZapD share overlapping functions with ZapA in stabilizing the FtsZ-ring structure, and yet zapC zapD double mutants do not exhibit marked changes in cell lengths compared to cells lacking zapC alone. These data therefore suggest that ZapC and ZapD do not have completely redundant roles in stabilizing the FtsZ ring. While the widely conserved ZapA has been shown to promote both cross-linking and bundling of FtsZ protofilaments (11), roles for ZapC and ZapD in cross-linking of FtsZ protofilaments remain to be explored. Additionally, while ZapC and ZapD both bind and bundle FtsZ in vitro, the FtsZ GTPase activity is markedly reduced in ZapC-mediated bundles in contrast to ZapD-mediated FtsZ bundles at substoichiometric ratios (25). This difference in FtsZ GTPase activities can be due to a first-order effect where ZapC directly reduces the GTPase activity of FtsZ or due to a second-order effect where the decrease in GTPase activity can be attributed to ZapC being a more robust FtsZ bundler than ZapD. Taken together, the genetics and bundling data suggest that ZapC and ZapD may employ distinct molecular mechanisms in achieving FtsZ-ring stability. While the precise ZapC docking site in FtsZ has not yet been determined, results suggest that it is not likely to be in the conserved C-terminal tail of FtsZ (S. Chaudhry and A. Janakiraman, unpublished results). Hence, it may be hypothesized that since ZapC and ZapD appear to bind different regions of FtsZ, they are also likely to be distinct in the precise molecular mechanisms by which they achieve FtsZ bundling. Such variations in modulating the establishment of a stable FtsZ ring, a critical step during division, likely serve as “fail-safe” mechanisms for the host in the natural environment.
Roles for Zap-mediated FtsZ bundling.
Similarly to FtsZ stabilizers ZipA, ZapB, and ZapC in E. coli, ZapD is largely restricted to the Gammaproteobacteria class of Gram-negative bacteria. With the characterization of ZapD as a division factor, the number of proteins that have been shown to interact directly with FtsZ and enhance FtsZ assembly in E. coli division has risen to five. Why E. coli would retain so many proteins with a similar apparent function of promoting FtsZ bundling is not clear. Perhaps, one lead in answering this question comes from a recent study that proposes a new role for the FtsZ C-terminal tail in mediating stabilizing lateral interactions among FtsZ protofilaments independent of FtsZ-associated proteins such as ZapA (7). Species-specific differences in residues at the very end of the FtsZ conserved C-terminal tail are proposed to enhance the innate bundling capacity of B. subtilis FtsZ compared to that of E. coli FtsZ (7). In light of these findings and prior studies (10, 36), which support a critical role for FtsZ lateral interactions in vivo, we suggest that perhaps the abundance of FtsZ bundlers in E. coli compensates for a relatively weak inherent bundling capacity of E. coli FtsZ and thereby affords greater division efficiency in vivo.
Furthermore, an interesting possibility is that Z-associated proteins such as ZapD enhance FtsZ-ring stability under specific environmental conditions where the fitness of the established division apparatus becomes especially critical to the host. While FtsZ appears to be regulated at the level of assembly/disassembly, little is known of whether and, if so, how Zap proteins may themselves be regulated under various environmental conditions. While each of the Z-associated proteins ZapA, ZapB, and ZapC is located as a single gene unit on the E. coli chromosome, the gene arrangement of zapD in the coaE-zapD and perhaps in the yacG operon presents an intriguing scenario where zapD may itself be regulated by an as-yet-unknown mechanism.
Ultimately, the identification and characterization of ZapD underscore the diversity in regulatory proteins that modulate FtsZ assembly and should aid in developing a more comprehensive understanding of FtsZ structure and dynamics during the early stages of cell division.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Bernhardt, Joe Lutkenhaus, and Stewart Shuman for generous gifts of strains, Ronnie Ghose for helpful suggestions with protein purification, the New York Structural Biology Center for use of transmission electron microscopy facilities, Helen Yu for technical assistance, and Manjula Reddy and Petra Levin for critical reading of the manuscript.
This work was supported by NIGMS SC2GM082336 (to A.J.) and NIH/NCRR/RCMI infrastructural grant 2 G12 RR003060-26A1 (to CCNY).
