The catalytic domain of human ADAM-8 was expressed, purified and crystallized in complex with a hydroxamic acid inhibitor, batimastat. The crystal structure of the enzyme–inhibitor complex was refined to 2.1 Å resolution.
Keywords: ADAM-8, metalloproteases, inhibitors, batimastat, inflammation
Abstract
The role of ADAM-8 in cancer and inflammatory diseases such as allergy, arthritis and asthma makes it an attractive target for drug development. Therefore, the catalytic domain of human ADAM-8 was expressed, purified and crystallized in complex with a hydroxamic acid inhibitor, batimastat. The crystal structure of the enzyme–inhibitor complex was refined to 2.1 Å resolution. ADAM-8 has an overall fold similar to those of other ADAM members, including a central five-stranded β-sheet and a catalytic Zn2+ ion. However, unique differences within the S1′ binding loop of ADAM-8 are observed which might be exploited to confer specificity and selectivity to ADAM-8 competitive inhibitors for the treatment of diseases involving this enzyme.
1. Introduction
ADAM (a disintegrin and metalloproteinase) proteins are transmembrane and secreted proteins involved in cellular adhesion, signaling and migration (Wolfsberg et al., 1995 ▶). Roughly half are catalytically active proteases and are involved in the proteolysis of adhesion molecules, growth factors, receptors and cytokines (Yamamoto et al., 1999 ▶; Naus et al., 2004 ▶). These are all important biological control processes to maintain proper cell growth and function. Consequently, dysregulation of ADAMs is associated with pathophysiological states, including inflammation, asthma, cancer, cardiovascular dysfunction and Alzheimer’s disease (Fourie et al., 2003 ▶; Matsuno et al., 2008 ▶; Levula et al., 2009 ▶; Mahoney et al., 2009 ▶). Therefore, efforts are currently under way to develop small-molecule inhibitors to selectively target ADAMs for therapeutic purposes. One such dual-specific inhibitor of ADAM-10 and ADAM-17 has been developed by Incyte (INCB7839) and has shown promising results in treating metastatic breast cancer in phase II clinical trials (Newton et al., 2010 ▶).
ADAM-8 has been shown to participate in a wide array of cellular functions (Fourie et al., 2003 ▶; Levula et al., 2009 ▶; Mahoney et al., 2009 ▶). It has been associated with diseases including allergic asthma, tumorigenesis, aberrant neural cell signaling and arthritis, and has been demonstrated to be up-regulated in various tumors (Ishikawa et al., 2004 ▶; Naus et al., 2004 ▶; Foley et al., 2007 ▶; Gómez-Gaviro et al., 2007 ▶; Valkovskaya et al., 2007 ▶; Zack, Malfait et al., 2009 ▶). Transgenic mice harboring a catalytically inactive form of ADAM-8 demonstrated protection against experimental inflammatory arthritis, implicating ADAM-8 catalytic activity in joint disease (Zack, Melton et al., 2009 ▶).
A primary goal in studying the proteolytic activity of ADAM-8 is to discover inhibitors that bind the active site of the enzyme with high affinity and selectivity. Although potent inhibitors of ADAM-8 have been developed, they are nonselective and as such have documented toxicity-related issues that prevent rigorous human physiological studies from being performed (Dormán et al., 2010 ▶). The major obstacles towards developing a selective ADAM-8 inhibitor can be attributed to the similar enzyme fold and structurally similar binding cavities. Significant resources have been deployed to identify structural features within the matrix metalloproteinase (MMP) family that might be exploited in inhibitor design, specifically within the S1′ pocket that is critical for compound selectivity (Lovejoy et al., 1999 ▶; Shieh et al., 2008 ▶).
Full-length ADAM-8, like other ADAMs, contains a pro-domain and catalytic, disintegrin, cysteine-rich, EGF-like and transmembrane domains. Moreover, it contains the canonical histidine motif (HEX X H XXGXX HD) that coordinates the active-site zinc ion required for catalytic activity (Stöcker & Bode, 1995 ▶; Wolfsberg et al., 1995 ▶; Primakoff & Myles, 2000 ▶; Becherer & Blobel, 2003 ▶). The activation mechanism is autolytic and was recently determined in vitro using recombinant proteins to yield a final product consisting of only the catalytic domain (Hall et al., 2009 ▶). Inhibitors such as batimastat can bind the pro-form of the enzyme to inhibit all steps of this process (Schlomann et al., 2002 ▶). Thus, a selective inhibitor of ADAM-8 could have potentially far-reaching effects beyond those related to proteolysis.
