Abstract
The guinea pig cytomegalovirus (GPCMV) co-linear gene and potential functional homolog of HCMV UL84 (GP84) was investigated. The GP84 gene had delayed early transcription kinetics and transient expression studies of GP84 protein (pGP84) demonstrated that it targeted the nucleus and co-localized with the viral DNA polymerase accessory protein as described for HCMV pUL84. Additionally, pGP84 exhibited a transdominant inhibitory effect on viral growth as described for HCMV. The inhibitory domain could be localized to a minimal peptide sequence of 99 aa. Knockout of GP84 generated virus with greatly impaired growth kinetics. Lastly, the GP84 ORF was capable of complementing for the loss of the UL84 coding sequence in a chimeric HCMV. Based on this research and previous studies we conclude that GPCMV is similar to HCMV by encoding single copy co-linear functional homologs of HCMV UL82 (pp71), UL83 (pp65) and UL84 genes.
Introduction
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that causes asymptomatic infection in normal healthy individuals. However, it has emerged as a serious infection in immune suppressed individuals, including organ transplant and AIDS patients as well as the fetus during pregnancy (Pass, 2001). Indeed, congenital infection of the fetus by HCMV (1–2% of live births in the US) is a major cause of mental retardation and hearing loss in surviving newborns in the developed world (Ross & Boppana, 2004; Griffiths & Walter, 2005). Although there are candidate vaccines in clinical trials there is currently no effective vaccine to HCMV (Schleiss & Heineman, 2006). Antivirals are available but these act at late stages of the virus life cycle and result in the development of resistant strains emerging during prolonged antiviral therapy (Biron, 2006; Chou, 2008; McGregor 2010). Consequently, there is a need for the development of new vaccine strategies as well as the development of novel antiviral strategies that act at an earlier stage of the virus life cycle and less likely to lead to the development of resistant strains of virus.
In human cytomegalovirus (HCMV) the UL82, UL83 and UL84 genes are thought to have evolved from a common ancestor by gene duplication, with subsequent differentiation to fulfill unique functions associated with the virus life cycle (Davison & Stow, 2005). Both UL82 and UL83 encode tegument proteins whereas UL84 encodes a non-structural protein (He et al., 1992; Hensel et al., 1992; Schmolke et al., 1995). Both tegument proteins, pp71 (UL82) and pp65 (UL83), have roles in initiating viral life cycle in the infected cell as well as avoiding the cellular innate immune response (Abate et al., 2004; Baldick et al., 1997; Cantrel et al., 2006). The function of UL84 protein in the HCMV viral life cycle is not entirely understood but it has an essential role insofar as knockout HCMV UL84 mutants are non viable (Dunn et al., 2003; Xu et al., 2004; Yu et al., 2003). The UL84 proteins are unique to betaherpesviruses and in HCMV it is required in an auxiliary role for HCMV DNA replication (Sarisky & Hayward, 1996; Xu et al., 2004). Specific interaction of pUL84 with the DNA polymerase accessory protein (pUL44) has been demonstrated (Strang et al., 2009). However, Reid et al., (2003) using HSV-1 DNA replication fork proteins showed that replication from the HCMV origin site is independent of UL84 protein. Initially identified as a nuclear protein, pUL84 can shuttle between the nucleus and the cytoplasm and multimerize with itself (Colletti et al., 2004; He et al., 1992; Lischka et al., 2003; Lischka et al., 2006; Xu et al., 2002). The purpose of pUL84 shuttling has yet to be defined but involves the alpha beta importin pathway (Lischka et al., 2003). Potentially, shuttling may be linked to pUL84 homology with DExD/H box family of helicases, which shuttle between the nucleus and cytoplasm (Colletti et al., 2005; Lischka et al., 2006). The viral protein is also suggested to have UTPase activity (Colletti et al., 2005) and other pUL84 homologs, including GPCMV, carry UTPase motifs (Davison & Stow, 2005). Additionally, pUL84 exhibits a transdominant inhibitory growth effect on HCMV and interaction studies determined that the inhibitory effect is linked to specific interaction with IE2-p86 that impairs transactivation (Gebert et al., 1997; Colletti et al, 2004). Since IE2-p86 is important for the initiation of HCMV viral replication, this transdominant inhibitory effect could potentially be developed as a novel antiviral strategy, which would impact on virus replication at an earlier stage than current antivirals. Therefore in addition to identifying GPCMV GP84 as a functional homolog we sought to identify a minimal peptide domain with a transdominant inhibitory effect on GPCMV growth for potential development as an antiviral in the guinea pig model.
The strict species specificity of HCMV precludes studying this virus in a non-human animal model. Consequently, animal CMVs are used in their respective hosts for viral pathogenicity or candidate vaccine studies. Various animal models have been explored in the study of CMV (Schleiss, 2006; Barry et al., 2006). The guinea pig model of CMV is uniquely useful for the study of congenital infection and the development of intervention strategies that prevent infection of the fetus (Kumar & Nankervis, 1978; Kern, 2006; Schleiss, 2002). One focus of vaccines against HCMV has been directed towards the pp65 viral tegument protein (UL83), the major CTL target (Gonczol & Plotkin, 2001). The homolog protein, GP83, in GPCMV has been successfully used as protective vaccine in the guinea pig model (McGregor et al., 2004; Schleiss et al., 2007). In contrast, the M84 gene in murine cytomegalovirus (MCMV), while co-linear to UL84, encodes a functional homolog to pp65 (UL83) and is a major CTL target (Ye et al., 2004). Accordingly, M84 vaccines provide protection against MCMV challenge in mice (Holtappels et al., 2001; Morello et al., 2000). Hence, from the standpoint of vaccine development studies in the guinea pig model it was important to more properly define the function of the co-linear homolog to UL84, GP84, in the GPCMV life cycle. We previously identified proteins encoded by the GPCMV genes GP82 and GP83 as tegument proteins and direct functional homologs of HCMV encoded proteins pp71 (UL82) and pp65 (UL83) (McGregor et al., 2004; Schleiss et al., 1999). Preliminary studies suggest that the predicted pGP84 is potentially a functional homolog of pUL84 based on the presence of conserved UTPase motifs not found in the MCMV homolog (Davison & Stow, 2005). Therefore in this paper we attempted to demonstrate that pGP84 is a functional homolog pUL84.
Results
GP84 expression and cellular location of pGP84 protein
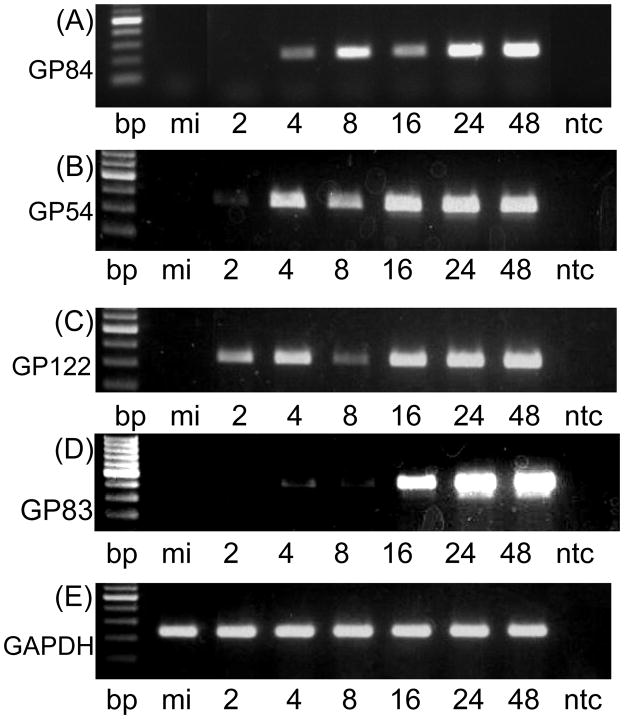
The GP84 ORF encodes a protein of 483 aa. in length that exhibits the highest homology to HCMV UL84 protein (22% identity by BLAST analysis). Analysis of the kinetics of gene expression in wild type virus infected cells reveals the GP84 gene expression to be delayed early kinetics (Figure 1), which is similar to HCMV UL84 (He et al., 1992). This conclusion is based on comparative RT-PCR assays also performed for different classes of GPCMV genes previously characterized (Figure 1): immediate early, IE2, unique exon GP122 (Yamada et al., 2009); early, GP54, the viral polymerase (Schleiss, 1995); late, GP83, encoding the homolog to pp65 (Schleiss et al., 1999). Additionally, the class of kinetics was also confirmed by the inclusion of protein inhibitor cycloheximide (for IE transcripts) or DNA replication inhibitor phosphonoacetic acid, PAA (for detection of IE and E transcripts) in virus infected cultures. GP84 transcripts were detected in the presence of PAA but not in the presence of cycloheximide which confirmed GP84 as an E transcript, see supplementary Figure S1.
Figure 1. Expression studies of GP84.
1(i) RT-PCR assay of GP84 mRNA expression in virus infected cells. Wild type virus infected GPL cells (moi=3 pfu/cell) were harvested at different time points for RNA extraction and mRNA analysis by RT-PCR using GP84, GP54 (viral polymerase), GP122 (IE2 unique exon) and cellular GAPDH specific primers as described in materials and methods. Viral gene classes: immediate early, IE2, unique exon GP122 (Yamada et al., 2009); early, viral polymerase GP54 (Schleiss, 1995); and late, GP83, encoding pp65 homolog (Schleiss et al., 1999). RT-PCR products were analyzed by agarose gel electrophoresis: GP84 (gel A); GP54 (gel B); GP122 (gel C); and GAPDH control (gel D). In all gels lanes are as the same. 100 bp ladder (NEB) followed by mock infected cells (mi), time samples 2–48 hr post infection (hpi) and no template control (ntc). Time samples 2, 4, 8, 16, 24 and 48 hr post infection.
