Abstract
Oxalate catabolism is conducted by phylogenetically diverse organisms, including Methylobacterium extorquens AM1. Here, we investigate the central metabolism of this alphaproteobacterium during growth on oxalate by using proteomics, mutant characterization, and 13C-labeling experiments. Our results confirm that energy conservation proceeds as previously described for M. extorquens AM1 and other characterized oxalotrophic bacteria via oxalyl-coenzyme A (oxalyl-CoA) decarboxylase and formyl-CoA transferase and subsequent oxidation to carbon dioxide via formate dehydrogenase. However, in contrast to other oxalate-degrading organisms, the assimilation of this carbon compound in M. extorquens AM1 occurs via the operation of a variant of the serine cycle as follows: oxalyl-CoA reduction to glyoxylate and conversion to glycine and its condensation with methylene-tetrahydrofolate derived from formate, resulting in the formation of C3 units. The recently discovered ethylmalonyl-CoA pathway operates during growth on oxalate but is nevertheless dispensable, indicating that oxalyl-CoA reductase is sufficient to provide the glyoxylate required for biosynthesis. Analysis of an oxalyl-CoA synthetase- and oxalyl-CoA-reductase-deficient double mutant revealed an alternative, although less efficient, strategy for oxalate assimilation via one-carbon intermediates. The alternative process consists of formate assimilation via the tetrahydrofolate pathway to fuel the serine cycle, and the ethylmalonyl-CoA pathway is used for glyoxylate regeneration. Our results support the notion that M. extorquens AM1 has a plastic central metabolism featuring multiple assimilation routes for C1 and C2 substrates, which may contribute to the rapid adaptation of this organism to new substrates and the eventual coconsumption of substrates under environmental conditions.
INTRODUCTION
In nature, oxalate is found in many higher plant families in the form of calcium oxalate crystals (29). Some plants can accumulate these crystals at levels of up to 80% (wt/wt) of their dry weight; in the majority of plant organs, they are found as intracellular or extracellular deposits. Various functions, including defense mechanisms, the regulation of calcium levels in tissue and organs, and the detoxification of aluminum and other heavy metals, have been described for oxalate (29). Oxalate is produced in large quantities not only by plants but also by different classes of fungi (12, 23). The functions of oxalate in fungi include pathogenesis during plant infection, competition between fungi, and control of environmental nutrients and toxins (23). Oxalate is likely to be available as a nutrient for plant-associated microorganisms and particularly for those microorganisms found in the soil during the decay of plant material. Although calcium oxalate, which is likely to predominate in nature, is less soluble than potassium oxalate, it supports growth of bacteria in soil (11). With an oxidation number of plus three, oxalate is the most highly oxidized two-carbon compound known, and only two electrons are available. Although oxalate is a rather “poor” substrate, a number of oxalotrophic bacteria have been isolated from various ecological niches, including terrestrial (10) and aquatic (64) habitats and the gastrointestinal tract, under both aerobic and anaerobic conditions. Phylogenetically, oxalotrophic bacteria belong to distinct groups (33, 55–57).
The utilization of oxalate has been studied in Oxalobacter formigenes and Cupriavidus oxalaticus (formerly Pseudomonas oxalaticus [67]), both members of the Burkholderiales. O. formigenes was found in the human gut and those of other warm-blooded animals (3), and its presence is known to inhibit the formation of kidney stones (2, 60). C. oxalaticus was isolated from the intestine of an Indian earthworm (32), and its oxalate metabolism was investigated by Quayle and coworkers in the 1960s (48–54). The two organisms convert oxalate to formate by using oxalyl-coenzyme A (oxalyl-CoA) decarboxylase and formyl-CoA transferase; both enzymes have been previously characterized and identified (6, 6a, 42, 48, 49, 61, 65). Although C. oxalaticus is able to oxidize formate to carbon dioxide with the generation of redox equivalents (51), it remains unclear how the generation of reductant is accomplished in the anaerobe O. formigenes (20). Both organisms use the glycolate pathway for the assimilation of oxalate into biomass (21, 53, 55), a pathway that is also referred to as the glycerate pathway (20). By this pathway, oxalate is reduced to glyoxylate and then converted into glycerate by glyoxylate carboligase and tartronic semialdehyde reductase.
The pink-pigmented facultative methylotrophs comprise another group of oxalotrophs that has been less well studied regarding oxalotrophic growth. These bacteria are known as major plant colonizers (22, 37) but have also been isolated from soil, dust, and lake sediments (31). Plant-associated methylotrophs utilize methanol, which is released by plants during cell wall synthesis, as a source of carbon and energy. Methylotrophy has been well studied in the past 50 years (4, 14); however, methanol is not the only carbon substrate available for Methylobacterium during phyllosphere colonization (66). In fact, oxalate may function as an important carbon source in plant-associated habitats and/or in soil during the decay of plant material. In the model strain Methylobacterium extorquens AM1, Blackmore and Quayle detected oxalyl-CoA decarboxylase and oxalyl-CoA reductase activities in cell extracts, but glyoxylate carboligase was absent. Instead, serine cycle enzymatic activities, i.e., serine glyoxylate aminotransferase and hydroxypyruvate reductase, were found, and oxalate assimilation via some variant of the serine cycle has been suggested (8). Due to the specific activity of oxalyl-CoA reductase in M. extorquens AM1 being 20-to-30-fold lower than in oxalate-grown C. oxalaticus, it remained unclear whether oxalate reduction is the major pathway for glyoxylate generation (8). The recently discovered ethylmalonyl-CoA (EMC) pathway (1, 24–28), which operates in methylotrophs during the assimilation of C1 compounds (45) and acetyl-CoA (59) allowing for glyoxylate regeneration, was unknown when M. extorquens AM1 was first studied during oxalotrophic growth (8), and its function during oxalate utilization has not yet been investigated. This was the first study using a systems-level approach, including metabolomics, proteomics, and flux balance analysis in parallel with complementary mutant characterization, to elucidate the central metabolism of M. extorquens AM1 grown on oxalate.
MATERIALS AND METHODS
Chemicals.
[13C]sodium oxalate (99%) was purchased from Cambridge Isotope Laboratories; all other chemicals were purchased from Sigma.
Medium composition and cultivation conditions.
M. extorquens AM1 was grown on minimal medium (34) supplemented with 20 mM potassium oxalate in shake flasks, or cultures were grown in a 500-ml bioreactor (Infors-HT) with a working volume of 400 ml at 28°C, an aeration rate of 0.2 liters/min, stirring at 1,000 rpm, and 5 mM potassium oxalate. The pH of the bioreactor medium was maintained at 7.0 by the addition of oxalic acid (1 M), which allowed simultaneous substrate feeding. Cultures pregrown with succinate in a shake flask were centrifuged (3,000 × g for 2 min), and the cell pellets were resuspended in fresh medium containing oxalate and used for inoculation. Alternatively, glycerol stocks from oxalate-grown wild-type cells were used for inoculation. Cells were harvested by centrifugation at 6,000 × g at room temperature and frozen in liquid nitrogen until analysis. Mutant growth characterization was performed in shake flasks inoculated with a low cell density corresponding to an initial optical density of 0.03 at 600 nm. After an overnight incubation, the optical density was measured every 2 h.
Quantification of oxalate and formate.
The quantification of formate and oxalate was performed using high-performance liquid chromatography (HPLC) and a UV light-visible light (UV-VIS) detector at 210 nm as described before (59). Succinic acid was added to the cell-free supernatant as an internal standard. In addition, formate identification was confirmed enzymatically using NAD+-dependent formate dehydrogenase (Fdh) (15). The oxidation of formate was spectroscopically monitored at 365 nm and 37°C using 0.5 ml of a reaction mixture containing 200 mM Tris-HCl buffer (pH 7.6), 0.75 mM NADP+, and 2 U of formate dehydrogenase (Candida boidinii). The reaction was started by the addition of 100 μl of supernatant.
Proteome analysis.
For proteome analysis, cells were grown in a bioreactor and processed as described previously (22, 59). Briefly, proteins were separated by 1-dimensional (1D) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and, after tryptic digestion, were analyzed using reverse-phase HPLC coupled with high-accuracy mass spectrometry. Tandem mass spectroscopy (MS/MS) spectra were searched against a database using Mascot (Matrix Science) and X!tandem. A database containing all annotated proteins in the M. extorquens AM1 genome was downloaded from the Genoscope website (68). To determine significant changes in protein levels during growth on acetate compared with methanol and acetate, the spectral counts for each protein were normalized to the sum of all spectra detected for each sample. Average values for each substrate were obtained from three biological replicate experiments and used to calculate the fold changes in the normalized spectral counts (spectral counts of a protein obtained from oxalate samples divided by spectral counts of the protein obtained from the reference sample). A one-way analysis of variance with log2 transformation was used to evaluate the statistical significance of observed changes (47). Prior to normalization, zero values were set to 1. MS/MS data have been deposited in the PRIDE database (accession no. 17697 to 17699).