Here, we present the expression and purification of ADAM-8, which allowed a 2.1 Å resolution cocrystal structure of the enzyme with its competitive inhibitor batimastat [(2R,3S)-N 4-hydroxy-N 1-[(1S)-2-(methylamino)-2-oxo-1-(phenylmethyl)ethyl]-2-(2-methylpropyl)-3-(2-thienylthio)-methylbutanediamide] to be determined. Comparison of the ADAM-8 catalytic site with that of the homologous ADAM-33 revealed striking differences in the S1′ pocket. Such differences may allow structure-based rational drug design of selective inhibitors and yield insights into ADAM-8 substrate and inhibitor preferences.
2. Materials and methods
2.1. Expression and purification of human ADAM-8 catalytic domain
ADAM-8 consisting of the pro-domain and the catalytic domain (Met1–Ser403) was expressed, purified and activated as described previously (Hall et al., 2009 ▶). Briefly, ADAM-8 undergoes activation via autolytic removal of the pro-domain, thus producing mature protein (Ser196–Ser403). The enzymatic activity of this product was confirmed by an activity assay as described elsewhere (Hall et al., 2009 ▶). The enzyme used for crystallization was fully activated as analyzed by PAGE analysis and was determined to be monomeric by size-exclusion chromatography, with a purity of greater than 98% as determined by SDS–PAGE. N-terminal sequencing by mass spectrometry determined that the mature catalytic domain begins at Ser196. The inhibitor batimastat (synthesized at Pfizer) was added to activated ADAM-8 to a final concentration of 1 mM. The complex was concentrated to 5–10 mg ml−1 (in 50 mM Tris pH 7.6, 100 mM NaCl, 5 mM CaCl2), flash-cooled on dry ice and stored at 193 K.
2.2. Crystallization
Purified and activated ADAM-8 protein complex was formed by adding batimastat in a tenfold molar excess from a 100 mM stock in 100% dimethylsulfoxide (DMSO) such that the final concentration of DMSO did not exceed 2%. Over 400 unique crystallization conditions were screened prior to optimization. In the optimized conditions, purified ADAM-8–batimastat complex at 10 mg ml−1 was crystallized using the sitting-drop vapor-diffusion method by mixing 1 µl complex solution with 1 µl 0.1 M Tris pH 8.0, 1.0 M sodium formate at 291 K. Crystal growth was optimized by the addition of 10% methanol, generating small needle-like crystals of usable size (approximately 80 × 10 × 10 µm) in 10 d. The resulting crystals were harvested and flash-cooled in liquid nitrogen for data collection. Crystals were transferred to 10% methanol, 1.3 M sodium formate, 10% glycerol and then plunged into liquid nitrogen for cryogenic storage.
2.3. Data collection and structure determination
Diffraction data were collected using an ADSC Q315R detector on beamline 5.0.1 at the Advanced Light Source (Berkeley, California, USA). Data were integrated and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The ADAM8–batimastat complex crystallized in space group P21, with unit-cell parameters a = 91.6, b = 50.9, c = 93.5 Å, β = 102.4° and four protein molecules in the asymmetric unit. The structure was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▶) from the CCP4 suite (Winn et al., 2011 ▶) using ADAM-33 as the search model (PDB entry 1r55; Orth et al., 2004 ▶). The structure was refined by iterative cycles of model building in O (Jones et al., 1991 ▶) and refinement in REFMAC5 (Murshudov et al., 2011 ▶) and PHENIX (Adams et al., 2010 ▶), using an optimized twin fraction of 0.262 along (l, −k, h) and noncrystallographic symmetry restraints. Data statistics for the final structure (R work = 18.9%, R free = 25.8%) can be found in Table 1 ▶.
Table 1. Data-collection and refinement statistics for ADAM-8–batimastat (PDB entry 4dd8).
Values in parentheses are for the last shell.
| Data-collection statistics | |
| Reservoir solution | 0.1 M Tris pH 8.0, 1 M sodium formate, 10% methanol |
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 91.6 |
| b (Å) | 50.9 |
| c (Å) | 93.5 |
| α (°) | 90 |
| β (°) | 102.4 |
| γ (°) | 90 |
| Resolution range (Å) | 50–2.1 |
| No. of observations | 176092 |
| No. of unique reflections | 49479 |
| Average multiplicity | 3.6 (2.8) |
| Completeness (%) | 99.3 (95.9) |
| Rmerge† (%) | 10.6 (36.5) |
| Average I/σ(I) | 10.9 (2.9) |
| Refinement statistics | |
| Resolution (Å) | 44.7–2.1 (2.18–2.10) |
| No. of reflections | 47689 |
| Rcryst‡ (%) | 18.8 |
| Rfree§ (%) | 25.5 |
| No. of non-H atoms | |
| Protein | 4016 |
| Ligand | 128 |
| Ions | 22 |
| Solvent | 415 |
| Mean B factor (Å2) | |
| Protein (molecule A, B, C, D) | 23.32, 22.43, 24.21, 26.25 |
| Ligand (molecule A, B, C, D) | 25.40, 26.41, 23.43, 27.72 |
| Ions | 27.55 |
| Solvent | 26.32 |
| R.m.s. deviations from ideal values | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.682 |
| Ramachandran statistics¶ | |
| Most favored regions (%) | 97.5 |
| Disallowed regions (%) | 0.2 |
R
merge = 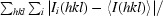
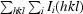 , where I
i(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflection.