Footnotes
Published ahead of print 13 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adams D, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 2. Addinall SG, Lutkenhaus J. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 39:D289–D294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Badger J, et al. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60:787–796 [DOI] [PubMed] [Google Scholar]
- 6. Bernhardt T, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buske PJ, Levin PA. 1 February 2012, posting date The extreme C-terminus of the bacterial cytoskeletal protein FtsZ plays a fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. [Epub ahead of print.] doi:10.1074/jbc.M111.330324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camberg JL, Hoskins JR, Wickner S. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen JC, Beckwith J. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395–413 [DOI] [PubMed] [Google Scholar]
- 10. Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18:235–244 [DOI] [PubMed] [Google Scholar]
- 11. Dajkovic A, Pichoff S, Lutkenhaus J, Wirtz D. 2010. Cross-linking FtsZ polymers into coherent Z rings. Mol. Microbiol. 78:651–668 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Boer PA. 2010. Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13:730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68:720–735 [DOI] [PubMed] [Google Scholar]
- 16. Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74:504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galli E, Gerdes K. 2010. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 76:1514–1526 [DOI] [PubMed] [Google Scholar]
- 18. Galli E, Gerdes K. 2012. The FtsZ-ZapA-ZapB interactome of Escherichia coli. J. Bacteriol. 194:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geissler B, Elraheb D, Margolin W. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 100:4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goehring NW, Gonzalez MD, Beckwith J. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61:33–45 [DOI] [PubMed] [Google Scholar]
- 21. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52:801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185 [DOI] [PubMed] [Google Scholar]
- 24. Hale CA, Rhee AC, de Boer PA. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hale CA, et al. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193:1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haney SA, et al. 2001. Genetic analysis of the Escherichia coli FtsZ. ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980–11987 [DOI] [PubMed] [Google Scholar]
- 27. Janakiraman A, Fixen KR, Gray AN, Niki H, Goldberg MB. 2009. A genome-scale proteomic screen identifies a role for DnaK in chaperoning of polar autotransporters in Shigella. J. Bacteriol. 191:6300–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janakiraman A, Goldberg MB. 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. U. S. A. 101:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirkpatrick CL, Viollier PH. 2011. New(s) to the (Z-)ring. Curr. Opin. Microbiol. 14:691–697 [DOI] [PubMed] [Google Scholar]
- 30. Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 31. Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 93:12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma X, Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mingorance J, Rivas G, Vélez M, Gómez-Puertas P, Vicente M. 2010. Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18:348–356 [DOI] [PubMed] [Google Scholar]
- 34. Mishra P, Park PK, Drueckhammer DG. 2001. Identification of yacE (coaE) as the structural gene for dephosphocoenzyme A kinase in Escherichia coli K-12. J. Bacteriol. 183:2774–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohammadi T, et al. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monahan L, Robinson A, Harry E. 2009. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 74:1004–1017 [DOI] [PubMed] [Google Scholar]
- 37. Mossessova E, Lima CD. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865–876 [DOI] [PubMed] [Google Scholar]
- 38. Mukherjee A, Lutkenhaus J. 1999. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181:823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukherjee A, Lutkenhaus J. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pacheco-Gómez R, Roper DI, Dafforn TR, Rodger A. 2011. The pH dependence of polymerization and bundling by the essential bacterial cytoskeletal protein FtsZ. PLoS One 6:e19369 doi:10.1371/journal.pone.0019369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722–1734 [DOI] [PubMed] [Google Scholar]
- 42. Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pichoff S, Shen B, Sullivan B, Lutkenhaus J. 2012. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 83:151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. RayChaudhuri D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. RayChaudhuri D, Park JT. 1994. A point mutation converts Escherichia coli FtsZ septation GTPase to an ATPase. J. Biol. Chem. 269:22941–22944 [PubMed] [Google Scholar]
- 46. Romberg L, Simon M, Erickson HP. 2001. Polymerization of FtsZ, a bacterial homolog of tubulin, is assembly cooperative? J. Biol. Chem. 276:11743–11753 [DOI] [PubMed] [Google Scholar]
- 47. Rueda S, Vicente M, Mingorance J. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitz A, Morrison M, Agunwamba A, Nibert ML, Lesser C. 2009. Protein interaction platforms: visualization of interacting proteins in yeast. Nat. Methods 6:500–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sengupta S, Nagaraja V. 2008. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 36:4310–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen B, Lutkenhaus J. 2009. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol. Microbiol. 72:410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Silhavy TJ, Berman ML, Enquist LW. 1984. Genetic transduction using P1vir, p 107–112 In Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U. S. A. 99:3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weart RB, Levin PA. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185:2826–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu XC, Margolin W. 2000. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol. 182:6203–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zaslaver A, et al. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3:623–628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.