Dynamic labeling experiments.
The 13C-labeling experiments were performed as described previously (45), with adaptations. We performed three technical replicate experiments. Cells were pregrown on 5 mM oxalate at natural 13C abundance. The incubation of cells with labeled oxalate was performed in 50-ml Falcon tubes containing minimal medium with 3.4 mM [U-13C]oxalate. After the addition of exponentially growing culture to fresh medium containing [U-13C]oxalate (at a final concentration of 0.1 mg of cell dry weight [CDW]/ml), the samples were continuously mixed and incubated for various durations. Over 10 min, the pH increased from 6.7 to 7.0, but the intracellular metabolite concentrations were stable. Quenching and metabolite extraction were performed as detailed below. The calculation of 13C-label incorporation was conducted as described elsewhere (45); the 13C-labeled metabolite fractions were normalized to the 13C fraction of oxalate in the medium, which was between 70% and 80%.
Sampling, quenching, and metabolite extraction.
To determine label incorporation by mass spectrometry sampling, the quenching and extraction of CoA thioesters were performed as described previously (45), with several adaptations (59). For this analysis, sample volumes containing 0.5 mg of CDW were added to −20°C 95% acetonitrile containing 25 mM formic acid. After incubation for 10 min on ice with occasional mixing, samples were frozen in liquid nitrogen and lyophilized. The sampling of central metabolites was performed by fast filtration for salt reduction as described by Bolten et al. (9), with some modifications. Cell suspensions of 0.1 mg of CDW were harvested by vacuum filtration without washing. Subsequently, each filter was transferred to a vessel containing a mixture of acetonitrile (60% [vol/vol]), methanol (20% [vol/vol]), and 0.5 M formic acid (20% [vol/vol]) at −20°C for quenching and metabolite extraction.
HPLC-MS analysis.
All HPLC-MS analyses of CoA thioesters were performed with an HPLC system coupled to an LTQ-Orbitrap mass spectrometer equipped with an electrospray ionization probe (45), including online desalting (59). The analysis of central metabolites was performed using nanoflow ion-pair reverse-phase HPLC coupled with nanospray high-resolution MS as described previously (35) with a split-free nano-LC Ultra system connected to an LTQ-Orbitrap mass spectrometer.
Enzyme assays.
The activity of oxalyl-CoA reductase (glyoxylate-NADP+ oxidoreductase) was determined using the method of Quayle and Taylor (54). To measure oxalyl-CoA reductase activity in M. extorquens AM1, 400 to 600 mg of frozen cells was resuspended in 0.5 ml of 200 mM Tris-HCl (pH 8.6) containing 4 mM dithiothreitol. After the addition of glass beads (0.1 mm in diameter), the cell solution was treated in a tissue lyser (Retsch, Haan, Germany) for 10 min at 30 Hz. Cell debris and glass beads were removed by centrifugation at 4°C (58). Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Thermo Scientific [63]) according to the manufacturer's instructions and bovine serum albumin (BSA) as a standard. The oxidation of glyoxylate was spectroscopically monitored at 365 nm (εNADPH = 3,400 M−1 cm−1). The reaction mixture contained 100 mM Tris-HCl buffer (pH 8.6), 2 mM dithiothreitol, 3 mM CoA, 0.5 mM NADP+, and 0.1 to 1.5 mg/ml of protein. The reaction was started by the addition of 100 mM glyoxylate to the mixture. Enzyme activity is given in units (U) corresponding to micromoles per minute.
Mutant construction.
Kanamycin (Km) insertion mutations in panE2, oxs, frc1, frc2, and oxc were constructed using the pCM184 allelic exchange suicide vector and mated into M. extorquens AM1 as described previously (43). Deletion mutations in the strains described above were generated by introduction of the pCM157 cre expression vector in each mutant strain followed by plasmid curing and screening for Km sensitivity as described previously (43). Mutations were confirmed by diagnostic PCR.
Flux balance analysis.
The previously described iRP911 genome-scale metabolic model of M. extorquens AM1 (46), with the addition of oxalyl-CoA synthetase, was used to balance the energetic constraints on metabolism during growth with oxalate as the carbon source (see Table S3 in the supplemental material). The computations were performed with the CellNetAnalyzer (36) and MATLAB (Mathworks, Inc.) programs, as described previously (46). The determination of biomass composition is a prerequisite to performing an accurate flux balance analysis. During growth on different carbon sources, the biomass composition of M. extorquens AM1 showed only minor changes, with the exception of polyhydroxybutyrate (59). Polyhydroxybutyrate constituted <0.5% of the CDW of oxalate-grown M. extorquens AM1, which is a content similar to that of methanol-grown cells (∼2%); thus, the biomass composition of cells grown on methanol was implemented in the metabolic model for flux calculation (46).
RESULTS
M. extorquens AM1 exhibited a doubling time of 6 h when grown on 5 mM potassium oxalate in a bioreactor (Table 1), which is lower than that observed on methanol (4 h [46]) but is significantly increased compared to the doubling time on acetate (10 h [59]). The biomass yield was 0.07 ± 0.01 (g of [C] of CDW) of (g of [C] from oxalate), which is six times less than that from acetate. The oxalate uptake rate was 34 ± 4 mmol · g−1 (CDW) · h−1 as determined from three independent cultivations in the bioreactor.
Table 1.
Growth rates, oxalate uptake rates, and yields of M. extorquens AM1 wild-type and mutant strains grown on oxalatea
| Strain or mutation(s) | Gene inactivated | Growth (h−1)b | Uptake (mmol · g−1 (CDW) · h−1)c | Yield (g of [C] of CDW) of (g of [C])d | Reference |
|---|---|---|---|---|---|
| Wild type | 0.106 ± 0.005 | 33.8 ± 4.4 | 0.070 ± 0.006 | ||
| Formyl-CoA transferase 1 | Δfrc1 | − | (+) | ND | This study |
| Formyl-CoA transferase 2 | Δfrc2 | ++ | ++ | ND | This study |
| Oxalyl-CoA decarboxylase | Δoxc | − | − | ND | This study |
| Formate dehydrogenase 3 | Δfdh3 | ++ | ++ | ND | 16 |
| Formate dehydrogenase 4 | Δfdh4 | ++ | ++ | ND | 16 |
| Formate dehydrogenase 1 and 2 | Δfdh1 Δfdh2 | (+) | +e | ND | 16 |
| Formate dehydrogenase 1, 2, and 3 | Δfdh1 Δfdh2 Δfdh3 | − | (+)e | ND | 16 |
| Formate dehydrogenase 1, 2, 3, and 4 | Δfdh1 Δfdh2 Δfdh3 Δfdh4 | − | − | ND | 15 |
| Oxalyl-CoA reductase | ΔpanE2 | − | − | ND | Skovran et al., unpublished |
| Oxalyl-CoA synthetase | Δoxs | ++ | ++ | ND | Skovran et al., unpublished |
| Oxalyl-CoA synthetase and oxalyl-CoA reductase | Δoxs ΔpanE2 | 0.089 ± 0.001 | 28.4 ± 1.5 | 0.069 ± 0.004 | Skovran et al., unpublished |
| Glycerate kinase | Δgck | − | − | ND | 17 |
| PEP carboxylase | Δppc | + | ++ | ND | 5 |
| Malate thiokinase | ΔmtkA | ++ | ++ | ND | 18 |
| Crotonyl-CoA carboxylase/reductase | Δccr | 0.117 ± 0.007 | 37.6 ± 0.6 | 0.071 ± 0.001 | 19 |
| Ethylmalonyl-CoA/methylmalonyl-CoA epimerase | Δepm | ++ | ++ | ND | 38 |
| Ethylmalonyl-CoA mutase | Δecm | ++ | ++ | ND | 62 |
| Methylsuccinyl-CoA dehydrogenase | Δmsd | + | + | ND | 38 |
| Mesaconyl-CoA hydratase | Δmcd | − | − | ND | 40 |
| Malyl-CoA/β-methylmalyl-CoA lyase 1 and 2 | ΔmclA1 ΔmclA2 | − | − | ND | 44 |
| Propionyl-CoA carboxylase | ΔpccA | (+)* | (+) | ND | 38 |
| Methylmalonyl-CoA mutase | ΔmcmA | − | (+) | ND | 38 |
Cultivations were carried out either in shake flasks or in a bioreactor (wild type, Δoxs ΔpanE2, Δccr, or ΔpccA) for determination of growth rates and oxalate uptake rates. All cultivations were carried out in three independent replicate experiments.
++, growth similar to that seen with the wild type; +, growth rate lower than that seen with the wild type; (+) almost no growth; −, no growth; *, growth rate increased during cultivation.
Oxalate uptake rate. ++, uptake rate similar to that seen with wild type; +, uptake rate lower than wild type; (+), almost no oxalate uptake; −, no oxalate utilization detected.