, where I
i(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of the reflection.
R
cryst = 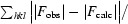
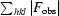 calculated from the working data set.
calculated from the working data set.
R free was calculated from 5% of data randomly chosen not to be included in refinement.
The Ramachandran results were determined by MolProbity.
Ramachandran statistics show that while 97.5% of all residues in the asymmetric unit lie within the most favored regions, there are 16 outliers. It should be noted that similar residues are marked as outliers in each of the four molecules in the asymmetric unit. Cys310 is an outlier in each molecule, likely owing to its involvement in a disulfide bond with Cys395. Residue Glu358 is also an outlier in each molecule and is constrained by tight lattice packing with symmetry-related molecules. Gln354 also participates in packing interactions. Residues Gln278–His285 lie near a disordered loop and are therefore highly mobile, which may explain their poor geometry.
Several metal ions were identified and refined in the structure. The specific ions used in refinement were chosen after thorough evaluation of biochemical evidence, anomalous scattering, coordination geometry, bond distances and electron density (Primakoff & Myles, 2000 ▶; Schlomann et al., 2002 ▶; Becherer & Blobel, 2003 ▶).
2.4. T m data collection
The protein-unfolding temperature (T m) of ADAM-8 in the presence and absence of 1 µM batimistat was assessed using an iCycler monitoring SYPRO Orange fluorescence at 540 nm. All measurements were obtained in 50 mM Tris pH 7.5, 150 mM NaCl, 5 mM CaCl2. The T m values were collected and the curve shapes were compared.
3. Results and discussion
3.1. Structural interpretation
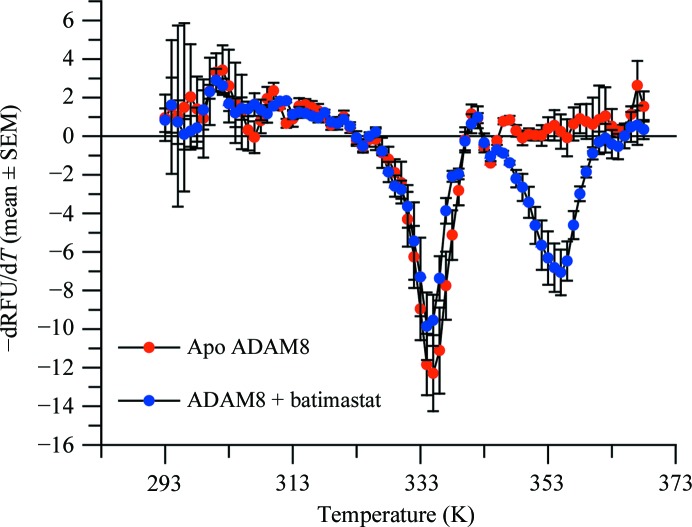
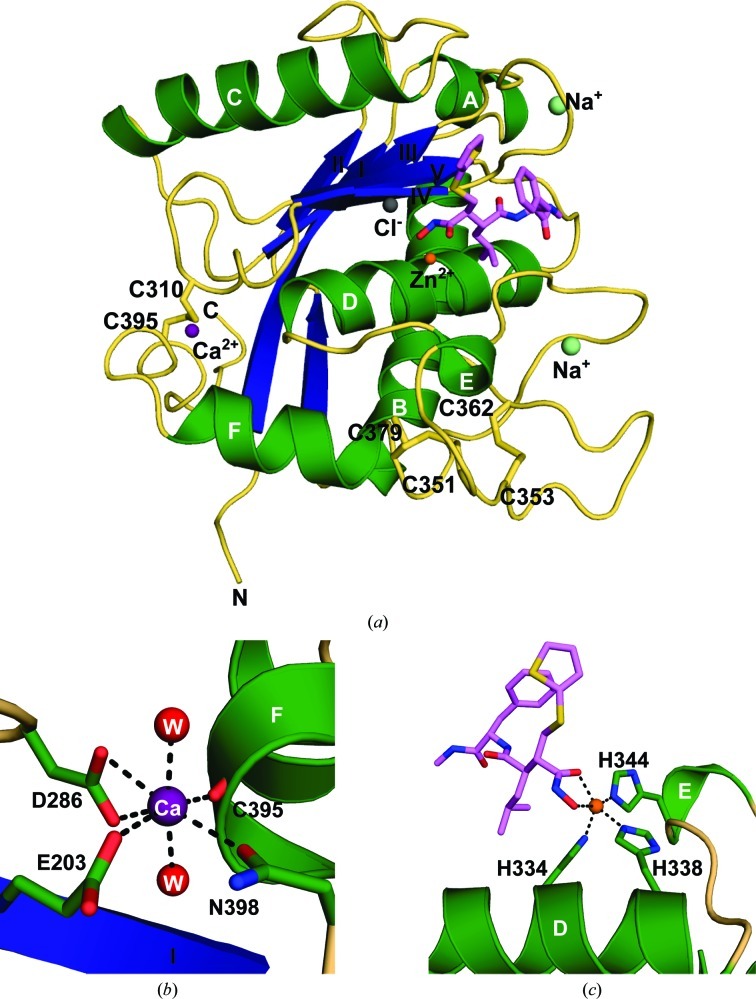
The presence of batimastat, a broad-spectrum competitive inhibitor of metalloproteinases (MPs; Wojtowicz-Praga et al., 1997 ▶), increased the overall stability of ADAM-8, as shown by the 21 K increase in the protein-unfolding temperature (Fig. 1 ▶). The presence of two inflection points at 334 and 355 K suggests that some molecules of ADAM-8 were not complexed with batimastat, possibly owing to misfolding of the protein. Despite this observation, the complex was successfully crystallized, allowing a 2.1 Å resolution structure to be determined by X-ray crystallography. The overall fold of the ADAM-8 protein resembles the α/β structure of other MPs (Grams et al., 1995 ▶; Botos et al., 1996 ▶; Orth et al., 2004 ▶; García-Castellanos et al., 2007 ▶; Gerhardt