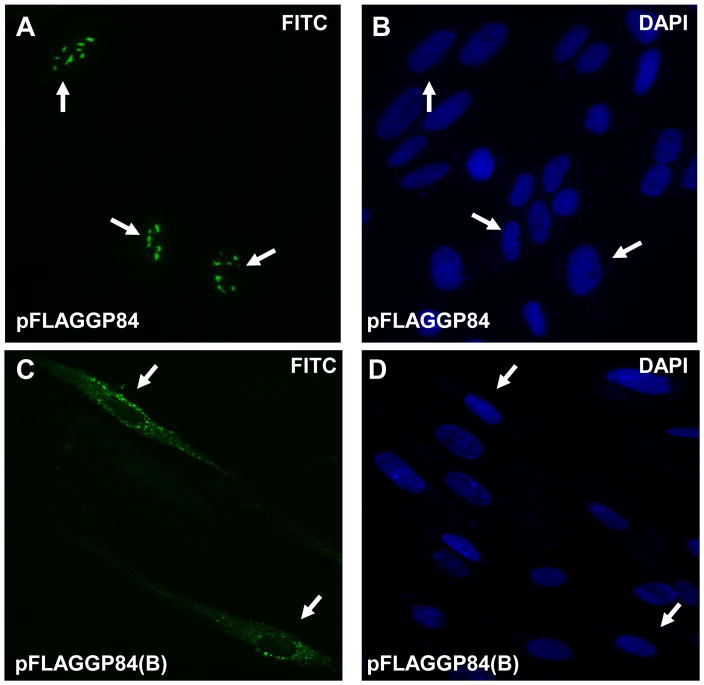
1(ii) Transient expression studies of full length and N-terminal truncated GP84. Immunofluorescence assay of FLAG-tagged GP84 protein transiently expressed in GPL cells transfected with pFLAGGP84 or pFLAGGP84(B). Assay performed at 10 hr post transfection. Primary antibody anti-FLAG and secondary antibody anti-mouse IgG conjugated to FITC (images A and C). Cells counterstained with DAPI (images B and D). Arrow indicates same cell under FITC and DAPI filters. Images A and B, full length pGP84 (pFLAGGP84 plasmid). Images C and D, N-terminal truncated pGP84 (pFLAGGP84(B) plasmid).
The full length GP84 ORF was cloned in frame into the expression vector pCMV2A (Promega), which tagged the N-terminal domain of the ORF with an epitope FLAG tag. Transient expression studies of pGP84 were carried out on GPL cells. The cellular distribution of pGP84 was determined at various times between 3 and 48 hr post transfection via immunofluorescence assay. Figure 1(ii), image A, indicates that pGP84 is a nuclear protein and this result is consistent with the cellular location reported for HCMV pUL84 (Xu et al., 2002). Using convenient restriction enzyme sites a series of collapses were made to the full length GP84 ORF to generate N- and C-terminal deletions as described in materials and methods (and shown in Supplementary Figure S2). In transient cellular expression assays, the N-terminal collapsed pGP84 (lacking the first 211 aa of full length pGP84) encoded on plasmid pFLAGGP84(B) appeared only in the cytoplasm and not in the nucleus (Figure 1(ii), image C). In contrast, the C-terminal and combined N- and C-terminal truncated versions of pGP84 (encoded on plasmids pFLAGGP84(S) and pFLAGGP84(B) respectively) were found in both nuclear and cytoplasmic compartments (Supplementary Figure S3). This indicated that pGP84 carries a N-terminal NLS as described for HCMV pUL84 (Xu et al., 2002). However, further study would be required to determine if pGP84 is dependent upon the alpha and beta importin pathway for nuclear targeting as described for HCMV (Lischka et al., 2006).
pGP84 co-localizes with the viral DNA polymerase accessory protein (pGP44)
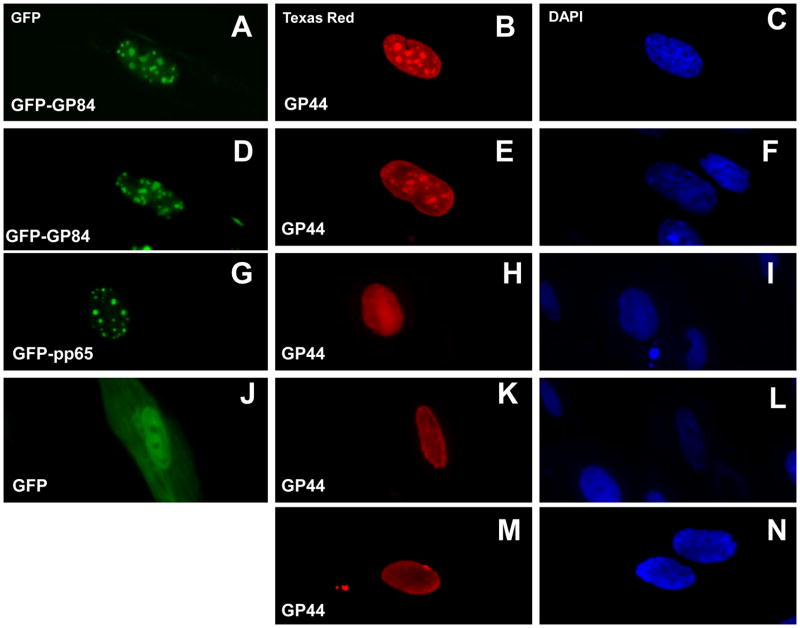
HCMV pUL84 plays an accessory role in viral DNA replication and has recently been shown to interact with the viral DNA polymerase accessory protein, pUL44, (Strang et al., 2009). The UL44 homolog gene (GP44) in GPCMV (Schleiss et al., 2008) encodes a protein of 407 amino acids that exhibits 57% identity with HCMV pUL44 by BLAST analysis (McGregor, unpublished data). In an effort to demonstrate similar interactions between GPCMV homolog proteins (pGP84 and pGP44) the GP44 coding sequence was cloned in frame into a FLAG tag expression plasmid to epitope tag the GP44 ORF. Transient expression studies were carried out in the presence of GFP tagged pGP84 expression plasmid or control GFP plasmids (expressing GFP only or a HCMV pp65 GFP fusion). Interactions were followed by cellular co-localization studies via immunofluorescence assay. Figure 2, panels A–F, show the co-localization of pGP84 and pGP44 in the nucleus of cells expressing both proteins. Identical patterns of nuclear aggregates were observed under GFP filter for pGP84 (panels A and D) or Texas red filter for pGP44 (panels B and E). Co-expression of pGP44 and GFP protein did not indicate any specific co-localization (Figure 2, panels J and K). The GFP protein was seen evenly distributed in both nuclear and cytoplamic compartments, whereas pGP44 was only located in the nucleus and was uniform in the nucleus without aggregates seen in the presence of pGP84. In another control experiment, co-expression of pGP44 with HCMV pp65-GFP fusion protein did not give rise to pGP44 nuclear aggregates (Figure 2, panels G and H). The pp65 protein produced characteristic punctate nuclear fluorescence but the pGP44 protein was evenly distributed in the nucleus. The expression pattern of pGP44 only (Figure 2, panel M) is the same pattern seen in GFP and GFP-pp65 co-transfection cells (K and H respectively). Overall the transient GP84 expression studies suggest that the pGP84 has similar cell localization properties to pUL84 and potentially interacts with the viral DNA replication machinery as demonstrated for HCMV pUL84 (Strang et al., 2009; Sarisky & Hayward, 1996).
Figure 2. Co-localization studies of transiently expressed pGP84 and pGP44.
Transient plasmid expression assay of FLAG tagged pGP44 (pFLAGGP44 plasmid) in the presence of GFP tagged vectors: GFP tagged pGP84 (images A–F); GFP tagged pp65 (G–I); GFP control (J–L). Images M and N correspond to transient expression of pGP44 only cells. pGP44 detected by anti-FLAG primary antibody and secondary antibody anti-mouse IgG conjugated to TRITC. pGP44 images B, E, H and M. Cells counterstained with DAPI to identify cell nucleus (right column). Image A and D, pGP84 expressed as N-terminal tagged GFP protein (pGFPGP84). Image G, pGFPpp65 (HCMV pp65 N-terminal GFP tagged). Image J, control GFP plasmid pGFP-C1 (Clontech). Protein cellular localization determined at 12 hr post transfection as described in materials and methods.
Transdominant Inhibitory effect of overexpression of GP84 on GPCMV growth
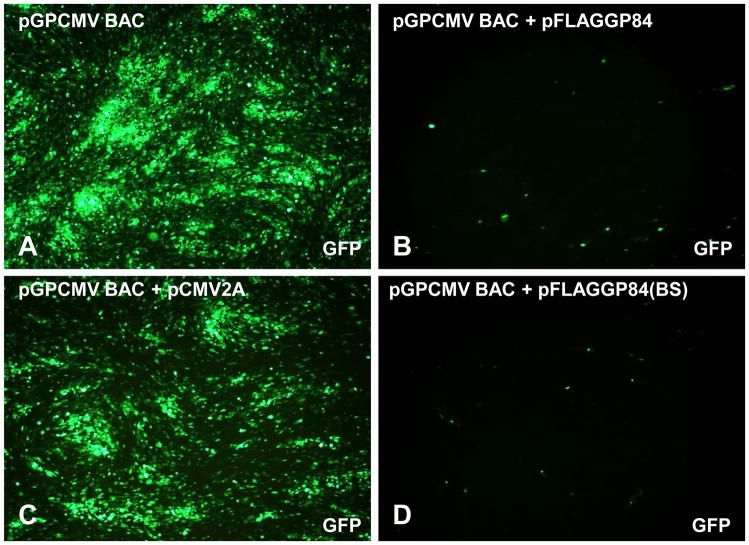
Gerbert et al., (1997) demonstrated that over expression of pUL84 in a human cell line resulted in subsequent impairment of HCMV growth, indicating that pUL84 has a transdominant inhibitory effect on HCMV when over expressed under HCMV IE promoter control. The specific interaction of pUL84 with the immediate early transactivation protein IE2 is the basis for the inhibitory effect (Gerbert et al., 1997). A GPCMV homolog to IE2 has recently been identified with the encoded protein sequence in the unique GPCMV IE2 exon (GP122) showing 62% identity to HCMV pUL122 (McGregor, unpublished data; Schleiss et al., 2008; Yamada et al., 2009). Therefore it seemed probable that a similar transdominant inhibitory effect for GPCMV could be demonstrated. Two transdominant inhibitory GPCMV growth assays were developed. In the first assay, infectious GPCMV BAC DNA (McGregor & Schleiss, 2001) was co-transfected onto confluent monolayers of GPL cells in the presence or absence of GP84 expression plasmids or control plasmids. Virus development was allowed to proceed for a similar length of time and final viral titers from individual wells determined at the end of the experiment (day 18). During the experiment GFP positive transfected cells, GPCMV replication and viral spread could easily be followed in real time by virtue of the GFP gene incorporated into the GPCMV genome. Transfection of the GPCMV BAC DNA by itself or with the empty expression vector resulted in rapid generation of viral plaques and GFP viral spread across the monolayer (Figure 3, panels A and C). The presence of the empty expression vector (pCMV2A) did not affect the development of viral spread and only slightly impaired viral yield, 1.5 × 106 pfu/ml in the absence of pCMV2A (empty vector) compared to 1.4 × 106 pfu/ml in the presence of pCMV2A. In contrast, co-transfection of GPCMV BAC DNA with the GP84 expression vector (pFLAGGP84) severely impaired development of virus and viral spread across the monolayer (Figure 3, panel B) with an average virus yield of 1x 102 pfu/ml. N-terminal or C-terminal truncated versions of GP84 were equally as effective at inhibiting GPCMV virus growth (average virus yield 1x 102 pfu/ml). Surprisingly, the double truncated N- and C-terminal GP84 collapse that encodes 99 aa (codons 212–311) of pGP84 sequence (pFLAGGP84BS) was equally capable of inhibiting GPCMV (average virus yield 1x 102 pfu/ml) as full length pGP84 (pFLAGGP84), Figure 3 (image B and D). The use of VP16 (HSV-1) or GP83 (GPCMV pp65 homolog) expression plasmids (McGregor et al., 2004) produced similar results to GPCMV BAC co-transfection with empty vector (1.2 × 106 pfu/ml and 1.3 × 106 pfu/ml respectively) and had no effect on virus growth. These results indicate that pGP84 has a trans-dominant inhibitory activity against GPCMV similar to that described for pUL84 against HCMV (Gerbert et al., 1997). Overall the pGP84 transdominant inhibitory assay impaired virus yield by approximately 10,000 fold. However, only a small 99 aa domain (codons 212–311) would appear to be sufficient for inhibitory growth effect. A similar minimal inhibitory domain has not been identified for pUL84.