Yield from oxalate. ND, not determined.
Formate secretion.
Identification of proteins enriched during oxalotrophic growth.
To identify the enzymatic makeup of oxalate-grown cells and to find proteins that are enriched during oxalotrophic growth, we performed a label-free semiquantitative proteome analysis. As a reference, we used the proteomes previously characterized during growth on methanol and acetate (59). Proteins were identified by searching MS/MS spectra in a database containing all annotated proteins in the M. extorquens AM1 genome. Although fold changes in spectral counts are not linearly correlated with fold changes in protein amounts, this approach is suitable for the determination of significant variations in protein levels among different sample sets (7). Statistical significance was tested using a one-way analysis of variance. The majority of proteins (total, approximately 2,800; see Table S1 in the supplemental material) were detected in cells grown on oxalate, methanol, and acetate. The six most abundant proteins detected only in oxalate samples were the following (Fig. 1): (i) oxalyl-CoA decarboxylase (oxc; META1_0990), (ii and iii) formyl-CoA transferases (frc1 and frc2; META1_0988 and META1_0999, respectively), and (iv, v, and vi) oxalate formate antiporter (oxlT; META1_1925, META1_0993, and META1_0992, respectively). The protein annotated as oxalyl-CoA decarboxylase has an amino acid sequence identity of 62% to oxalyl-CoA decarboxylase of O. formigenes (42) and catalyzes oxalyl-CoA decarboxylation to formyl-CoA. Two paralogous formyl-CoA transferases with sequence identities of 62% (Frc1) and 56% (Frc2) to the homologue of O. formigenes (61) were detected. In O. formigenes, substrate uptake is accomplished by an oxalate/formate antiporter (OxlT) (30); three proteins with approximately 35% sequence identity were found in oxalate-grown M. extorquens AM1 cells.
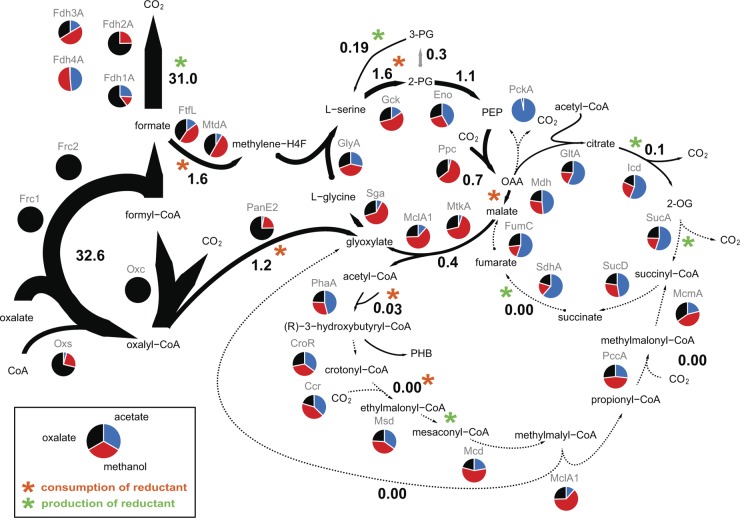
Fig 1.
Proposed network topology of the central metabolism operating during growth of M. extorquens AM1 on oxalate, including flux values calculated using flux balance analysis and proteome data. Theoretical flux distribution was calculated for a constrained oxalate uptake rate of 33.8 mmol · g−1 (CDW) · h−1 and a growth rate of 0.11 h−1; fluxes are given in mmol · g−1 (CDW) · h−1. Results of comparative proteome analysis are graphically included. Red, methanol; blue, acetate; black, oxalate samples. Normalized spectral counts are represented relative to total numbers of spectra from the three different sample sets. Consumption (orange) and production (red) of reductant are indicated by asterisks. For a better overview, not all intermediates and enzymes are shown; all enzymes of the central metabolism are listed in Table S2 in the supplemental material. Abbreviations used for metabolites: PG, phosphoglycerate; PEP, phosphoenolpyruvate; 2-OG, 2-oxoglutarate; OAA, oxaloacetate; H4F, tetrahydrofolate. Abbreviations used for proteins: Fdh1a, Fdh2a, Fdh3a, and Fdh4a, formate dehydrogenase; Frc1 and Frc2, fromyl-CoA transferase; Oxc, oxalyl-CoA decarboxylase; PanE2, oxalyl-CoA reductase; Oxs, oxalyl-CoA synthetase; FtfL, formate-tetrahydrofolate ligase; MtdA, methylene-tetrahydrofolate dehydrogenase; Gck, glycerate kinase; Eno, enolase; Ppc, PEP carboxylase; MtkA, malate thiokinase; MclA1, malyl-CoA/β-methylmalyl-CoA lyase; Sga, serine glyoxylate aminotransferase; GlyA, serine hydroxymethyltransferase; PhaA, β-ketothiolase; CroR, crotonase; Ccr, crotonyl-CoA carboxylase/reductase; Msd, methylsuccinyl-CoA dehydrogenase; Mcd, mesaconyl-CoA hydratase; PccA, propionyl-CoA carboxylase; McmA, methylmalonyl-CoA mutase; GltA, citrate synthase; Icd, isocitrate dehydrogenase; SucA, 2-oxoglutarate dehydrogenase; SucD, succinyl-CoA synthetase; SdhA, succinate dehydrogenase; FumC, fumarase; Mdh, malate dehydrogenase.
Based on the protein identifications, we predict that oxalate uptake in M. extorquens AM1 is enabled by an oxalate/formate antiporter. The import of the 2-fold negatively charged oxalate by simultaneous export of the monoanion formate contributes to an electrochemical and proton gradient across the membrane that may then be coupled to ATP synthesis (65). Oxalate is converted by formyl-CoA transferase and oxalyl-CoA decarboxylase to formate, which can then be oxidized to carbon dioxide by formate dehydrogenase. M. extorquens AM1 was previously reported to contain four different formate dehydrogenases (Fdh) (15). Tungsten-containing Fdh1 is NAD+ dependent and located in the cytoplasm (41). Fdh2 is a molybdenum enzyme, which uses NAD as a cosubstrate. The two enzymes are known to be functionally equivalent (16). Fdh3 is predicted to contain a TAT pathway signal, is probably located in the periplasm, and uses a cytochrome as an electron acceptor. Little is known about Fdh4; in vivo activity of the enzyme has been demonstrated only after the inactivation of Fdh1, -2, and -3. However, the electron acceptor remains unknown (15). Cells grown on oxalate displayed an approximately 4-fold-higher abundance (based on normalized spectral counts) of Fdh1 and Fdh2 than methanol-grown cells (Fig. 1). Based on these data, we propose that the oxidation of formate to carbon dioxide proceeds with the formation of NADH in the cytoplasm. Therefore, formate must be transferred back into the cell, in a manner similar to that seen during growth on formate, by either passive diffusion or an unknown formate transporter. NAD(P)+ transhydrogenase (Pnt) was approximately 10-fold more abundant in cells grown on oxalate than in those grown on methanol and may thus contribute to the conversion of NADH into NADPH. In similarity to the results seen with cells grown on methanol, we detected all enzymes of the tricarboxylic acid (TCA) cycle in oxalate-grown cells but only at low abundances (Fig. 1). Thus, we predict that the TCA cycle is not involved in oxidative processes and operates in an incomplete and purely anabolic manner.
Detection of specific metabolites present during oxalotrophic growth and their rapid labeling upon the addition of [U-13C]oxalate.