et al., 2007 ▶; Takeda et al., 2007 ▶; Mazzola et al., 2008 ▶; Mosyak et al., 2008 ▶), with a characteristic central five-stranded β-sheet (Fig. 2 ▶ a) formed by four β-strands (II, I, III and V) in a parallel configuration and a short fifth β-strand (IV) antiparallel to the rest of the sheet. The central β-sheet is flanked by two long α-helices, B and D, on its concave side and another long α-helix, C, on its convex side. Although α-helices B and D are shared with other MPs, such as ADAMTS (a disintegrin and metalloproteinase with thrombospondin motif; Gerhardt et al., 2007 ▶) and snake-venom MPs (Takeda et al., 2007 ▶), other known MMP structures lack the long α-helix C (Grams et al., 1995 ▶). Two short α-helices, A and E, are located at both edges of the β-sheet. One additional short α-helix, F, lies near the C-terminus and constitutes part of the Ca2+-binding site identified in the ADAM-33 structure (Orth et al., 2004 ▶). The calcium coordinates to the side chains of Glu203, Asp286 and Asn398, the carbonyl group of Cys395 and two waters (Fig. 2 ▶ b). Each molecule contains a Zn2+ ion in the catalytic center, which is coordinated by three histidines (His334, His338 and His344) and by the O1 and O2 atoms of batimastat (Fig. 2 ▶ c).
Figure 1.
Thermal unfolding assay monitoring ADAM-8 stabilization in the presence of batimastat. The thermal unfolding behavior of ADAM-8 was analyzed at pH 7.5 in the presence (blue) or absence (red) of 1 µM batimastat. The data are presented as the negative first derivative of the change in relative fluorescence units (RFU) against temperature (T). Error bars are the standard error of the mean of three replicates. The inflection point, indicative of unfolding, is observed at 334 K in the absence of batimastat. However, in the presence of batimastat a new inflection point is observed at 355 K.
Figure 2.
Crystal structure of ADAM-8. (a) Ribbon diagram of the ADAM-8–batimastat complex. Molecule D was used for this diagram and therefore for most of the discussion. Secondary-structural elements are shown with α-helices in green (labeled A–F), β-strands in blue (labeled I–IV) and coils in wheat. The inhibitor batimastat is shown in lavender. Ions observed in this structure are represented as spheres. (b) Coordination of the Ca2+ ion (purple). Secondary-structural elements of ADAM-8 are displayed as in (a), with side chains shown in green. Two water molecules (red) complete the coordination sphere. (c) Coordination of the catalytic Zn2+ ion (orange). Histidine residues from the canonical zinc-binding motif are shown in green and batimastat is shown in lavender.