Figure 3. GPCMV pGP84 transdominant growth inhibition assay.
GFP tagged GPCMV BAC DNA was transfected onto GPL cells alone or co-transfected with GP84 expression vectors (pFLAGGP84 or pFLAGGP84BS) or empty expression vector (pCMV2A). Development of viral plaques could be detected by GFP reporter gene expression in infected cells. Viral spread was monitored between 7 and 18 days post transfection. At day 18 all wells were harvested and viral titer determined. Data shown is representative GFP virus spread in wells of a six well dish at 16 days post transfection. Images: GPCMV BAC DNA only well, panel A; GPCMV BAC DNA plus empty vector (pCMV2A), panel C; B. GPCMV BAC DNA plus pFLAGGP84 (full length pGP84), panel B; GPCMV BAC DNA plus pFLAGGP84BS (truncated pGP84), panel D. Virus inhibition was obtained in the presence of full length or truncated pGP84 (images B and D).
The GPCMV BAC based transdominant inhibitory assay did not demonstrate a direct inhibitory effect on live virus infection as virus was derived from GPCMV BAC transfected cells. Consequently, a second inhibitory growth assay was developed that tested the ability of pGP84 to impair live GPCMV infection of GPL cells. Initially GPL cell lines were established that expressed the pGP84 minimal inhibitory domain fused to the C-terminal domain of GFP (pEGFP-C1) as described in materials and methods. As a control, GPL cell lines were established that only expressed GFP. Clonal cell lines were designated GPL-GFPGP84ID and GPL-GFP respectively and were tested for their ability to inhibit virus growth at different input virus moi. The pGP84 inhibitory effect on virus growth was most pronounced at the lowest moi (0.001–0.01 pfu/cell) with a 3 log drop in virus yield in comparison to the equivalent infection on control GPL–GFP cells (Table 1). The results demonstrate that the minimal pGP84 inhibitory domain when expressed in trans was effective at inhibiting live virus infection and verifies the results seen in the initial inhibitory assay. The pGP84 inhibitory effect was specific to GPCMV as no viral inhibition was seen with HSV-1 (data not shown). In HCMV the transdominant effect is thought to occur by pUL84 interacting with IE2-p86 to inhibit transactivation of early genes by IE2 protein (Gerbert et al., 1997). Although a homolog to IE2 protein has recently been identified in GPCMV (Schleiss et al., 2008; Yamada et al., 2009) further studies are required to characterize the IE2 transactivation function as well as IE2 protein-protein interactions.
Table 1. Transdominant inhibitory effect on GPCMV growth by the pGP84 minimal inhibitory domain expressed by permanently transduced cell.
Input virus infection is indicated and final virus yield determined by titration of infected cells at a set time point post infection as described in materials and methods. GPL-GFP, control cells expressing GFP. GPL GFP-GP84ID, cells expressing the pGP84 99 aa inhibitory domain fused to the C-terminal of GFP.
| Input GPCMV (pfu/cell) | Viral titer GFP cells | Viral titer GFPGP84ID cells |
|---|---|---|
| 0.1 | 2.5 × 106 pfu/ml | 3 × 104 pfu/ml |
| 0.01 | 1 × 106 | 4 × 103 |
| 0.001 | 2 × 106 | 5 × 103 |
Targeted knockout of GP84 in GPCMV
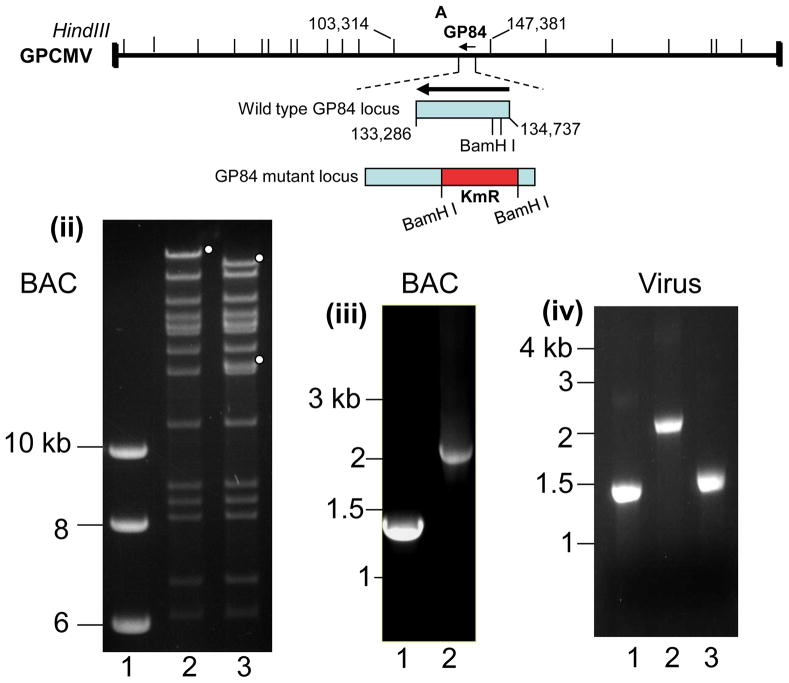
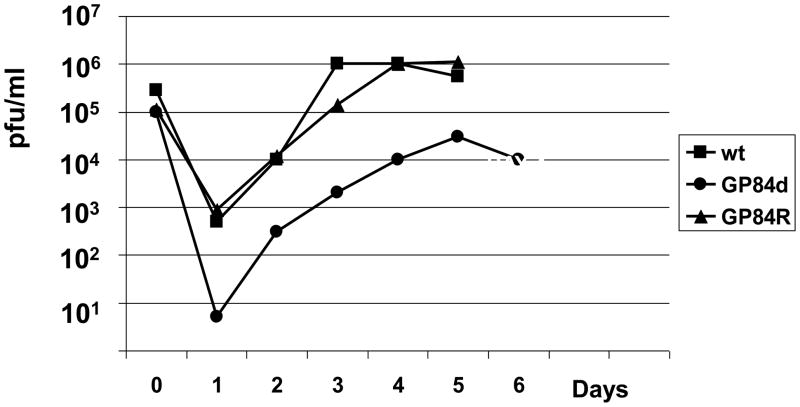
In HCMV the UL84 gene is essential for virus replication (Dunn et al., 2003; Xu et al., 2004; Yu et al., 2003) we sought to determine the essential nature of GP84 by specific gene knockout in an infectious GPCMV BAC (McGregor et al., 2001). An insertion deletion was introduced into the GP84 ORF by the insertion of a kanamycin cassette between two BamH I sites close to the start of the GP84 ORF in a GP84 shuttle vector for targeted knockout of GP84. Figure 4(i), shows the location of the GP84 gene knockout on the GPCMV genome. Full length mutant GPCMV BACs were screened by Hind III restriction profile analysis with the correct insertion of the Km cassette resulting in the disruption of the GPCMV Hind III ‘A’ band, co-ordinates 103,313–147,381 bp, (Schleiss et al., 2008; Gao and Isom et al., 1984). Figure 4(ii) shows the modified restriction pattern of the GP84 GPCMV BAC mutant under Hind III restriction enzyme profile analysis. The modified GP84 locus was further verified by PCR analysis using common PCR primers flanking the modified GP84 locus. PCR analysis of wild type and GP84 mutant GPCMV BAC DNA verified the GP84 locus was correctly modified by insertion of a Km cassette and small deletion to the GP84 coding sequence, see Figure 4(iii). Transfection of the GP84 mutant GPCMV BAC onto GPL cells resulted in the generation of infectious virus, designated vGP84d. PCR analysis of viral DNA confirmed that the mutant GP84 locus was retained in the virus unless rescued back to wild type, see Figure 4(iv). A multi-step growth curve of vGP84d, Figure 4(iv), demonstrated that vGP84d had impaired growth kinetics in comparison to wild type virus, or rescue virus (GP84R). Additionally, the mutant produced greatly enlarged cells in comparison to wild type virus (data not shown). The ability to generate a viable GP84 knockout virus contrasts with an absolute requirement for UL84 to generate viable HCMV (Dunn et al., 2003; Yu et al., 2003; Xu et al., 2002). However, there is a greater level of impairment associated with a GP84 knockout in comparison to knockout of the co-linear UL84 gene homolog in MCMV (M84), which does not exhibit any impaired growth kinetics in tissue culture (Morello et al., 1999).
Figure 4. Characterization of GP84 knockout GPCMV mutant (GP84d).
(i) GPCMV genome map of Hind III sites and the location of the knockout GP84 gene. Shown is the GPCMV Hind III ‘A’ locus (co-ordinates 103,314–147,381 bases) encoding GP84 CDS (complement 133,286–134,737 bases). Insertion of a kanamycin resistance cassette into the GP84 coding sequence between two BamH I sites (134,097 and 134,314 bases) results in an insertion deletion disruption of the GP84 coding sequence.