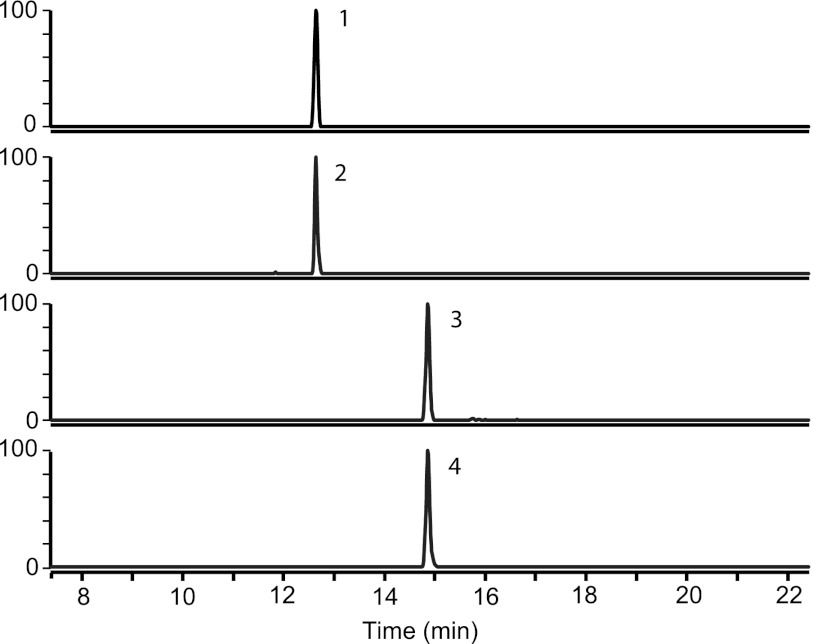
The above-described identification of oxalyl-CoA decarboxylase and formyl-CoA transferase in oxalate-grown cells rather than during growth on methanol or acetate suggests that their substrates and/or products are present in oxalate-grown cells. To verify the presence of these metabolites during oxalotrophic growth, we analyzed CoA thioesters from M. extorquens AM1 grown in the bioreactor. After metabolite extraction and analysis using HPLC-MS, we identified oxalyl-CoA and formyl-CoA in cells grown with oxalate as the carbon source (Fig. 2) but not in the previously characterized methanol- and acetate-grown cells. To validate the idea of the conversion of oxalate through oxalyl-CoA and formyl-CoA, we monitored the incorporation of [U-13C]oxalate. Only 3 s after the addition of labeled oxalate, we detected a mass shift of plus two in oxalyl-CoA and of plus one in formyl-CoA. The labeled fraction of oxalyl-CoA was 0.8 after 3 s and 1.0 after 1 min. However, the labeled fraction of formyl-CoA was 0.6 after 3 s and 0.8 after 1 min; no further increase of labeling in formyl-CoA was observed during an additional 10 min. This apparent equilibrium may be related to the reversibility of formyl-CoA transferase activity and the pool of unlabeled intra- and extracellular formate.
Fig 2.
LC-MS analysis of CoA thioesters in cell extracts of M. extorquens AM1 during growth on naturally labeled oxalate and after incubation with [U-13C]oxalate. Peak 1, oxalyl-CoA (M0); peak 2, oxalyl-CoA (M + 2); peak 3, formyl-CoA (M0); peak 4, formyl-CoA (M + 1).
Growth characterization of mutants involved in oxalate conversion to CO2.
To study the role and function of the formyl-CoA transferase, knockout mutants were generated for the paralogues (frc1 and frc2) and for oxalyl-CoA decarboxylase (oxc). Growth characterization of these mutants in the presence of oxalate as the sole source of carbon and energy was performed in shake flasks (Table 1), and succinate was used as a control. All the strains grew normally with succinate, but the Δoxc and Δfrc1 mutants were unable to grow with oxalate as the sole carbon source; only the Δfrc2 mutant grew similarly to the wild-type strain on oxalate. To investigate the oxidation of formate and to identify which formate dehydrogenase catalyzes the reaction, we tested various mutant strains (Δfdh1 Δfdh2, Δfhd3, Δfdh1 Δfdh2 Δfdh3, Δfdh4, and Δfdh1 Δfdh2 Δfdh3 Δfdh4 mutants) (Table 1). With the exception of the triple and quadruple mutants, all of these strains were able to grow on formate (15, 16). Whereas the single mutants (Δfdh3 and Δfdh4 mutants) grew in the presence of oxalate, the double mutant (Δfdh1 Δfdh2 mutant) displayed a severe growth defect and grew only slowly on oxalate. Oxalate concentration in the supernatant decreased over time, and formate secretion was detected in parallel (at a level of approximately 1 mM after 24 h). The Δfdh1 Δfdh2 Δfdh3 triple mutant, like the Δfdh1 Δfdh2 Δfdh3 Δfdh4 quadruple mutant, was unable to grow on oxalate; however, a minor decrease in the oxalate concentration (approximately 1 to 2 mM within 24 h) was observed with a concomitant formate buildup (1 to 2 mM) in the triple mutant that was not observed in the Δfdh1 Δfdh2 Δfdh3 Δfdh4 mutant. These results demonstrate that oxalate decarboxylation to formate by oxalyl-CoA decarboxylase and formyl-CoA transferase and its subsequent oxidation by formate dehydrogenase are crucial steps in generation of reductant.
Oxalate conversion to glyoxylate.
Next, we aimed at the identification of proteins involved in the assimilation of the C2 substrate. As mentioned above, glyoxylate carboligase is a common enzyme among the oxalotrophic bacteria (21, 53, 55) characterized thus far (13) that initiates the assimilation of the C2 substrate into biomass in these organisms. However, based on activity measurements of cell extracts (8) and the complete genome sequence of the methylotroph (68), glyoxylate carboligase is absent in M. extorquens AM1. In 1970, Blackmore and Quayle detected oxalyl-CoA reductase activity in cell extracts and proposed that oxalyl-CoA reductase operates in conjunction with serine cycle enzymes during oxalotrophy (8). To confirm the activity of oxalyl-CoA reductase, we quantified the enzyme activity based on the reverse reaction, i.e., glyoxylate oxidation (8), in cell extracts from oxalate-, methanol-, and acetate-grown M. extorquens AM1 cells. The activities were 0.54 ± 0.06 U/mg of protein in cells grown with oxalate as the carbon source and 0.07 ± 0.02 and 0.02 ± 0.01 U/mg of protein in the methanol- and acetate-grown cells, respectively. Based on these different activities, we searched the proteome for a reductase with the highest abundance in oxalate-grown cells relative to cells grown in the presence of methanol and acetate. One protein annotated as a putative ketopantoate reductase (panE2) met the requirements. This protein was 3-fold more abundant on oxalate than on methanol and 25-fold more abundant on oxalate than on acetate (Fig. 1). Furthermore, the protein levels were correlated with the enzyme activities measured in cell extracts; the activity of an oxalate-grown cell extract was eight times higher than that of an extract of methanol-grown cells and 27 times higher than that of an extract of an acetate-grown culture. Analysis using a ΔpanE2 mutant indeed confirmed this finding and led to the identification of oxalyl-CoA reductase (E. Skovran, S. Yang, A. D. Palmer, and M. E. Lidstrom, unpublished data). Furthermore, a neighboring gene annotated as a putative acyl-CoA synthetase was identified as oxalyl-CoA synthetase (oxs) (Skovran et al., unpublished). Additionally, oxalyl-CoA synthetase was found to be more abundant in oxalate-grown cells than in those grown on methanol or acetate. These enzymes allow glyoxylate generation directly from oxalate conversion to oxalyl-CoA and its reduction. Inactivation of oxalyl-CoA reductase (ΔpanE2 mutant) resulted in the inability of M. extorquens AM1 to grow on oxalate, whereas a mutant of oxalyl-CoA synthetase (Δoxs mutant) grew normally on oxalate. Notably, however, a double mutant of oxalyl-CoA synthetase and oxalyl-CoA reductase (Δoxs ΔpanE2 mutant) grew in the bioreactor, albeit at a rate reduced by 15% in comparison with the wild-type strain (Table 1). These results suggest that the lack of growth of the ΔpanE2 mutant was due to toxicity effects rather than implying that oxalyl-CoA reductase activity is essential. Thus, we concluded that an alternative entry into central metabolism must exist, as described below.
Metabolic network of central metabolism.
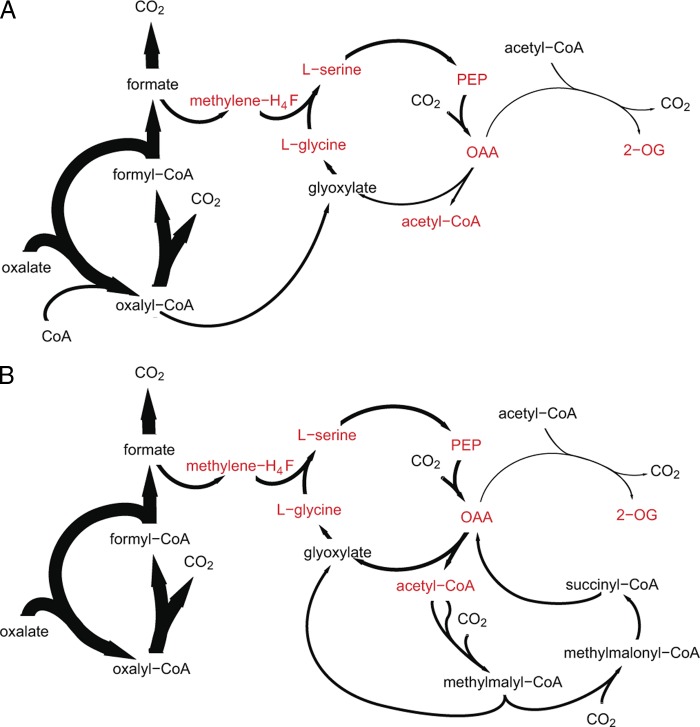
As mentioned above, oxalyl-CoA synthetase and oxalyl-CoA reductase may operate in conjunction with the serine cycle (8). All serine cycle enzymes, i.e., serine hydroxymethyltransferase, serine glyoxylate aminotransferase, hydroxypyruvate reductase, glycerate kinase, enolase, phosphoenolpyruvate (PEP) carboxylase, malyl-CoA/β-methylmalyl-CoA lyase, and malate thiokinase, were detected in the proteome of M. extorquens AM1, displaying approximately half their intensities during growth on oxalate relative to the levels on methanol (Fig. 1). Uniquely, the abundance of the serine cycle enolase was equal to that seen on methanol. Additionally, we detected all the enzymes of the tetrahydrofolate-dependent pathway, which are required for formate assimilation, showing fold changes of between 0.5 and 1 relative to levels seen with the methanol samples. These findings are in accordance with the assimilation route proposed by Blackmore and Quayle (8), wherein C3 unit synthesis from C1 units plus glyoxylate and carboxylation to C4 are feasible based on the presence of C1 assimilation and serine cycle enzymes (Fig. 3A). Malate thiokinase and malyl-CoA lyase may be involved in acetyl-CoA and glyoxylate production. However, the entire set of EMC pathway enzymes was also detected in oxalate-grown cells, with fold changes of between 0.5 and 0.9 for the oxalate versus the methanol- and acetate-grown cells. The latter pathway may thus represent a second potential route for glyoxylate generation by the conversion of acetyl-CoA via the EMC pathway and may explain the above-mentioned phenotype of the Δoxs ΔpanE2 double mutant, which grows in the presence of oxalate (Fig. 3B).