ADAM-8 is stabilized by three disulfide bridges, similar to ADAM-17 (Mazzola et al., 2008 ▶) and ADAM-33 (Orth et al., 2004 ▶). In ADAM-8, these disulfide bonds are formed by the following pairs of cysteine residues: Cys310–Cys395, Cys351–Cys379 and Cys353–Cys362 (Fig. 2 ▶ a).
There are four independent molecules in the asymmetric unit of the ADAM-8 crystal structure. These four independent molecules overlay well, with an r.m.s. deviation of less than 0.148 Å, which suggests that the folding of ADAM-8 is quite rigid and is only minimally affected by crystal packing. The connection loop between α-helix C and β-strand III (residues 277–285) shows some flexibility in molecules B and C and is partially disordered in molecules A (residues 278–281) and C (residues 280–282). This region is distal to the catalytic center and should not affect catalytic function.
3.2. Catalytic site definition and inhibitor interactions
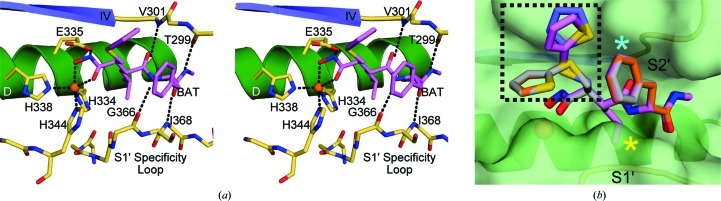
The active site of ADAM-8 is enclosed by four key structural elements: α-helix D (residues 327–335), β-strand IV (residues 301–304), the entrance loop to β-strand IV (residues 298–301) and the S1′ specificity loop (residues 365–375). These structural features define a cleft into which the target-peptide sequence would bind in order for cleavage to occur. The canonical trihistidine zinc-binding motif forms the base of the cleft and the catalytic zinc ion is clearly visible. The most relevant interactions of ADAM-8 and the inhibitor batimastat are shown in Fig. 3 ▶. All potential hydrogen-bond donor and acceptor atoms in the batimastat molecule are involved in binding to ADAM-8 (Fig. 3 ▶ a). The methyl amide end of batimastat binds to the backbone carbonyl of Thr299 (2.6 Å) and the amide linkage of Ile368 (2.8 Å). The middle amide group of batimastat interacts with the backbone carbonyl of Gly366 (2.8 Å) and may be involved in a long hydrogen bond to the amide of Val301 (3.2 Å). Interestingly, the only putative hydrogen bond involving side-chain atoms is between Glu335 and the hydroxamate group of the inhibitor (2.7 Å). Glu335 is conserved in all MPs, and is not specific to ADAM-8, which is likely to explain why batimastat serves as a nonspecific inhibitor across all MP families (Wojtowicz-Praga et al., 1997 ▶).
Figure 3.
ADAM-8–batimastat interactions. (a) Stereo diagram displaying interactions between the catalytic center of ADAM-8 and the competitive inhibitor batimastat (lavender). The catalytic zinc is shown in orange. Protein residues interacting with the inhibitor are shown in khaki. (b) Conformational flexibility of batimastat bound to ADAM-8. ADAM-8 secondary-structural elements are shown as in Fig. 2 ▶(a). The molecular surface covering this region is shown in light green. Batimastat molecules are bound to molecule A (light blue), molecule B (gray), molecule C (orange) and molecule D (lavender) within the asymmetric unit. Differences in the 2-thienylthiomethyl group are marked within the dashed box. The S1′ specificity loop and the S2′ pocket are labeled. Secondary-structural elements are shown for orientation and are colored as in Fig. 2 ▶(a).
3.3. Ligand conformational flexibility
The four independently bound batimastat molecules in the asymmetric unit exhibit some interesting conformation changes from one molecule to another. These conformational deviations of the batimastat molecules are clearly seen when the four independent chains in the ADAM-8 structure are superimposed (Fig. 3 ▶ b). These deviations are localized to the 2-thienylthiomethyl group. This group appears to point into the solvent region and has the freedom to adopt any low-energy conformation. The motility observed in this region indicates that this group is not likely to contribute strongly to the molecular interactions with the protein. It remains to be seen whether this group might be removed without compromising binding affinity when considering future inhibitor design. In order to increase inhibitor specificity and activity, further interactions should be exploited. Such interactions might benefit from extension of the 2-methypropyl group (Fig. 3 ▶ b, yellow asterisk) more deeply into the S1′ specificity pocket to connect with polar residues such as Ser367, Ser370, Ser371 and Arg374 and with nonpolar residues such as Ile368, Phe372 and Phe373. The phenyl group P2′ moiety (Fig. 3 ▶ b, cyan asterisk) could also be modified/expanded to accommodate additional interactions with the side chains of Thr299 and Thr300 on the entrance loop to β-strand IV.