(ii) Wild type and GP84 mutant GPCMV BAC Hind III restriction profile analysis.Agarose gel of the DNA restriction profile analysis of mutant and wild type GPCMV BACs. Lanes: 1. kb ladder (NEB). 2. wt GPCMV BAC; 3. GP84d GPCMV BAC. Insertion of a kanamycin cassette into the GP84 gene introduces a new Hind III site into the GPCMV genome and disrupts the Hind III ‘A’ band encoding the GP84 gene. Full length (lane 2) and modified bands encoding part of the Hind III ‘A’ band (lane 3) are indicated with circles.
(iii) GPCMV BAC Diagnostic PCR analysis of the GP84 locus.Common primers flanking the site of modification in the GP84 locus were used to verify the modification to the GP84 locus in comparison to the wild type locus. Shown is an agarose gel of GP84 PCR products from different BAC DNA templates. Lanes: 1. wt GPCMV BAC; 2. GP84d GPCMV BAC mutant. Shift in the size of the locus confirms correct insertion of the kanamycin cassette plus deletion of GP84 sequence between BamH I sites.
(iv) Viral DNA Diagnostic PCR analysis of the GP84 locus. Common diagnostic GP84 locus primers were used to confirm the status of the GP84 locus in viral genomes: 1. wt GPCMV; 2. GP84d mutant; 3. GP84d rescue virus. The mutant virus retains the same modification generated in the GPCMV BAC mutant and the rescue virus has a wild type sized GP84 locus.
(v) Multi step growth curve of GPCMV GP84 mutant (GP84d)Growth curve performed as described in materials and methods. GPCMV GP84 knockout mutant (GP84d) was compared to wild type GPCMV (wt) and GP84d rescue virus (GP84R). Input virus moi was 0.1 pfu/cell with time points taken at 24 hr. intervals between 0–6 days post infection.
Substitution of GP84 in place of UL84 in a chimeric HCMV
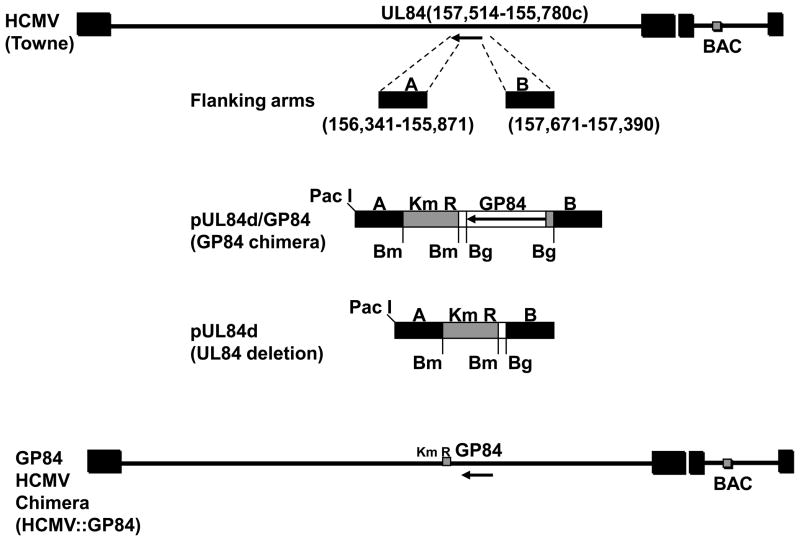
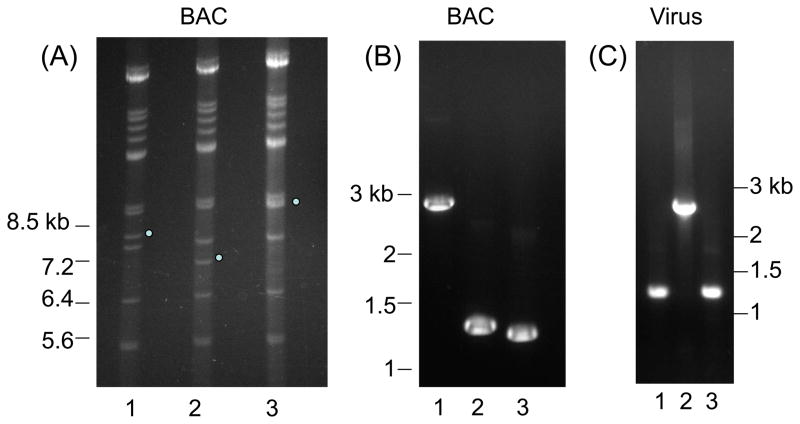
In an ultimate test of similarity of function between UL84 and GP84 we sought to determine if the GP84 coding sequence could functionally replace the UL84 coding sequence in a chimeric HCMV. Mutant HCMV were generated using a Towne strain HCMV BAC (Hahn et al., 2003). Two types of UL84 HCMV mutants were generated: a UL84 deletion mutant (HCMV::UL84d); a GP84 chimeric HCMV, encoding GP84 in place of UL84 (HCMV::GP84). A shuttle vector was generated for the UL84 locus for targeted modifications as described in materials and methods and is summarized in Figure 5(i). For generation of the chimeric HCMV, the FLAG tagged GP84 ORF was cloned into the pUL84d shuttle vector to generate pUL84d/GP84, which placed the GP84 ORF under UL84 promoter control. In separate recombination reactions the linearised plasmid DNA (pUL84d or pUL84d/GP84) was electroporated into recombination induced DH10B bacteria carrying the HCMV (Towne strain) BAC. Full length HCMV mutants (HCMV::UL84d or HCMV::GP84 BACs) were isolated and screened by Hind III restriction digestion to verify modification of the 7.8 kb subgenomic Hind III fragment (HCMV Towne co-ordinate nucleotides 120,817 – 128,615) predicted to encode UL84. Figure 5(ii)A shows the modifications to the UL84 locus, which were a result of the introduction of a novel Hind III site present in the Km cassette. The deletion mutant removes the majority of the UL84 coding sequence (codons 59-401) but introduces a Km cassette. The chimera removes the majority of the UL84 coding sequence and introduces the GP84 ORF and a Km cassette. The modifications to the UL84 locus were further verified by diagnostic PCR using common primers flanking the modified UL84 locus which confirmed the specific modifications to the UL84 locus, see Figure 5(ii)B.
Figure 5. Generation and characterization of GP84 chimeric HCMV.
(i) Overall strategy for the generation of UL84 knockout and GP84 chimera HCMV BAC mutants. Construction of UL84 knockout shuttle vectors. UL84 flanking sequence was generated by PCR (right arm left arm, A and B). For UL84 deletion HCMV BAC a BamH I Km cassette was cloned into the shuttle vector to generate pUL84d. For generation of the GP84 chimeric HCMV the FLAG tagged GP84 ORF from pFLAGGP84 was PCR cloned as a Bgl II cassette 5’ to the Km cassette in plasmid pUL84d to generate construct pUL84d/GP84. Shuttle vectors were linearised by Pac I digestion prior to recombination in bacteria with HCMV Towne BAC following protocol described by McGregor et al. (2004). Structure of GP84 HCMV chimeric virus is shown in the bottom of the figure.
(ii). Analysis of HCMV wild type and UL84 mutant structures. (A) HCMV BAC DNA Hind III restriction profiles. Lanes: 1. wt HCMV BAC DNA; 2. UL84 deletion mutant HCMV BAC DNA; 3. GP84 chimera HCMV BAC DNA. Dots mark wild type or modified 7.8 kb Hind III subgenomic fragment (co-ordinates 120,817–128,615 HCMV Towne) encoding the UL84 wild type locus (lane 1) and modified by the deletion of the majority of the UL84 coding sequence and insertion of a Km cassette (UL84 deletion mutant HCMV BAC, lane 2) plus the insertion of the GP84 coding sequence (GP84 HCMV chimera BAC, lane 3).
(B) PCR analysis of wild type and mutant UL84 loci from HCMV BAC clones. Common PCR primers amplified the modified UL84 locus from the HCMV BAC constructs and PCR products were analysed by agarose gel electrophoresis. Lane: 1. GP84 chimeric HCMV BAC; 2. wild type HCMV BAC; 3. UL84 deletion HCMV BAC.
(C) PCR analysis of wild type and mutant UL84 loci from viral DNA. Common PCR primers amplified the modified UL84 locus from wild type and GP84 chimeric HCMV viral DNA and PCR products were analysed by agarose gel electrophoresis. Note that the rescue virus was generated to the lethal UL84 knockout. Lane: 1. wild type HCMV; 2. GP84 HCMV chimera; 3. UL84 deletion rescue virus. Diagnostic PCR confirms that the chimeric virus encodes a modified UL84 locus similar to the GP84 HCMV chimeric BAC (B, lane 1).
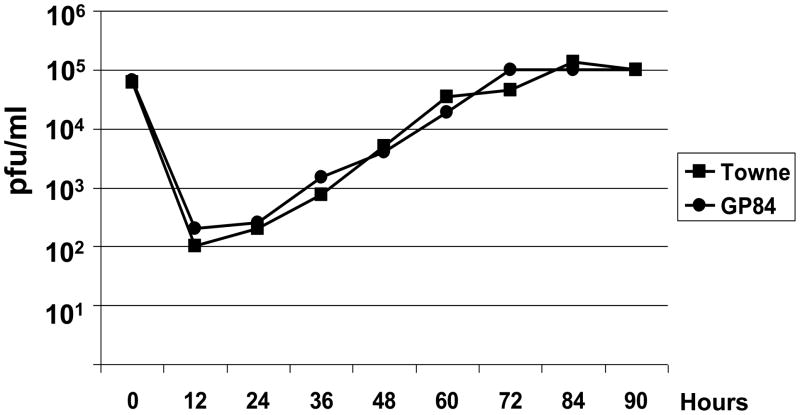
(iii) One step growth curve of wild type HCMV (Towne) and GP84 chimeric HCMV. Growth curve carried out as described in materials and methods with an initial input of virus of 3 pfu/cell (moi) with time point samples taken every 12 hr (0–90 hr post infection) as described in materials and methods.