Fig 3.
Schematic view of the two different oxalate assimilation stategies. (A) Oxalate assimilation by a variant of the serine cycle, including operation of oxalyl-CoA synthetase and oxalyl-CoA reductase. (B) Oxalate assimilation via C1 units by the serine cycle and EMC pathway, without oxalyl-CoA synthetase and oxalyl-CoA reductase. Precursor metabolites are labeled in red; for abbreviations, see Fig. 1 legend.
A number of mutants were used to identify the enzymes required during oxalotrophic growth. Among the mutants of serine cycle enzymes, we tested glycerate kinase, PEP carboxylase, and malate thiokinase mutants (Table 1). A mutant with a defect in glycerate kinase did not grow on oxalate, and thus, no alternative for C3 unit synthesis exists, a determination similar to what has been reported for methylotrophic growth (17). A mutant deficient in PEP carboxylase, which is indispensable during growth on methanol (5), exhibited a reduced growth rate on oxalate, suggesting that an alternative route for the synthesis of C4 units, e.g., the condensation of glyoxylate with acetyl-CoA, must exist and is sufficient to allow growth on oxalate. A malate thiokinase mutant unable to grow on methanol (18) grew normally on oxalate, which indicates that malate cleavage is not required for glyoxylate and acetyl-CoA generation during oxalate assimilation. However, an alternative pathway for acetyl-CoA synthesis in the mutant, possibly consisting of pyruvate decarboxylation, must exist.
In addition to the serine cycle enzymes, we tested mutants with defects in the EMC pathway. The pathway is required during C1 and C2 assimilation for the conversion of acetyl-CoA to glyoxylate (19, 38, 39). Mutant strains lacking the key enzyme of the pathway, crotonyl-CoA carboxylase/reductase, or ethylmalonyl-CoA/methylmalonyl-CoA epimerase or ethylmalonyl-CoA mutase grew with oxalate as the carbon source in a manner similar to that seen with the wild type (Table 1). This observation indicates that the EMC pathway is dispensable during oxalotrophy. Remarkably, however, the inactivation of enzymes catalyzing reactions downstream of the irreversible methylsuccinyl-CoA dehydrogenase, i.e., mesaconyl-CoA hydratase, malyl-CoA/β-methylmalyl-CoA, propionyl-CoA carboxylase, and methylmalonyl-CoA mutase, resulted in severe growth defects on oxalate. A methylsuccinyl-CoA dehydrogenase mutant grew on oxalate but did so more slowly than the wild type. We hypothesized that the growth phenotype of mutants deficient in the latter group of enzymes may be caused by toxic effects as a consequence of the “trapping” of CoA thioesters, and this prediction was subsequently confirmed. The phenotypes of the mutants in the lower part of the EMC pathway indicate the operation of this pathway in the wild-type strain.
13C-labeling experiment to demonstrate the operation of assimilatory pathways in vivo.
The results of the above-described mutant analysis suggest the operation of a variant serine cycle without EMC pathway operation for glyoxylate regeneration during oxalate assimilation (Fig. 3A). However, oxalyl-CoA reduction is dispensable, and the phenotypes of mutants in the lower part of the EMC pathway (which were not growing on oxalate) suggest flux through the EMC pathway. A possible alternative to oxalyl-CoA reduction as a route for carbon assimilation consists of oxalate assimilation via formate and its conversion via the tetrahydrofolate-dependent pathway and the serine cycle. Glyoxylate is converted by serine glyoxylate aminotransferase to glycine, which is condensed with methylene-tetrahydrofolate to form serine. The latter is then converted to hydroxypyruvate by serine glyoxylate aminotransferase, reduced to glycerate, and converted to phosphoglycerate and then PEP, which is carboxylated to oxaloacetate. If glyoxylate is not produced from oxalate, it can alternatively be synthesized from oxaloacetate by its reduction to malate and conversion to malyl-CoA, which is cleaved in acetyl-CoA and glyoxylate. Acetyl-CoA can be converted by EMC pathway enzymes to glyoxlate (Fig. 3B). To test this hypothesis of two distinct putative assimilation strategies and to elucidate the metabolic route used for glyoxylate generation, we performed dynamic 13C-labeling experiments in the wild-type and in the Δoxs ΔpanE2 double mutant. To this end, we grew the M. extorquens AM1 wild-type strain and the double mutant in bioreactors on naturally labeled oxalate, mixed each culture with fresh medium containing [U-13C]oxalate (at a final concentration of approximately 70% [U-13C]oxalate), and followed the 13C-labeling incorporation in central metabolites and CoA thioesters in parallel using two designated sampling protocols, as described in Materials and Methods.
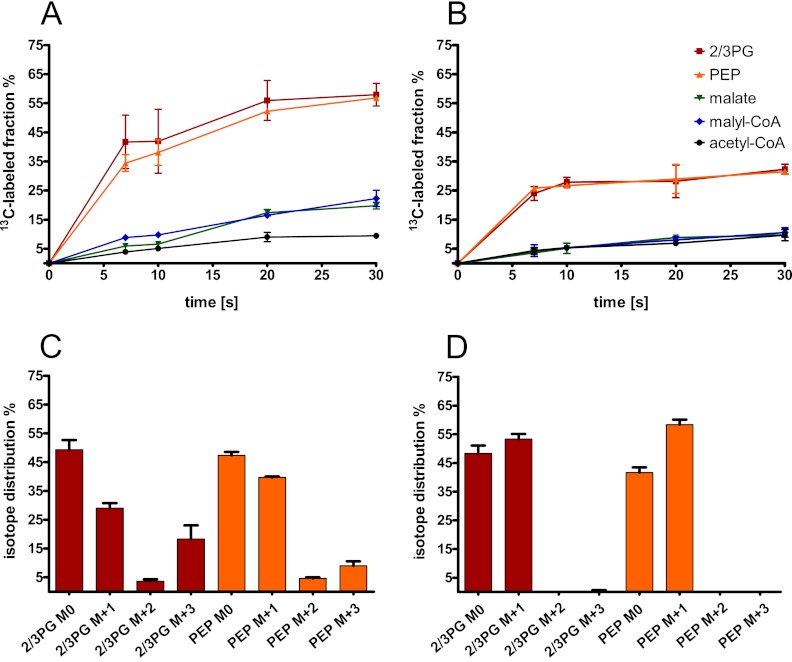
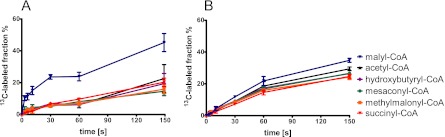
Upon sampling the central metabolites, we detected the following serine cycle intermediates: 2/3-phosphoglycerate, PEP, malyl-CoA, and acetyl-CoA. During incubation of wild-type cells with [U-13C]oxalate, we found that 2/3-phosphoglycerate and PEP were labeled first followed by malate and malyl-CoA and then acetyl-CoA (Fig. 4A). Based on detection of malyl-CoA and the label incorporation into this metabolite before acetyl-CoA, we suggest that acetyl-CoA synthesis results from malyl-CoA cleavage in wild-type cells rather than pyruvate decarboxylation. The isotope distributions of 2/3-phosphoglycerate after 7 s of incubation with 13C-labeled oxalate in the wild-type strain were 30% with one carbon-13 atom incorporated (M + 1), approximately 5% with two 13C atoms (M + 2) incorporated, and 20% with three 13C atoms (M + 3) incorporated. The isotope distributions of PEP were similar: 40% M + 1, 5% M + 2, and 10% M + 3 (Fig. 4C). The mass shift of 1 can be explained by the condensation of 13C-labeled methylene-tetrahydrofolate with unlabeled [U-12C]glyoxylate. The fraction with two labeled carbon atoms originates from [U-13C]glycine (derived from [U-13C]oxalate converted via glyoxylate to glycine) and its condensation with [12C]methylene-tetrahydrofolate. In the case of the uniformly labeled C3 compound, the carbon atoms of glyoxylate and the C1 unit are both predicted to be labeled. For the Δoxs ΔpanE2 double mutant, we found that, 7 s after label addition, a maximum of one 13C atom was incorporated in both 2/3-phosphoglycerate and PEP (55% and 60%, respectively) (Fig. 4D). These results thus demonstrate oxalyl-CoA reduction in the M. extorquens AM1 wild-type strain and its absence in the double mutant.