3.4. Sequence and structural comparison of ADAM-8 with other metalloproteinases
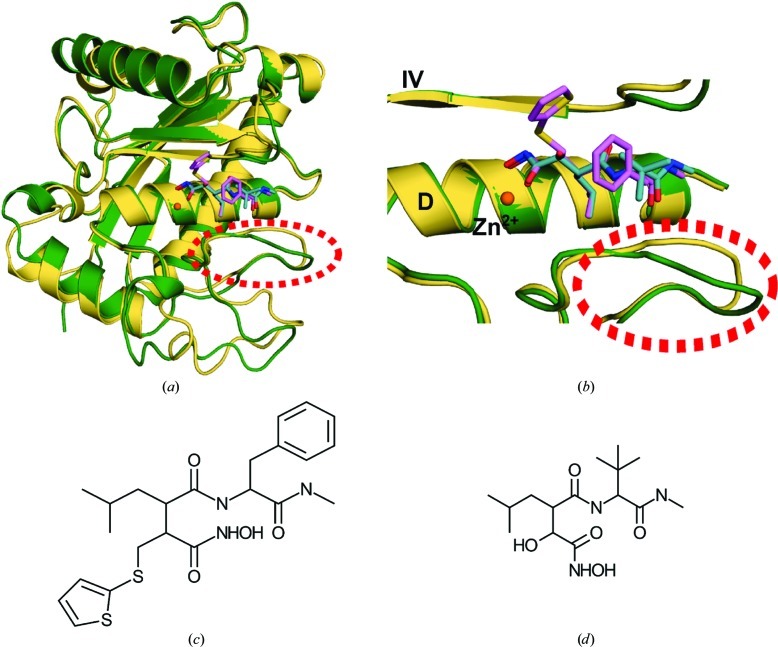
Comparison of the amino-acid sequences of various members of the metalloprotease family shows high variability (Table 2 ▶). With respect to sequence, the closest homolog of ADAM-8 is ADAM-33 (Figs. 4 ▶ a and 4 ▶ b; Orth et al., 2004 ▶). Interestingly, the next most homologous proteins are not members of the ADAM subfamily, but rather the snake-venom MPs adamalysin II (Cirilli et al., 1997 ▶) and atrolysin C (Botos et al., 1996 ▶), followed by human ADAMTS-1, ADAMTS-4 and ADAMTS-5 (Gerhardt et al., 2007 ▶; Mosyak et al., 2008 ▶). Although ADAM-8 is most similar in sequence to ADAM-33, its structure is most similar to that of adamalysin II (Table 2 ▶). These enzymes exhibit marked conservation of secondary-structural elements (with the exception of α-helix C) and zinc-binding residues in the catalytic center. However, various loop regions exhibit large deviations which become evident proximal to the active sites in which the peptide substrate or a competitive inhibitor might bind (Figs. 4 ▶ c and 4 ▶ d). The S1′ specificity loop is a key region for such deviations, since it determines the length, depth and breadth of the substrate-binding cleft. The beginning of this loop is generally marked by a well conserved methionine residue (Met364 in ADAM-8) tucked under the zinc-binding site. Beyond this methionine residue, metalloproteinases show poor sequence homolgy. It seems probable that the sequence of the S1′ specificity loop could produce local conformational differences that could contribute to substrate selectivity and, by association, to the development of more selective inhibitory compounds.
Table 2. Percentage identity of the amino-acid sequences of selected metalloproteinases.
| Protein (No. of residues) | Sequence identity† (%) | R.m.s. deviation (Å)/No. of Cα atoms |
|---|---|---|
| hADAM-8 (201) | 100 | — |
| hADAM-33 (252) | 44 | 0.744/144 |
| Adamalysin II (197) | 38 | 0.695/157 |
| Atrolysin C (203) | 35 | 0.835/154 |
| hADAMTS-1 (210) | 27 | 0.806/118 |
| hADAMTS-4 (211) | 25 | 0.693/126 |
| hADAMTS-5 (210) | 25 | 0.780/140 |
| hADAM-17 (200) | 21 | 1.906/124 |
| hMMP-12 (182) | 10 | ND‡ |
| hMMP-13 (186) | 9 | ND‡ |
| hMMP-8 (184) | 4 | ND‡ |
Sequence alignment was performed using ClustalW (Thompson et al., 2002 ▶).