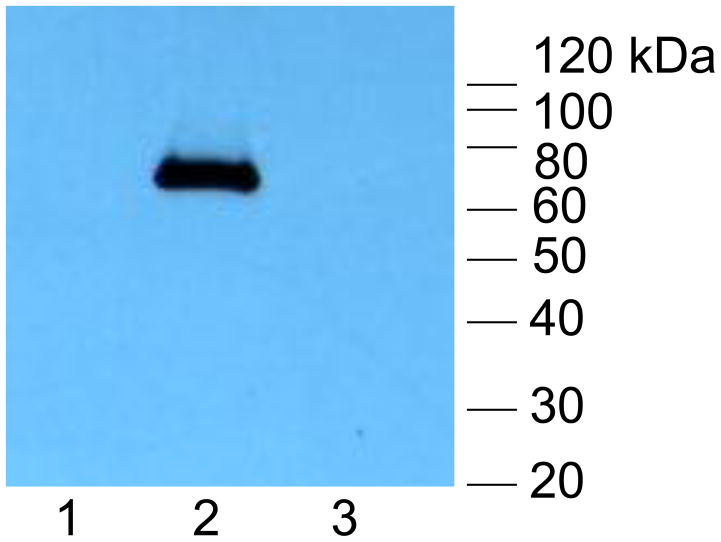
(iv) Western blot of FLAG-tagged GP84 protein expressed in chimeric virus infected cells. Lanes: (1) Mock infected HFF cells; (2) GP84 chimeric virus infected cells; (3) HCMV (Towne) infected cells. Protein ladder (Invitrogen). GP84 protein detected with primary antibody mouse anti-FLAG and secondary anti-mouse IgG conjugated to HRP as described in materials and methods.
Two independent HCMV BAC clones for each mutant were transfected separately onto HFF cells to generate virus. Despite repeated transfections of the UL84 deletion knockout HCMV BAC DNA (HCMV::UL84d) no virus was generated. This confirmed the essential nature of the UL84 gene. The lethal UL84 knockout could be rescued by co-transfection of the HCMV::UL84d BAC mutant with a rescue PCR product (3.7 kb fragment) encoding the wild type UL83-UL85 locus to generate virus (see material and methods). The chimeric HCMV BAC encoding GP84 (HCMV::GP84) generated viable virus, designated HCMV::GP84. A one step growth curve of HCMV::GP84 indicated that the chimera grew with similar kinetics to wild type HCMV (Towne) on HFF cells, see Figure 5(iii). Expression of FLAG tagged GP84 protein in chimeric HCMV infected HFF cells was confirmed by Western blot of infected cell lysate which detected a band of approximately 70 kDa, the predicted size of the GP84 protein, see Figure 5(iv). This data suggests that GP84 can functionally replace the loss of UL84 in a chimeric HCMV and provides the strongest evidence that GP84 is a functional homolog of UL84.
Discussion
In conclusion our studies on GP84 protein demonstrate that it is most likely the functional homolog of pUL84 in GPCMV as it shares a number of common features with pUL84. Furthermore, the GP84 ORF can substitute for the loss of the essential UL84 coding sequence in a chimeric HCMV. Further, studies are required to characterize the pGP84 NLS and the interactions of pGP84 with the GPCMV IE2 homolog. Preliminary studies indicate that the pGP84 NLS cannot be easily transplanted to another protein by the simple grafting of only the N-terminal domain to a reporter gene (McGregor, unpublished data). This suggests that the pGP84 carries a more complex bipartite NLS as described for HCMV pUL84 (Lischka et al., 2006). Analysis of the GPCMV IE2 unique exon GP122 indicates that it has high homology with HCMV UL122 exon, which suggests that GPCMV IE2 may function in a similar manner to HCMV IE2. However, no transactivation studies have been performed with the full length IE2 protein. Previous success has been achieved for analysis of transactivation activity associated with the GPCMV pp71 homolog, encoded in GP82 (McGregor et al., 2004). Additionally, the GPCMV genome has recently been fully sequenced (Schleiss et al., 2008; Yamada et al., 2009), which should allow the development of suitable IE2 transactivation/pGP84 interference assays. Since pUL84 is linked to virus replication it is curious that a MCMV M84 mutant is not impaired in tissue culture. This result certainly confuses the issue of MCMV being the primary animal model for pathogenicity, vaccine strategies against CMV. However, the GPCMV model is not a perfect substitute. The GP84 knockout mutant was capable of generating viable virus despite it being impaired. Possibly rodent CMV have evolved a DNA replication strategy that is more dependent upon a cellular protein rather than the viral pUL84 homolog. Reid et al., (2003) have demonstrated HCMV origin dependent DNA replication in the absence of pUL84, which supports an alternative DNA replication strategy not dependent on pUL84. This area awaits further investigation. The transient expression protein localization studies were performed with FLAG epitope and GFP tagged ORFs. This is a common strategy to study protein cellular localization in the absence of specific antibodies to the respective protein(s). Similar results were obtained for the same protein (pGP84 and pGP44) under both GFP or FLAG tagging and it is therefore unlikely that these tags contributed to cellular localization patterns. However, ideally future pGP84 studies should additionally employ protein specific antibodies as well as the continued use of FLAG or GFP tagged constructs.
In HCMV, the UL83 protein, pp65, is the protective T cells target and not pUL84 (Wills et al., 1996). In MCMV the co-linear homolog gene to UL84, M84, encodes the protective T cell target antigen but not the UL83 homolog, M83 (Morello et al., 1999; Ye et al., 2004). It was important to define GP84 as a functional homolog of UL84 as this has direct implications for the development of CMV candidate vaccines in the guinea model. Our results demonstrate the potential problem associated with the study of species specific animal CMV in vaccine studies and necessitates the functional definition of homolog proteins in addition to their use as candidate vaccine targets. In earlier studies we had identified pGP83 as encoding a tegument protein with properties similar to HCMV pp65 that acted as a protective T cell target antigen (Schleiss et al., 1999; McGregor et al., 2004; Schleiss et al., 2007). The immune response to pGP84 is unknown but does not seem to be as dominant as the T cell response to pGP83 in wild type virus infected animals (Schleiss et al., 2007). Therefore pGP84 is not likely to be a good vaccine target antigen as is the case for HCMV pUL84. Presumably, identification of pGP84 as a functional homolog to pUL84 will allow the future study of the role of pUL84 homolog in an animal model or the development of novel pUL84 based antiviral strategies in the guinea pig model.
Antivirals against HCMV are an important concern due to the development of resistant strains under current antiviral therapies (Biron et al., 2006). The finding that pGP84 has a transdominant inhibitory effect on viral growth, as does pUL84 on HCMV, is of potential interest from the standpoint of development of a novel antiviral drug strategy that could be tested in the guinea pig animal model. The ability to minimize the pGP84 inhibitory domain to 99 aa, or perhaps even smaller, could allow the future development of novel compounds that mimic this domain and which are also inhibitory towards the virus (Robinson et al., 2008). Potentially, an antiviral that inhibits IE2 protein could be more successful in combating CMV infection rather than current antivirals that inhibit late stages of the virus life cycle (Biron, 2006; McGregor, 2010) and allow emergence of resistant strains. Antiviral peptide strategies have been demonstrated to be effective against herpesvirus infections (Loregian et al., 2005). However, the further development an antiviral strategy in GPCMV awaits characterization of pGP84/IE2 homolog protein interactions. Stamminger and colleagues recently tested the ability of a peptide aptamer antiviral that targets the NLS of pUL84 (Kaiser et al., 2009). This could also be investigated in GPCMV but would first require further characterization of the pGP84 NLS.
Previously, in our attempts to increase the relevance of the GPCMV antiviral model we successfully generated a chimeric GPCMV encoding HCMV UL97 in place of the homolog gene GP97. This chimeric virus grew with wild type kinetics and was viable in vivo (McGregor et al., 2008; 2010). The generation of a viable HCMV chimera encoding GP84 in place of the essential UL84 is a reverse approach that also demonstrates similarity of gene function between GPCMV and HCMV. In conclusion, our findings on GPCMV GP84 and our previous studies on GP83 indicate a divergence of functions between co-linear homologs to UL83 and UL84 in MCMV and GPCMV. In GPCMV the co-linear homologs appear to strictly retain function similar to HCMV but in MCMV these functions are less conserved and appear to be shared between both proteins. Overall, these results in conjuction with our previous studies on GP83 and GP82, encoding homologs to pp65 and pp71 respectively, further increases the significance of the GPCMV animal model for pathogenicity, vaccine and antiviral studies.
Materials and Methods
Cells, viruses and oligonucleotides
GPCMV (strain 22122, ATCC VR682), GPCMV BAC derived mutant virus, and vAM403, a green fluorescent protein (GFP) tagged wild-type GPCMV (McGregor and Schleiss, 2001) were propagated on guinea pig fibroblast lung cells (GPL; ATCC CCL 158) as previously described (McGregor et al., 2004). HCMV was propagated on human foreskin fibroblast cells in DMEM (Gibco-BRL) with 10% FCS supplemented with 1 × anti-anti (Invitrogen). Virus titrations were carried out on six-well plates. Plaques were stained with 10% Giemsa stain or visualized by fluorescence microscopy. All oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, TX).
Cloning of GPCMV GP84 and generation of GP84 expression vectors and GP84 knockout shuttle vector
The full length GP84 ORF was PCR cloned from GPCMV viral DNA as a EcoR I /Hind III fragment into plasmid pNEB193 (New England Biolabs) using the PCR primers FGP84Ec (5’ GAATTCGATGATCGAAGAGACGGCCGAAC) and RGP84Hd (5’ AAGCTTACACGGATTTGGGCGCGGGATCGC) to generate pNEBGP84 and sequenced to verify the GP84 coding sequence. The GP84 ORF was cloned in frame as a EcoR I/Hind III fragment into the expression vector pCMV2A (Promega) to generate the plasmid pFLAGGP84, expressing the full length GP84 ORF (483 aa) tagged at the N -terminus with a FLAG epitope tag. Using convenient restriction enzyme sites (BamH I and Sal I) a series of collapses were made to the full length GP84 ORF to generate N- and C-terminal deletions (see supplementary Figure S1). A Sal I collapse at the 3’ end of the ORF generated a C-terminal deletion mutant encoding codons 1–311, designated pFLAGGP84(S). A 5’ BamH I collapse generated an inframe deletion at the N-terminus of the GP84 coding sequence between the FLAG tag and codon 212, designated pFLAGGP84(B). Collapse of both termini generated pFLAGGP84(BS). The complete GP84 ORF was also PCR cloned in-frame into the C-terminal polycloning site of pEGFP-C1 (Clontech) as an EcoR I fragment using the primers FGP84Ec and RGP84Ec (GAATTCACACGGATTTGGGCGCGGGATCGC). Additionally, a truncated version of the GP84 coding sequence (codons 170–309) was PCR cloned in frame into pEGFP-C1 (Clontech) as a Bgl II /Sal I fragment to generate pGFPGP84ID using the primers FGP84Bg (5’ AGATCTAAGAACGGCGACGAGCGACACAGCATC) and RGP84Sal (5’ GTCGACTTAGGGCGCCGTCACGATCAAGTG). For generation of a GP84 knockout shuttle vector the plasmid pNEBGP84 was cut with BamH I to create an internal deletion in the GP84 coding sequence (codons 140–213). A kanamycin (Km) cassette was PCR amplified from pACYC177 (NEB) using the primers using primers previously described (McGregor et al., 2004) and cloned into the pNEBGP84 BamH I collapse to generate the GP84 knockout vector pNEBGP84km(Bm). The GPCMV DNA polymerase accessory protein, GP44, coding sequence (Schleiss et al., 2008) was PCR cloned as a BamH I fragment in-frame into the expression vector pCMV2C to FLAG epitope the N terminus of the GP44 protein. GP44 PCR was performed using the primers F44BmXh (5’ GGATCCTCTCGAGCTTCTGCGACGCCGGAGGGTATCG) and R44BmXh (5’ GGATCCTCGAGTCAGGCGGAACATTTCTGCTTCTTGACGTTAGAC).