Fig 4.
Incorporation of the 13C label in central metabolites over time after incubation with [U-13C]oxalate. M. extorquens AM1 wild-type (A and C) and oxalyl-CoA synthetase and oxalyl-CoA reductase (Δoxs ΔpanE2) double mutant (B and D) cells were grown on naturally labeled oxalate and mixed with fresh medium containing [U-13C]oxalate; label incorporation was measured in central metabolites at different times. The 13C-labeled fractions of central metabolites (A and B) were normalized to the 13C fraction of oxalate in the medium. The isotope distributions of 2/3-phosphoglycerate (2/3-PG) and PEP (C and D) were determined after 7 s of incubation with [U-13C]oxalate. The medium contained 70% to 80% [U-13C]oxalate. Averages and standard deviations of the results of three technical replicate experiments are given.
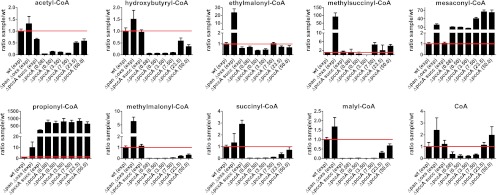
Sampling for CoA thioesters revealed the presence of the following EMC pathway intermediates in both the wild-type strain and the Δoxs ΔpanE2 double mutant during growth on oxalate: acetyl-CoA, hydroxybutyryl-CoA, ethylmalonyl-CoA, methylsuccinyl-CoA, mesaconyl-CoA, propionyl-CoA, methylmalonyl-CoA, succinyl-CoA, and malyl-CoA. The concentrations of the following CoA thioesters were significantly lower in the wild-type strain than in the Δoxs ΔpanE2 double mutant (with reductions ranging from 5- to 90-fold; see below): ethylmalonyl-CoA, methylsuccinyl-CoA, mesaconyl-CoA, propionyl-CoA, and methylmalonyl-CoA. The reduced pool sizes may indicate a lower flux through the EMC pathway, as a lower substrate concentration is consistent with lower enzyme activity when the substrate concentration is not greater than twice the Km value. After incubation with [U-13C]oxalate, we detected label incorporation in CoA thioesters in both the wild-type and Δoxs ΔpanE2 cells; first malyl-CoA, followed by acetyl-CoA, and then all other CoA thioesters were labeled in parallel (Fig. 5). Malyl-CoA was labeled sooner in the wild-type than in the double mutant cells; however, acetyl-CoA was labeled later in the wild-type strain than in the double mutant. Acetyl-CoA, the product of the serine cycle, is derived from one C1 unit and one carbon dioxide. The labeled fractions of C1 units in the two strains were identical (see the labeling patterns of the C3 compounds in Fig. 4), and levels of carbon dioxide production were expected to be similar in the two strains, given by their similar growth rates. With respect to the similar intracellular concentrations of acetyl-CoA (see below) in the wild-type strain and the double mutant, the delayed labeling of acetyl-CoA is related to decreased malate cleavage in the wild-type compared with the Δoxs ΔpanE2 cells.
Fig 5.
Incorporation of the 13C label in CoA thioesters over time after incubation with [U-13C]oxalate. M. extorquens AM1 wild-type (A) and oxalyl-CoA synthetase and oxalyl-CoA reductase (Δoxs ΔpanE2) double mutant (B) cells were grown on naturally labeled oxalate and mixed with fresh medium containing [U-13C]oxalate; label incorporation was measured at different times. The 13C-labeled fractions of CoA thioesters were normalized to the 13C fraction of oxalate in the medium. Averages and standard deviations of the results of three technical replicate experiments are given.
Additionally, we investigated EMC pathway mutants for effects on intermediates to test the hypothesis that intermediates of the pathway accumulate upon a blockage of the lower part of the pathway, which may explain the observed growth defects in these mutants. A propionyl-CoA carboxylase (ΔpccA) mutant was chosen to monitor the pool sizes of EMC pathway intermediates. The EMC pathway intermediate pool sizes were compared for the ΔpccA mutant grown on oxalate versus the wild-type cells and versus the ΔpccA mutant grown on succinate to determine whether the block is absent during growth on succinate (Fig. 6). Due to the growth defect of the ΔpccA mutant (Table 1), the mutant cultures were pregrown on succinate before being transferred to oxalate-containing medium. CoA thioester samples were taken at various times after the transfer to oxalate. During the first 8 h after the transfer of succinate-grown ΔpccA mutant to oxalate, the intracellular concentrations of acetyl-CoA, hydroxybutyryl-CoA, methylmalonyl-CoA, succinyl-CoA, and malyl-CoA were decreased by 10- to 100-fold compared with the concentrations seen with the exponentially growing wild type. However, the mesaconyl-CoA concentration was 10 times higher and propionyl-CoA concentration was 700 times higher in ΔpccA cells than in the wild type. The ΔpccA mutant began to grow slowly after 24 h on oxalate and continuously increased its growth rate (with a doubling time of 9 h seen at 50 h after the transfer to oxalate). After 24 h, the pool sizes of mesaconyl-CoA and propionyl-CoA were still higher (70-fold and 700-fold, respectively) than that of the wild-type cells. All other CoA thioesters (upstream of methylsuccinyl-CoA dehydrogenase) displayed less than a 3-fold difference in intracellular pool size compared to that seen with the wild type. Additionally, as shown by comparisons of the CoA thioester pool size of the ΔpccA mutant grown on oxalate to that seen with the same strain grown on succinate, the intracellular concentrations of mesaconyl-CoA and propionyl-CoA were significantly increased on oxalate (10-fold and 4-fold, respectively, during the first 8 h and 60-fold for mesaconyl-CoA after 24 h). These data indicate that the growth inhibition of the mutants in the lower part of the EMC pathway is caused by a toxicity effect due to a trapping of CoA thioesters between the irreversible methylsuccinyl-CoA dehydrogenase and the downstream enzyme knockouts. The conversion of acetyl-CoA in the EMC pathway results in a loss of reductant and a failure to provide sufficient C2 units for biosynthesis. Additionally, the hydrolysis of highly concentrated CoA thioesters may lead to acidification of the cytoplasm. During succinate assimilation, less carbon is “trapped” in the EMC pathway (there are lower intracellular concentrations of mesaconyl-CoA and propionyl-CoA), which is likely related to a lower flux through the EMC pathway that allows the mutant to grow on succinate.
Fig 6.
Changes of intracellular CoA thioester concentrations of M. extorquens AM1 oxalyl-CoA synthetase and oxalyl-CoA reductase (Δoxs ΔpanE2) double mutant grown on oxalate versus the wild type (wt), propionyl-CoA carboxylase (ΔpccA) mutant grown on succinate versus the wt, and ΔpccA mutant versus the wt during different incubation times on oxalate. A preculture of ΔpccA mutant was grown on succinate; after centrifugation, cells were resuspended in fresh medium containing oxalate as the sole carbon source. Samples for CoA thioester quantification were taken at various times (given in parentheses in hours); for quantification, an internal standard was added. Samples of wt, Δoxs ΔpanE2 double mutant, and ΔpccA mutant cells were taken during exponential growth (exp). Ratios of intracellular CoA thioester concentrations were calculated using three technical replicate experiments; averages and standard deviations are given.
Metabolic network topology of central metabolism based on flux balance analysis.