No significant structural similarity was detected.
Figure 4.
Structural comparison of the inhibitor-bound catalytic domains of ADAM-8 and ADAM-33. (a) Superposition of ADAM-8 (green) and ADAM-33 (PDB entry 1r55; Orth et al., 2004 ▶; wheat). The inhibitors are shown in lavender (batimastat, bound to ADAM-8) and cyan (marimastat, bound to ADAM-33). The S1′ specificity loop region is highlighted by a dashed red circle. (b) Detailed comparison of the ADAM-8 and ADAM-33 catalytic centers, in particular the differences in the S1′ specificity loop (dashed red circle). (c) Chemical structure of batimastat. (d) Chemical structure of marimastat.
Supplementary Material
PDB reference: ADAM-8–batimastat, 4dd8
Acknowledgments
The authors would like to thank M. Marino, T. Benson and J. Monahan for aiding in the completion of this study, P. Adams and P. Zwart for technical expertise during data collection and R. Gosavi and R. Petrovich for critical reading of the manuscript. This research was supported in part by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health (1 ZIA ES102645-03). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Becherer, J. D. & Blobel, C. P. (2003). Curr. Top. Dev. Biol. 54, 101–123. [DOI] [PubMed]
- Botos, I., Scapozza, L., Zhang, D., Liotta, L. A. & Meyer, E. F. (1996). Proc. Natl Acad. Sci. USA, 93, 2749–2754. [DOI] [PMC free article] [PubMed]
- Cirilli, M., Gallina, C., Gavuzzo, E., Giordano, C., Gomis-Rüth, F. X., Gorini, B., Kress, L. F., Mazza, F., Paradisi, M. P., Pochetti, G. & Politi, V. (1997). FEBS Lett. 418, 319–322. [DOI] [PubMed]
- Dormán, G., Cseh, S., Hajdú, I., Barna, L., Kónya, D., Kupai, K., Kovács, L. & Ferdinandy, P. (2010). Drugs, 70, 949–964. [DOI] [PubMed]
- Foley, S. C., Mogas, A. K., Olivenstein, R., Fiset, P. O., Chakir, J., Bourbeau, J., Ernst, P., Lemière, C., Martin, J. G. & Hamid, Q. (2007). J. Allergy Clin. Immunol. 119, 863–871. [DOI] [PubMed]
- Fourie, A. M., Coles, F., Moreno, V. & Karlsson, L. (2003). J. Biol. Chem. 278, 30469–30477. [DOI] [PubMed]
- García-Castellanos, R., Tallant, C., Marrero, A., Solà, M., Baumann, U. & Gomis-Rüth, F. X. (2007). Arch. Biochem. Biophys. 457, 57–72. [DOI] [PubMed]
- Gerhardt, S., Hassall, G., Hawtin, P., McCall, E., Flavell, L., Minshull, C., Hargreaves, D., Ting, A., Pauptit, R. A., Parker, A. E. & Abbott, W. M. (2007). J. Mol. Biol. 373, 891–902. [DOI] [PubMed]
- Gómez-Gaviro, M., Domínguez-Luis, M., Canchado, J., Calafat, J., Janssen, H., Lara-Pezzi, E., Fourie, A., Tugores, A., Valenzuela-Fernández, A., Mollinedo, F., Sánchez-Madrid, F. & Díaz-González, F. (2007). J. Immunol. 178, 8053–8063. [DOI] [PubMed]
- Grams, F., Crimmin, M., Hinnes, L., Huxley, P., Pieper, M., Tschesche, H. & Bode, W. (1995). Biochemistry, 34, 14012–14020. [DOI] [PubMed]
- Hall, T., Leone, J. W., Wiese, J. F., Griggs, D. W., Pegg, L. E., Pauley, A. M., Tomasselli, A. G. & Zack, M. D. (2009). Biosci. Rep. 29, 217–228. [DOI] [PubMed]
- Ishikawa, S., Takenaka, K., Yanagihara, K., Miyahara, R., Kawano, Y., Otake, Y., Hasegawa, S., Wada, H. & Tanaka, F. (2004). Clin. Cancer Res. 10, 6579–6585. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Levula, M. et al. (2009). Ann. Med. 41, 497–507. [DOI] [PubMed]