Generation of HCMV UL84 knockout shuttle vector and GP84/UL84 chimera shuttle vector
A shuttle vector was generated for the HCMV UL84 locus (UL84 CDS co-ordinates 157,514–155780c) based on HCMV Towne strain sequence (Genbank # AY3151972) for targeted modifications of the HCMV (Towne) BAC. Left and right arms of flanking homology around the UL84 locus were generated by PCR. Left arm (HCMV Towne coordinates nucleotides 157,671–157,390) and right arm (HCMV Towne co-ordinates 156,341–155,871) were cloned into the same pUC19 vector sequentially as EcoR I, BamH I and Hind III, BamH I fragments respectively (see Figure 5). The primers used for the right arm were FBBRFLKUL84 (5’ GGATCCATAAGATCTCGACCCTTCTTTTCGGACGC) and RHPRFLKUL84 (5’ AAGCTTGCATTAATTAAACAGTCTTGCGGTTCCGTCTCCG). The left arm primers were FELFLKUL84 (5’ GAATTCGCGACAGCTGGATGACCTCATCCGCGAAC) and RBLFLKUL84 (5’ GGATCCGGTCGTGGTGGTAGTGGTGGTGGCGAG). This construct deletes the majority of the UL84 coding sequence (codons 59 to 401) but retained the upstream UL84 promoter. Unique BamHI and Bgl II sites between the left and right flanking arms allowed cloning of a Km selection marker (McGregor et al., 2004) and the GP84 ORF. Insertion of the BamH I Km cassette generated plasmid pUL84d for deletion of the UL84 coding sequence. For generation of the chimeric GP84-HCMV, the FLAG tagged GP84 ORF was PCR amplified from pFLAGGP84 as a Bgl II fragment using primers FFLAGGP84Bg (5’ AGATCTGCCACCATGGATTACAAGGATGACGACGATAAG) and RGP84BgEcV (5’ AGATCTGATATCTATCTTACACGGATTTGGGCGCGGGATCGCTTC) and cloned into a unique Bgl II site in pUL84d to generate pUL84d/GP84, which placed the GP84 ORF under UL84 promoter control. The GP84 insertion also introduced a unique EcoR V site 3’ to the GP84 coding sequence. This allowed the insertion of a kanamycin cassette flanked by EcoR V restriction sites (McGregor et al., 2004) to generate pUL84d/GP84Km (Figure 4). This construct would be used to introduce the GP84 ORF into the HCMV Towne BAC.
Generation of recombinant BACmids and analysis of BAC structure
An inducible recombination system (ET system) was introduced into DH10B bacterial cells containing either the GPCMV BAC plasmid (McGregor and Schleiss, 2001) or the HCMV (Towne) BAC plasmid (Hahn et al., 2003) using a protocol previously described (McGregor et al., 2008). For knockout of the UL84 gene the plasmid pUL84d was linearized by a unique restriction enzyme (Pac I) outside the flanking homologous region. For generation of the chimeric GP84 HCMV BAC construct the chimeric shuttle vector pUL84d/GP84km was linearised by Pac I digestion. Linearized shuttle vector DNA was band isolated and resuspended at a concentration of 1 μg/ul, and electroporated into recombination induced HCMV BAC-containing cells in separate reactions for each mutant. Recombinant bacterial colonies of GPCMV BAC or HCMV mutants were isolated by chloramphenicol (12.5 μg/ml) and kanamycin (20 μg/ml) antibiotic selection in LB agar bacterial Petri dishes. Bacterial plates were initially incubated at 39° C to remove the ts ET recombination plasmid (Genebridges). Mutant GPCMV BAC DNA was analyzed by EcoR I and Hind III restriction digestion to verify the accuracy of the predicted genome configuration after mutation (McGregor and Schleiss, 2001). HCMV BAC mutants were analysed by Hind III restriction profile analysis. Insertion of the Km drug resistance cassette into the viral genome introduced a novel Hind III restriction. Full length HCMV mutants (HCMV::UL84d or HCMV::GP84 BACs) were further analyzed at the UL84 locus by diagnostic PCR using common primers (F84 5’CACTTTCCTCGCCACCACTAC; R84 5’GTAAAAGCGGCGGTAGATACG) flanking the modified UL84 locus. Diagnostic PCR reactions were carried out using conditions described in McGregor et al. (2008) except the extension time at 72° C was modified based on the size of each gene modification (based on an extension time of 30 sec. per 500 bases).
For knockout of the GP84 gene on the GPCMV BAC, the GP84 knockout shuttle vector pNEBGP84km(Bm) was linearized by EcoR I digestion which cut the plasmid in the polylinker sequence outside the GP84 coding sequence. DNA was band purified and 1 ug was electroporated into an aliquot of ET induced electrocompetent DH10B cells containing the GPCMV BAC as previously described (McGregor et al., 2004). GP84 knockout mutant GPCMV BAC colonies were selected under Km selection and screened by restriction profile analysis to identify full length mutants. The GP84 gene knockout was confirmed by comparative PCR analysis between wild type and mutant GPCMV BACs using common flanking primers FGP84g (5’ GAGATGATCGAAGAGACGGCCGAAC) and RGP84g (5’ GTCTACGCCCATAGCGTCGTAGTCG) to amplify either the complete GP84 gene (1.4 kb) or the modified GP84 gene locus (2.1 kb).
Generation of mutant GPCMV, chimeric HCMV and rescue of lethal HCMV knockout mutant
For generation of recombinant viruses, large-scale BAC DNA was purified from E. coli using a Qiagen maxi plasmid kit. GPCMV BAC DNA was used to transfect GPL cells in six well dishes using Lipofectamine 2000 (Invitrogen) transfection reagent as previously described (McGregor et al., 2004). HCMV BAC DNA was transfected onto HFF cells in six well dishes using supertransfect reagent (Qiagen) following manufacturer’s protocol. Transfections were followed for at least 3–4 weeks for the production of viral plaques. GPCMV GFP positive viral plaques were detected via microscopy using an inverted microscope equipped for GFP fluorescent imaging (McGregor and Schleiss, 2001). The HCMV BAC did not carry a GFP tag and formation of viral plaques was followed under bright field microscopy. Mutants were rescued back to wild type phenotype by mutant BAC co-transfection with the appropriate rescue plasmid (GPCMV GP84d knockout mutant) or PCR product (HCMV UL84 knockout mutant). HCMV rescue PCR product (UL83-UL85 PCR product) was generated by PCR amplification of Towne genomic DNA. PCR primers were generated from AD169 genomic sequence (UL83 forward primer 5’ GTTCATCAACAGGTTACCTGAGATGCT, UL85 reverse primer 5’GCTGCGCAATATGACGCTGAGGTTGATG). Amplifying a predicted 3.7 kb PCR product (121,003–124,762 AD169 co-ordinates). PCR product was band purified and used to rescue lethal HCMV UL84 knockout by co-transfection of PCR product (10 μg) with BAC DNA (1 μg) per transfection well. Rescued viral plaques were picked and subject to further rounds of plaque purification by limiting dilution. Viral DNA was extracted from virus-infected cells as previously described (McGregor and Schleiss, 2001). Rescue was confirmed by PCR analysis using common flanking gene primers for GP84 (GPCMV) or UL84 (HCMV) as described previously.
GPCMV multi-step growth curve and HCMV one step growth curve
GPCMV infection was performed on six well dishes as previously described (McGregor and Schleiss, 2001) except that the input virus (moi) was 0.1 pfu/cell per well. Virus time samples were harvested immediately after 1hr virus adsorption (day 0) and every 24 hrs (days 1–7). Samples were harvested, stored at −80° C and titrated after the final time point had been harvested as previously described. For HCMV a one step growth curve was performed on confluent monolayers of HFF cells in 6 well dishes with an input virus moi of 3 pfu/cell. One step growth curve was performed as previously described (McGregor and Schleiss, 2001). Virus time samples were taken every 12 hours from 0–96 hrs post infection. Time samples were stored at −80° C and titrated after the final time point had been harvested.
Immunofluorescence and Western blot analysis
Immunofluorescence assays were performed on full length and truncated GP84 proteins transiently expressed in GPL cells by transfection of GP84 FLAG tagged expression plasmids pFLAGGP84, pFLAGGP84(B), pFLAGGP84(S) and pFLAGGP84(BS). Assay was performed as previously described on GPL cover slips in six well dishes (McGregor et al., 2004) except that samples were fixed in −20° C methanol at various time points post transfection (3, 5, 6, 8, 12, 18, 24, 36 and 48 hr post transfection). The mouse anti-FLAG antibody (Sigma) was used at 1/1000 dilution and the secondary antibody anti-mouse IgG (Sigma) conjugated to FITC was used at 1/1000 dilution. For co-localization study of plasmid based transient expression of GFP tagged GP84 (pGFPGP84) and FLAG tagged GP44 (pFLAGGP44) plasmid transfections were performed as described above but cells were fixed with 4% paraformaldehyde for 5 min to maintain GFP fluorescence and post fixation treated with 0.2% tritonX100 to increase cell membrane permeability. Immunofluorescence assay was performed as described above except the secondary antibody was anti-mouse IgG conjugated to Texas red (Santa Cruz Biotech.) at 1/200 dilution. Cells were also counterstained with DAPI (Vector labs) to identify the cell nucleus. Co-localization experiments were harvested at 12 hr. post transfection to enable optimal detection of Texas red (conjugated to secondary antibody) tagged GP44 protein. Western blot was carried out as previously described (Schleiss et al., 1999) except that the primary antibody was mouse anti-FLAG (1/1,000 dilution) and the secondary was anti-mouse IgG conjugated to horseradish peroxidase (Santa Cruz Biotech.) used at a 1:10,000 dilution followed by detection by chemiluminescence using ECL substrate (Amersham) following the manufacturer’s protocol.