To identify the optimal flux distribution for oxalate metabolism, we performed a flux balance analysis using the iRP911 genome-scale metabolic model of M. extorquens AM1 (46). The theoretical maximal growth rate of the cell was 0.19 h−1 for the experimentally determined oxalate uptake rate. The model suggests oxalyl-CoA reduction to glyoxylate, conversion to glycine, and its condensation with methylene-tetrahydrofolate produced from formate reduction, which results in the formation of C3 units, and no EMC pathway operation. Acetyl-CoA is produced via cleavage of malyl-CoA, which is formed by PEP carboxylase, malate dehydrogenase, and malate thiokinase as part of the serine cycle. This metabolic network topology is in line with the results of 13C-labeling experiments and mutant analysis described above. However, the calculated growth rate was approximately one-third higher than the experimentally determined rate. The theoretical biomass yield obtained from the flux balance analysis was 0.11 (g of [C] of CDW) of (g of [C] from oxalate), which is also almost one-third higher than the experimentally determined value. With both the oxalate uptake rate and the growth rate constrained to their experimental values, the calculated yield was 0.07 (g of [C] of CDW)/(g of [C] from oxalate). The simulated flux distribution of the central metabolism for the growth rate experimentally determined for M. extorquens AM1 during growth with oxalate is represented in Fig. 1. Notably, the enzyme activity measured in vitro for oxalyl-CoA reductase (which is 19.4 mmol · g−1 [CDW] · h−1; see above) is sufficient for glyoxylate generation by oxalyl-CoA reduction.
DISCUSSION
Growth in the presence of the highly oxidized oxalate requires an oxidative branch for the generation of reductant following cleavage of the carbon-carbon bond and an assimilation process involving oxalate reduction. The reduction of carbon is mandatory for the incorporation of oxalate into biomass, as the oxidation state of oxalate is higher than that of cellular carbon (at an average of +0.5). Here, we confirm that M. extorquens AM1 assimilates oxalate in a cyclic process of CoA transfer from formyl-CoA to oxalate, whereby formyl-CoA is the product of oxalyl-CoA decarboxylation. Reductant is produced by formate oxidation via formate dehydrogenase (8). We identified two different assimilation strategies for oxalate in M. extorquens AM1, whereby either of the two allows growth. The strategy that predominates in the wild-type strain is based on a minimal set of enzymes to produce all precursor metabolites (Fig. 3A). Oxalyl-CoA is reduced to glyoxylate, which, upon condensation with C1 units deriving from formate by the operation of a variant of the serine cycle, results in the synthesis of C3 and C4 units (8). The serine cycle is modified, as it does not require malyl-CoA cleavage and EMC pathway operation for glyoxylate regeneration. However, acetyl-CoA for biosynthesis may be produced either by malyl-CoA cleavage or pyruvate decarboxylation. Three of the TCA cycle enzymes are required for the synthesis of C5-precursor metabolites.
A second assimilation strategy, which was used when the oxalyl-CoA synthetase and oxalyl-CoA reductase were inactivated, consists of oxalate assimilation exclusively occurring via C1 units (Fig. 3B). Here, oxalate is decarboxylated to formate and subsequently converted into glyoxylate via the tetrahydrofolate-dependent pathway in connection with the serine cycle and the EMC pathway. The second assimilation strategy was also found to operate in wild-type cells, although to a limited extent, and was dispensable.
The two separate oxalate assimilation strategies differ in their metabolic flux distributions of the assimilatory and oxidative branches. To obtain the same growth rate during oxalate assimilation exclusively via C1 units as occurs during oxalate assimilation by the variant of the serine cycle, the organism must operate the EMC pathway and generate higher fluxes through the operation of the serine cycle for the C1 assimilation. Moreover, higher rates operating through oxalyl-CoA decarboxylase, formyl-CoA transferase, and formate dehydrogenase are required to account for the additional synthesis of C1 units and reductant needed for assimilation. In fact, we observed a lower growth rate for the Δoxs ΔpanE2 double mutant than the wild-type strain, which implies that the M. extorquens AM1 double mutant is not able to generate the higher rates required in the oxidative and/or assimilatory branches and, thus, that one or more of these enzymes constitute metabolic bottlenecks during oxalate assimilation. Based on a flux analysis of methanol-grown cells, we conclude that neither the serine cycle nor the EMC pathway can be the rate-limiting step. The organism is able to generate fluxes through both pathways required for C1 assimilation, accounting for a growth rate of at least 0.17 h−1 (46). However, enzymes in the oxidative branch operate at very high rates, in the range of 30 to 32 mmol · g−1 (CDW) · h−1. This range is twice as high as the rates observed during methanol assimilation (e.g., 13 mmol · g−1 [CDW] · h−1 for formate dehydrogenase [46]). Therefore, we suggest that oxalyl-CoA decarboxylase, formyl-CoA transferase, and/or formate dehydrogenase is the metabolic bottleneck(s) in the C1 oxalate assimilation strategy and thus restrict(s) growth. As mentioned above, we found that the C1 assimilation strategy also operates in the wild-type strain to a limited extent. The C1 assimilation strategy is blocked when crotonyl-CoA carboxylase/reductase is inactivated; the Δccr mutant showed a slight tendency for an increased growth rate compared with the wild-type results (Table 1). This confirms the hypothesis that the operation of the EMC pathway reduces growth during oxalate assimilation. Taking these data together, the lifestyle of M. extorquens AM1 during oxalate utilization seems to be limited in the oxidative branch, and oxalyl-CoA reductase is required to overcome the bottleneck and thus allows a higher growth rate on oxalate.
Although oxalate is a low-energy carbon compound, the flux balance analysis suggests that central metabolism is not operating under optimal energetic conditions with this carbon source. A higher growth rate and higher yield were calculated by the flux balance analysis than were observed experimentally, which is likely related to the presence of futile cycles, as demonstrated in M. extorquens AM1 during methanol and acetate assimilation (46, 59). During growth on oxalate, we detected a carbon flux through the EMC pathway. This is one example of a pathway that operates during growth on oxalate being dispensable and decreasing the growth rate. It is likely that other futile cycles found during C1 and C2 assimilation, e.g., those involving PEP carboxylase/PEP carboxykinase (46, 59) and malate thiokinase/malyl-CoA thioesterase (46), are also functional during oxalate utilization and thus reduce biomass yield. The presence of such futile cycles and EMC pathway operation may allow the organism to quickly adapt its metabolism to the availability of different carbon compounds such as methanol and other C1 compounds, acetate, and various substrates entering the central metabolism at the level of acetyl-CoA. Rapid metabolic adaptations to changes in carbon source availability may contribute to the fitness of this bacterium and may provide a growth advantage in low-nutrient habitats such as the phyllosphere.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by ETH Zurich (ETH-09 09-2).
We thank Nathanaël Delmotte for his assistance in proteome analysis and Philipp Christen for his help in running the bioreactor. Furthermore, we thank Patrick Kiefer and Tobias Erb for helpful discussions and Alex Palmer for generating the Δoxc and Δfrc mutants.
Footnotes
Published ahead of print 6 April 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61:297–309 [DOI] [PubMed] [Google Scholar]
- 2. Allison MJ, Cook HM, Milne DB, Gallagher S, Clayman RV. 1986. Oxalate degradation by gastrointestinal bacteria from humans. J. Nutr. 116:455–460 [DOI] [PubMed] [Google Scholar]
- 3. Allison MJ, Dawson KA, Mayberry WR, Foss JG. 1985. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch. Microbiol. 141:1–7 [DOI] [PubMed] [Google Scholar]
- 4. Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom [Google Scholar]
- 5. Arps PJ, Fulton GF, Minnich EC, Lidstrom ME. 1993. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J. Bacteriol. 175:3776–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baetz AL, Allison MJ. 1989. Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J. Bacteriol. 171:2605–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a. Baetz AL, Allison MJ. 1990. Purification and characterization of fromyl-coenzyme A transferase from Oxalobacter formigenes. J. Bacteriol. 172:3537–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. 2007. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389:1017–1031 [DOI] [PubMed] [Google Scholar]
- 8. Blackmore MA, Quayle JR. 1970. Microbial growth on oxalate by a route not involving glyoxylate carboligase. Biochem. J. 118:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bolten CJ, Kiefer P, Letisse F, Portais JC, Wittmann C. 2007. Sampling for metabolome analysis of microorganisms. Anal. Chem. 79:3843–3849 [DOI] [PubMed] [Google Scholar]
- 10. Braissant O, Cailleau G, Aragno M, Verrecchia EP. 2004. Biologically induced mineralization in the tree Milicia excelsa (Moraceae): its causes and consequences to the environment. Geobiology 2:59–66 [Google Scholar]
- 11. Bravo D, et al. 2011. Use of an isothermal microcalorimetry assay to characterize microbial oxalotrophic activity. FEMS Microbiol. Ecol. 78:266–274 [DOI] [PubMed] [Google Scholar]
- 12. Casarin VP, Souche CG, Arvieu J-C. 2003. Quantification of oxalate ions and protons released by ectomycorrhizal fungi in rhizosphere soil. Agronomie 23:461–469 [Google Scholar]
- 13. Chang YY, Wang AY, Cronan JE., Jr 1993. Molecular cloning, DNA sequencing, and biochemical analyses of Escherichia coli glyoxylate carboligase. An enzyme of the acetohydroxy acid synthase-pyruvate oxidase family. J. Biol. Chem. 268:3911–3919 [PubMed] [Google Scholar]
- 14. Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chistoserdova L, et al. 2007. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J. Bacteriol. 189:9076–9081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chistoserdova L, Laukel M, Portais JC, Vorholt JA, Lidstrom ME. 2004. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 186:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chistoserdova L, Lidstrom ME. 1997. Identification and mutation of a gene required for glycerate kinase activity from a facultative methylotroph, Methylobacterium extorquens AM1. J. Bacteriol. 179:4946–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chistoserdova LV, Lidstrom ME. 1994. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification, sequence, and mutation of three new genes involved in C1 assimilation, orf4, mtkA, and mtkB. J. Bacteriol. 176:7398–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chistoserdova LV, Lidstrom ME. 1996. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology 142(Pt 6):1459–1468 [DOI] [PubMed] [Google Scholar]
- 20. Cornick NA, Allison MJ. 1996. Anabolic incorporation of oxalate by Oxalobacter formigenes. Appl. Environ. Microbiol. 62:3011–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornick NA, Allison MJ. 1996. Assimilation of oxalate, acetate, and CO2 by Oxalobacter formigenes. Can. J. Microbiol. 42:1081–1086 [DOI] [PubMed] [Google Scholar]
- 22. Delmotte N, et al. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dutton MV, Evans CS. 1996. Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 42:881–895 [Google Scholar]
- 24. Erb TJ, et al. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. U. S. A. 104:10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erb TJ, Brecht V, Fuchs G, Müller M, Alber BE. 2009. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc. Natl. Acad. Sci. U. S. A. 106:8871–8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erb TJ, Frerichs-Revermann L, Fuchs G, Alber BE. 2010. The apparent malate synthase activity of Rhodobacter sphaeroides is due to two paralogous enzymes, (3S)-malyl-coenzyme A (CoA)/{beta}-methylmalyl-CoA lyase and (3S)-malyl-CoA thioesterase. J. Bacteriol. 192:1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Erb TJ, Fuchs G, Alber BE. 2009. (2S)-Methylsuccinyl-CoA dehydrogenase closes the ethylmalonyl-CoA pathway for acetyl-CoA assimilation. Mol. Microbiol. 73:992–1008 [DOI] [PubMed] [Google Scholar]
- 28. Erb TJ, Retey J, Fuchs G, Alber BE. 2008. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283:32283–32293 [DOI] [PubMed] [Google Scholar]
- 29. Franceschi VR, Nakata PA. 2005. Calcium oxalate in plants: formation and function. Annu. Rev. Plant Biol. 56:41–71 [DOI] [PubMed] [Google Scholar]
- 30. Fu D, Sarker RI, Abe K, Bolton E, Maloney PC. 2001. Structure/function relationships in OxlT, the oxalate-formate transporter of Oxalobacter formigenes. Assignment of transmembrane helix 11 to the translocation pathway. J. Biol. Chem. 276:8753–8760 [DOI] [PubMed] [Google Scholar]
- 31. Green PN. 2006. Methylobacterium, 3rd ed. Springer, New York, NY [Google Scholar]
- 32. Khambata SR, Bhat JV. 1953. Studies on a new oxalate-decomposing bacterium, Pseudomonas oxalaticus. J. Bacteriol. 66:505–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khammar N, et al. 2009. Use of the frc gene as a molecular marker to characterize oxalate-oxidizing bacterial abundance and diversity structure in soil. J. Microbiol. Methods 76:120–127 [DOI] [PubMed] [Google Scholar]
- 34. Kiefer P, et al. 2009. Metabolite profiling uncovers plasmid-induced cobalt limitation under methylotrophic growth conditions. PLoS One 4:e7831 doi:10.1371/journal.pone.0007831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiefer P, Delmotte N, Vorholt JA. 2011. Nanoscale ion-pair reversed-phase HPLC-MS for sensitive metabolome analysis. Anal. Chem. 83:850–855 [DOI] [PubMed] [Google Scholar]
- 36. Klamt S, Saez-Rodriguez J, Gilles ED. 2007. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 1:2 doi:10.1186/1752-0509-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knief C, et al. 22 December 2011. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. [Epub ahead of print.] doi:10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Korotkova N, Chistoserdova L, Kuksa V, Lidstrom ME. 2002. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184:1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korotkova N, Lidstrom ME. 2001. Connection between poly-beta-hydroxybutyrate biosynthesis and growth on C(1) and C(2) compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Korotkova N, Lidstrom ME, Chistoserdova L. 2005. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 187:1523–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laukel M, Chistoserdova L, Lidstrom ME, Vorholt JA. 2003. The tungsten-containing formate dehydrogenase from Methylobacterium extorquens AM1: purification and properties. Eur. J. Biochem. 270:325–333 [DOI] [PubMed] [Google Scholar]
- 42. Lung HY, Baetz AL, Peck AB. 1994. Molecular cloning, DNA sequence, and gene expression of the oxalyl-coenzyme A decarboxylase gene, oxc, from the bacterium Oxalobacter formigenes. J. Bacteriol. 176:2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067 [DOI] [PubMed] [Google Scholar]
- 44. Okubo Y, Yang S, Chistoserdova L, Lidstrom ME. 2010. Alternative route for glyoxylate consumption during growth on two-carbon compounds by Methylobacterium extorquens AM1. J. Bacteriol. 192:1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peyraud R, et al. 2009. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U. S. A. 106:4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peyraud R, et al. 2011. Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1. BMC Syst. Biol. 5:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pham TV, Piersma SR, Warmoes M, Jimenez CR. 2010. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26:363–369 [DOI] [PubMed] [Google Scholar]
- 48. Quayle JR. 1963. Carbon assimilation by Pseudomonas oxalaticus (OX1). 6. Reactions of oxalyl-coenzyme A. Biochem. J. 87:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quayle JR. 1963. Carbon assimilation by Pseudomonas oxalaticus (Ox1). 7. Decarboxylation of oxalyl-coenzyme a to formyl-coenzyme A. Biochem. J. 89:492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quayle JR, Keech DB. 1960. Carbon assimilation by Pseudomonas oxalaticus (OX1). 343. Oxalate utilization during growth on oxalate. Biochem. J. 75:515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quayle JR, Keech DB. 1959. Carbon assimilation by Pseudomonas oxalaticus (OX 1). 1. Formate and carbon dioxide utilization during growth on formate. Biochem. J. 72:623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quayle JR, Keech DB. 1959. Carbon assimilation by Pseudomonas oxalaticus (OX 1). 2. Formate and carbon dioxide utilization by cell-free extracts of the organism grown on formate. Biochem. J. 72:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Quayle JR, Keech DB, Taylor GA. 1961. Carbon assimilation by Pseudomonas oxalaticus (OXI). 4. Metabolism of oxalate in cell-free extracts of the organism grown on oxalate. Biochem. J. 78:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quayle JR, Taylor GA. 1961. Carbon assimilation by Pseudomonas oxalaticus (OXI). 5. Purification and properties of glyoxylic dehydrogenase. Biochem. J. 78:611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sahin N. 2003. Oxalotrophic bacteria. Res. Microbiol. 154:399–407 [DOI] [PubMed] [Google Scholar]
- 56. Sahin N, Aydin S. 2006. Identification of oxalotrophic bacteria by neural network analysis of numerical phenetic data. Folia Microbiol. (Praha) 51:87–91 [DOI] [PubMed] [Google Scholar]
- 57. Sahin N, Kato Y, Yilmaz F. 2008. Taxonomy of oxalotrophic Methylobacterium strains. Naturwissenschaften 95:931–938 [DOI] [PubMed] [Google Scholar]
- 58. Schneider K, Asao M, Carter MS, Alber BE. 2012. Rhodobacter sphaeroides uses a reductive route via propionyl coenzyme A to assimilate 3-hydroxypropionate. J. Bacteriol. 194:225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schneider K, et al. 2012. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium extorquens AM1 during growth on acetate. J. Biol. Chem. 287:757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sidhu H, et al. 1998. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet 352:1026–1029 [DOI] [PubMed] [Google Scholar]
- 61. Sidhu H, et al. 1997. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol. 179:3378–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith LM, Meijer WG, Dijkhuizen L, Goodwin PM. 1996. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology 142:675–684 [DOI] [PubMed] [Google Scholar]
- 63. Smith PK, et al. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 64. Smith RL, Strohmaier FE, Oremland RS. 1985. Isolation of anaerobic oxalate-degrading bacteria from fresh-water lake-sediments. Arch. Microbiol. 141:8–13 [Google Scholar]
- 65. Svedruzić D, et al. 2005. The enzymes of oxalate metabolism: unexpected structures and mechanisms. Arch. Biochem. Biophys. 433:176–192 [DOI] [PubMed] [Google Scholar]
- 66. Sy A, Timmers AC, Knief C, Vorholt JA. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 71:7245–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vandamme P, Coenye T. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285–2289 [DOI] [PubMed] [Google Scholar]
- 68. Vuilleumier S, et al. 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.