Generation of GPL cell lines expressing GFP or GFP-GP84ID
A PCR cassette was generated that contained the minimal transdominant inhibitory domain for GP84 identified in the expression vector pFLAGGP84(BS) plus additional flanking sequence (codons 170–309). The PCR product was cloned as a Bgl II fragment inframe into the C-terminal domain of GFP expression plasmid pEGFPC-1 (Clontech) cut with Bgl II to generate pGFPGP84ID as described in plasmid constructs section. This plasmid expressed GFP under HCMV IE enhancer control but also encoded a neomycin drug resistance cassette under SV40 promoter control to allow selection of G418 (Invitrogen) antibiotic resistant mammalian cells. Plasmids pEGFP-C1 and pGFPGP84ID were separately transfected onto GPL cells in six well dishes (4 μg per well) as previously described (McGregor et al., 2004). At 36–48 hr post transfection the F-12 media was replaced with media containing G418 at a concentration of 600 μg/ml and the media was changed every two days for the first 7–10 days. At that stage the G418 concentration was dropped to 500ug/ml and the media changed every three days. Resistant colonies of cells were detected by about 18–24 days post transfection. GFP positive colonies were detected using an inverted microscope equipped for GFP fluorescent imaging. Between days 30–40, well separated, GFP positive cell colonies were individually isolated using cloning rings and transferred to 25cm2 flasks and expanded as cell lines. 10 cell lines were generated for each plasmid. 6 out of the 10 GFP cell lines maintained a strong level of GFP through multiple cell passages but only 3 out of 10 GFPGP84ID cell lines maintained consistent levels of GFP under multiple cell passages. Cells were maintained under G418 selection under passage in tissue culture (300 μg/ml). One GFP cell line per plasmid was selected for use in transdominant inhibitory growth assays.
Transdominant Inhibitory Growth Assays
(A) GPCMV BAC and GP84 expression plasmid transient inhibitory growth assay. Six well dishes of confluent monolayers of GPL cells were transfected with different combinations of GPCMV BAC DNA (1 μg) and GP84 FLAG tag expression vectors (4 μg). The different combinations were GPCMV BAC alone or plus: (1) pFLAGGP84; (2) pFLAGGP84(B); (3) pFLAGGP84(S); (4) pFLAGGP84(BS); (5) pTAG2A (empty vector). Transfected plates were transfected as previously described (McGregor et al., 2004). Assays were followed by GFP reporter gene expression for identification of GFP positive GPCMV BAC transfected cells and GFP viral spread across the cell monolayer. Assays were extended to day 18 where virus stocks were harvested and titrated out on GPL cells. Individual assays were repeated muliple times.
(B) GPCMV transdominant inhibitory growth assay on GFP and GFPGP84ID GPL cells. Clonal transformed GPL cells expressing GFP or GFPGP84ID (the minimal inhibitory domain from GP84 fused to the C-terminal domain of GFP) were used to seed six well dishes. Confluent monolayers were tested for their ability to support virus replication at different moi of GPCMV (vAM403), where moi was 0.1, 0.01 or 0.001 pfu/cell. Virus infections were carried out in duplicate wells and allowed to progress for 5 days at which time the wells were harvested and the viral titers determined by titration on regular GPL cells. Herpes simplex virus (type 1) at a moi 0.001 pfu/cell was used to infect both GFP and GFPGP84ID cells as an additional control to demonstrate specificity of the GP84 transdominant inhibitory growth effect to GPCMV.
RT-PCR GP84 expression
RT-PCR was performed as previously described (McGregor et al., 2008). The GP84 gene expression in ATCC wild type GPCMV infected GPL cells (input moi 1 pfu/cell) was determined using primers FGP84RT (5’ CTACGGCTTCATCAAGATG) and RGP84RT (5’ CATCGCGTCCTGGATGAAGTAC). Control cellular gene expression was for GAPDH as previously described (McGregor et al., 2008). IE2 (GP122) gene expression was determined using RT-PCR primers FGP122RT 5’GACATGGAGCGCCGGTAGCG and RGP122RT 5’CATGGCAGACCTGATGCGAC. The viral DNA polymerase gene (GP54) was assayed by primer set FGP54RT 5’GAGACGGAGGACGTTACTAAG and RGP54RT 5’CTCACAATCTATCTCGATGTC. For specific selection of IE transcripts only, cells were treated with cycloheximide (100 μg/ml) 1 hr. prior to virus infection and for 4 hr. post infection at which point the cells were harvested and RNA extracted for analysis. For exclusion of late gene transcripts, phosphonoacteic acid (PAA) was present (200 μg/ml) as previously described Schleiss et al. (1999) and cell monolayers were harvested at 12 hr. post infection for RNA extraction and analysis.
Supplementary Material
Supplementary Figure S1. Detection of GPCMV IE transcripts after cycloheximide treatment of infected cells.
Virus infected GPL cells (moi= 3pfu/cell) were treated with cycloheximide, for selction of IE transcripts, or phosphonoacetic acid (PAA), for exclusion of late (L) transcripts, as described in material and methods to determine if GP84 is an early (E) transcript. Cycloheximide treated cells were harvested at 4 hr. post infection and PAA treated cells harvested at 12 hr. post infection and samples processed for RNA extraction. Time point samples were separately subjected to RT-PCR assay for GP84 and GP122 gene expression as previously described and assay results analyzed by gel electrophoresis: (A) GP122 (IE2); (B) GP84. In the presence of cycloheximide, only GP122 expression is detected. In the presence of PAA, both GP122 and GP84 are detected. This confirms that GP122 (IE2) is an IE transcript and that GP84 is an E transcript. Gel samples 4 and 12 are time point samples post infection (4 hr. and 12 hr.), bp is the 100 bp marker (NEB) and ntc is a no template control sample for each assay.
Supplementary Figure S2. GP84 FLAG tagged expression constructs and cellular location of transiently expressed full length or truncated GP84. The GP84 ORF was cloned in frame into the FLAG tag expression vector pCMV2A. Subsequently, convenient restriction sites (BamH I and Sal I) were used to create N- and C-terminal truncations to the GP84 coding sequence. GP84 ORF codons present in the various FLAG tagged GP84 constructs are indicated along with the GP84 amino acid (aa) polypeptide length. Note that the FLAG epitope tag is 6 aa in length. Cellular location of GP84 protein detected by immunofluoresence assay for each plasmid is summarized as cellular (C) and/or nuclear (N). Figure 1 and supplemental Figure S3 show actual transient cellular expression patterns for the various GP84 proteins summarized in Figure S2.
Supplementary Figure S3. Cellular localization of truncated GP84 encoded on plasmids pFLAGP84(BS) and pFLAGGP84(S). Immunofluorescence assays of FLAG-tagged GP84 protein transiently expressed in GPL cells transfected with pFLAGGP84(BS) or pFLAGGP84(S). Assays performed at 10 hr. post transfection. Primary antibody anti-FLAG and secondary antibody anti-mouse IgG conjugated to FITC. Cells counterstained with DAPI. Arrows indicates the same cell under FITC and DAPI filters (images A and B or C and D). Images A and B, C-terminal truncated GP84 encoded by pFLAGGP84(S). Images C and D, N-and C-terminal truncated GP84 (pFLAGGP84(BS) plasmid). Truncated proteins are both cellular and nuclear in location.
Acknowledgments
We thank Joseph Sweet for technical assistance with the Western blot. We are grateful for the generous gift of the HCMV (Towne) BAC from Gabi Hahn (University of Munich, Germany). Research funding was supported by a MMF grant to AM and NIH grant to MS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78:10995–1006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldick CJ, Jr, Marchini A, Patterson CE, Shenk T. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol. 1997;71:4400–8. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry PA, Lockridge KM, Salamat S, Tinling SP, Yue Y, Zhou SS, Gospe SM, Jr, Britt WJ, Tarantal AF. Non-human primate models of intrauterine cytomegalovirus infection. ILAR J. 2006;47:49–64. doi: 10.1093/ilar.47.1.49. [DOI] [PubMed] [Google Scholar]
- Biron KK. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 2006;71:154–63. doi: 10.1016/j.antiviral.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Cantrell SR, Bresnahan WA. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J Virol. 2006;80:6188–91. doi: 10.1128/JVI.02676-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev Med Virol. 2008;18:233–46. doi: 10.1002/rmv.574. [DOI] [PubMed] [Google Scholar]
- Colletti KS, Xu Y, Cei SA, Tarrant M, Pari GS. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J Virol. 2004;78:9203–14. doi: 10.1128/JVI.78.17.9203-9214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Xu Y, Yamboliev I, Pari GS. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J Biol Chem. 2005;280:11955–60. doi: 10.1074/jbc.C400603200. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Stow ND. New genes from old: redeployment of dUTPase by herpesviruses. J Virol. 2005;79:12880–92. doi: 10.1128/JVI.79.20.12880-12892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. Functional profiling of a human cytomegalovirus genome. Proceedings of the National Academy of Science, USA. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Isom HC. Characterization of the guinea pig cytomegalovirus genome by molecular cloning and physical mapping. J Virol. 1984;52:436–47. doi: 10.1128/jvi.52.2.436-447.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997;71:7048–60. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczol E, Plotkin S. Development of a cytomegalovirus vaccine: lessons from recent clinical trials. Expert Opin Biol Ther. 2001;1:401–12. doi: 10.1517/14712598.1.3.401. [DOI] [PubMed] [Google Scholar]
- Griffiths PD, Walter S. Cytomegalovirus. Curr Opin Infect Dis. 2005;18:241–5. doi: 10.1097/01.qco.0000168385.39390.1b. [DOI] [PubMed] [Google Scholar]
- Hahn G, Rose D, Wagner M, Rhiel S, McVoy MA. Cloning of the genomes of human cytomegalovirus strains Toledo, TownevarRIT3, and Towne long as BACs and site-directed mutagenesis using a PCR–based technique. Virology. 2003;307:164–77. doi: 10.1016/s0042-6822(02)00061-2. [DOI] [PubMed] [Google Scholar]
- He YS, Xu L, Huang ES. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J Virol. 1992;66:1098–108. doi: 10.1128/jvi.66.2.1098-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel GM, Meyer HH, Buchmann I, Pommerehne D, Schmolke S, Plachter B, Radsak K, Kern HF. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J Gen Virol. 1992;77:3087–97. doi: 10.1099/0022-1317-77-12-3087. [DOI] [PubMed] [Google Scholar]
- Holtappels R, Podlech J, Grzimek NK, Thomas D, Pahl-Seibert MF, Reddehase MJ. Experimental preemptive immunotherapy of murine cytomegalovirus disease with CD8 T-cell lines specific for ppM83 and pM84, the two homologs of human cytomegalovirus tegument protein ppUL83 (pp65) Virology. 2001;75:6584–600. doi: 10.1128/JVI.75.14.6584-6600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser N, Lischka P, Wagenknecht N, Stamminger T. Inhibition of human cytomegalovirus replication via peptide aptamers directed against the nonconventional nuclear localization signal of the essential viral replication factor pUL84. J Virol. 2009;83:11902–13. doi: 10.1128/JVI.01378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar ML, Nankervis GA. Experimental congenital infection with cytomegalovirus: a guinea pig model. J Infect Dis. 1978;138:650–4. doi: 10.1093/infdis/138.5.650. [DOI] [PubMed] [Google Scholar]
- Kern ER. Pivotal role of animal models in the development of new therapies for cytomegalovirus infections. Antiviral Res. 2006;71:164–71. doi: 10.1016/j.antiviral.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Lischka P, Sorg G, Kann M, Winkler M, Stamminger T. A non-conventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J Virol. 2003;77:3734–48. doi: 10.1128/JVI.77.6.3734-3748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Rauh C, Mueller R, Stamminger T. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J Virol. 2006;80:10274–80. doi: 10.1128/JVI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian A, Marsden HS, Palù G. Protein-protein interactions as targets for antiviral chemotherapy. Rev Med Virol. 2005;12:239–62. doi: 10.1002/rmv.356. [DOI] [PubMed] [Google Scholar]
- McGregor A, Schleiss MR. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol Genet Metab. 2001;72:15–26. doi: 10.1006/mgme.2000.3102. [DOI] [PubMed] [Google Scholar]
- McGregor A, et al. Molecular, biological, and in vivo characterization of the guinea pig cytomegalovirus (CMV) homologs of the human CMV matrix proteins pp71 (UL82) and pp65 (UL83) J Virol. 2004;78:9872–89. doi: 10.1128/JVI.78.18.9872-9889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A. Current or new cytomegalovirus antivirals and novel animal model strategies. Inflammation & Allergy–Drug Targets. 2010 doi: 10.2174/187152810793358822. (in press) [DOI] [PubMed] [Google Scholar]
- McGregor A, Choi KY, Cui X, McVoy MA, Schleiss MR. Expression of the human cytomegalovirus UL97 gene in a chimeric guinea pig cytomegalovirus (GPCMV) results in viable virus with increased susceptibility to ganciclovir and maribavir. Antiviral Res. 2008;78:250–9. doi: 10.1016/j.antiviral.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A, Liebelt J, Bouska C, Schleiss M. A Chimeric Guinea Pig Cytomegalovirus (GPCMV) Encoding Wild Type or Mutant HCMV UL97 Renders GPCMV Susceptible or Resistant to Ganciclovir While Retaining An Ability To Disseminate In The Animal Host. 23rd Intl. Conference on Antiviral Research (2010) Antiviral Res. 2010;86:A23–26. [Google Scholar]
- Morello CS, Cranmer LD, Spector DH. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83) J Virol. 1999;73:7678–93. doi: 10.1128/jvi.73.9.7678-7693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello CS, Cranmer LD, Spector DH. Suppression of murine cytomegalovirus (MCMV) replication with a DNA vaccine encoding MCMV M84 (a homolog of human cytomegalovirus pp65) J Virol. 2000;74:3696–708. doi: 10.1128/jvi.74.8.3696-3708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass RF. Cytomegalovirus. In: Knipe DM, et al., editors. Fields Virology. 4. Vol. 2. Philadelphia: Lippincott, Williams & Wilkins; 2001. pp. 2675–705. [Google Scholar]
- Reid GG, Ellsmore V, Stow ND. An analysis of the requirements for human cytomegalovirus oriLyt-dependent DNA synthesis in the presence of the herpes simplex virus type 1 replication fork proteins. Virology. 2003;308:303–16. doi: 10.1016/s0042-6822(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Robinson JA, Demarco S, Gombert F, Moehle K, Obrecht D. The design, structures and therapeutic potential of protein epitope mimetics. Drug Discov Today. 2008;13:944–51. doi: 10.1016/j.drudis.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Ross SA, Boppana SB. Congenital cytomegalovirus infection: outcome and diagnosis. Semin Pediatr Infect Dis. 2005;16:44–9. doi: 10.1053/j.spid.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Sarisky RT, Hayward GS. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in co-transfection assays. J Virol. 1996;70:7398–413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiss MR, McGregor A, Jensen NJ, Erdem G, Aktan L. Molecular characterization of the guinea pig cytomegalovirus UL83 (pp65) protein homolog. Virus Genes. 1999;19:205–21. doi: 10.1023/a:1008136714136. [DOI] [PubMed] [Google Scholar]
- Schleiss MR. Sequence and transcriptional analysis of the guinea-pig cytomegalovirus DNA polymerase gene. J Gen Virol. 1995;76 ( Pt 7):1827–33. doi: 10.1099/0022-1317-76-7-1827. [DOI] [PubMed] [Google Scholar]
- Schleiss MR. Animal models of congenital cytomegalovirus infection: an overview of progress in the characterization of guinea pig cytomegalovirus (GPCMV) J Clin Virol. 2002;25:S37–49. doi: 10.1016/s1386-6532(02)00100-2. [DOI] [PubMed] [Google Scholar]
- Schleiss MR. Nonprimate models of congenital cytomegalovirus (CMV) infection: gaining insight into pathogenesis and prevention of disease in newborns. ILAR J. 2006;47:65–72. doi: 10.1093/ilar.47.1.65. [DOI] [PubMed] [Google Scholar]
- Schleiss MR, Heineman TC. Progress toward an elusive goal: current status of cytomegalovirus vaccines. Expert Rev Vaccines. 2006;4:381–406. doi: 10.1586/14760584.4.3.381. [DOI] [PubMed] [Google Scholar]
- Schleiss MR, Lacayo JC, Belkaid Y, McGregor A, Stroup G, Rayner J, Alterson K, Chulay JD, Smith JF. Pre-conceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J Infect Dis. 2007;195:789–98. doi: 10.1086/511982. [DOI] [PubMed] [Google Scholar]
- Schmolke S, Kern HF, Drescher P, Jahn G, Plachter B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol. 1995;69:5959–68. doi: 10.1128/jvi.69.10.5959-5968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang BL, Sinigalia E, Silva LA, Coen DM, Loregian A. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J Virol. 2009;83:7581–9. doi: 10.1128/JVI.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JG. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65–specific CTL. J Virol. 1996;70:7569–79. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Colletti KS, Pari GS. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J Virol. 2002;76:8931–8. doi: 10.1128/JVI.76.17.8931-8938.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Cei SA, Huete AR, Pari GS. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J Virol. 2004;78:10360–9. doi: 10.1128/JVI.78.19.10360-10369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Morello CS, Spector DH. Multiple epitopes in the murine cytomegalovirus early gene product M84 are efficiently presented in infected primary macrophages and contribute to strong CD8+–T–lymphocyte responses and protection following DNA immunization. J Virol. 2004;78:11233–45. doi: 10.1128/JVI.78.20.11233-11245.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Silva MC, Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proceedings of the National Academy of Sciences, USA. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Detection of GPCMV IE transcripts after cycloheximide treatment of infected cells.
Virus infected GPL cells (moi= 3pfu/cell) were treated with cycloheximide, for selction of IE transcripts, or phosphonoacetic acid (PAA), for exclusion of late (L) transcripts, as described in material and methods to determine if GP84 is an early (E) transcript. Cycloheximide treated cells were harvested at 4 hr. post infection and PAA treated cells harvested at 12 hr. post infection and samples processed for RNA extraction. Time point samples were separately subjected to RT-PCR assay for GP84 and GP122 gene expression as previously described and assay results analyzed by gel electrophoresis: (A) GP122 (IE2); (B) GP84. In the presence of cycloheximide, only GP122 expression is detected. In the presence of PAA, both GP122 and GP84 are detected. This confirms that GP122 (IE2) is an IE transcript and that GP84 is an E transcript. Gel samples 4 and 12 are time point samples post infection (4 hr. and 12 hr.), bp is the 100 bp marker (NEB) and ntc is a no template control sample for each assay.
Supplementary Figure S2. GP84 FLAG tagged expression constructs and cellular location of transiently expressed full length or truncated GP84. The GP84 ORF was cloned in frame into the FLAG tag expression vector pCMV2A. Subsequently, convenient restriction sites (BamH I and Sal I) were used to create N- and C-terminal truncations to the GP84 coding sequence. GP84 ORF codons present in the various FLAG tagged GP84 constructs are indicated along with the GP84 amino acid (aa) polypeptide length. Note that the FLAG epitope tag is 6 aa in length. Cellular location of GP84 protein detected by immunofluoresence assay for each plasmid is summarized as cellular (C) and/or nuclear (N). Figure 1 and supplemental Figure S3 show actual transient cellular expression patterns for the various GP84 proteins summarized in Figure S2.
Supplementary Figure S3. Cellular localization of truncated GP84 encoded on plasmids pFLAGP84(BS) and pFLAGGP84(S). Immunofluorescence assays of FLAG-tagged GP84 protein transiently expressed in GPL cells transfected with pFLAGGP84(BS) or pFLAGGP84(S). Assays performed at 10 hr. post transfection. Primary antibody anti-FLAG and secondary antibody anti-mouse IgG conjugated to FITC. Cells counterstained with DAPI. Arrows indicates the same cell under FITC and DAPI filters (images A and B or C and D). Images A and B, C-terminal truncated GP84 encoded by pFLAGGP84(S). Images C and D, N-and C-terminal truncated GP84 (pFLAGGP84(BS) plasmid). Truncated proteins are both cellular and nuclear in location.