- Lovejoy, B., Welch, A. R., Carr, S., Luong, C., Broka, C., Hendricks, R. T., Campbell, J. A., Walker, K. A., Martin, R., Van Wart, H. & Browner, M. F. (1999). Nature Struct. Biol. 6, 217–221. [DOI] [PubMed]
- Mahoney, E. T., Benton, R. L., Maddie, M. A., Whittemore, S. R. & Hagg, T. (2009). J. Comp. Neurol. 512, 243–255. [DOI] [PMC free article] [PubMed]
- Matsuno, O., Kumamoto, T. & Higuchi, Y. (2008). Inflamm. Allergy Drug Targets, 7, 108–112. [DOI] [PubMed]
- Mazzola, R. D., Zhu, Z., Sinning, L., McKittrick, B., Lavey, B., Spitler, J., Kozlowski, J., Neng-Yang, S., Zhou, G., Guo, Z., Orth, P., Madison, V., Sun, J., Lundell, D. & Niu, X. (2008). Bioorg. Med. Chem. Lett. 18, 5809–5814. [DOI] [PubMed]
- Mosyak, L. et al. (2008). Protein Sci. 17, 16–21. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Naus, S., Richter, M., Wildeboer, D., Moss, M., Schachner, M. & Bartsch, J. W. (2004). J. Biol. Chem. 279, 16083–16090. [DOI] [PubMed]
- Newton, R. C., Bradley, E. C., Levy, R. S., Doval, D., Bondarde, S., Sahoo, T. P., Lokanatha, D., Julka, P. K., Nagarkar, R. & Friedman, S. M. (2010). J. Clin. Oncol. (Meeting Abstracts), 28, 3025.
- Orth, P., Reichert, P., Wang, W., Prosise, W. W., Yarosh-Tomaine, T., Hammond, G., Ingram, R. N., Xiao, L., Mirza, U. A., Zou, J., Strickland, C., Taremi, S. S., Le, H. V. & Madison, V. (2004). J. Mol. Biol. 335, 129–137. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Primakoff, P. & Myles, D. G. (2000). Trends Genet. 16, 83–87. [DOI] [PubMed]
- Schlomann, U., Wildeboer, D., Webster, A., Antropova, O., Zeuschner, D., Knight, C. G., Docherty, A. J., Lambert, M., Skelton, L., Jockusch, H. & Bartsch, J. W. (2002). J. Biol. Chem. 277, 48210–48219. [DOI] [PubMed]
- Shieh, H. S., Mathis, K. J., Williams, J. M., Hills, R. L., Wiese, J. F., Benson, T. E., Kiefer, J. R., Marino, M. H., Carroll, J. N., Leone, J. W., Malfait, A. M., Arner, E. C., Tortorella, M. D. & Tomasselli, A. (2008). J. Biol. Chem. 283, 1501–1507. [DOI] [PubMed]
- Stöcker, W. & Bode, W. (1995). Curr. Opin. Struct. Biol. 5, 383–390. [DOI] [PubMed]
- Takeda, S., Igarashi, T. & Mori, H. (2007). FEBS Lett. 581, 5859–5864. [DOI] [PubMed]
- Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2002). Curr. Protoc. Bioinformatics, ch. 2, Unit 2.3. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Valkovskaya, N., Kayed, H., Felix, K., Hartmann, D., Giese, N. A., Osinsky, S. P., Friess, H. & Kleeff, J. (2007). J. Cell. Mol. Med. 11, 1162–1174. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wojtowicz-Praga, S. M., Dickson, R. B. & Hawkins, M. J. (1997). Invest. New Drugs, 15, 61–75. [DOI] [PubMed]
- Wolfsberg, T. G., Primakoff, P., Myles, D. G. & White, J. M. (1995). J. Cell Biol. 131, 275–278. [DOI] [PMC free article] [PubMed]
- Yamamoto, S., Higuchi, Y., Yoshiyama, K., Shimizu, E., Kataoka, M., Hijiya, N. & Matsuura, K. (1999). Immunol. Today, 20, 278–284. [DOI] [PubMed]
- Zack, M. D., Malfait, A. M., Skepner, A. P., Yates, M. P., Griggs, D. W., Hall, T., Hills, R. L., Alston, J. T., Nemirovskiy, O. V., Radabaugh, M. R., Leone, J. W., Arner, E. C. & Tortorella, M. D. (2009). Arthritis Rheum. 60, 2704–2713. [DOI] [PubMed]
- Zack, M. D., Melton, M. A., Stock, J. L., Storer, C. E., Barve, R. A., Minnerly, J. C., Weiss, D. J., Stejskal, J. A., Tortorella, M. D., Turk, J. R., Shevlin, K. M. & Malfait, A. M. (2009). Clin. Exp. Immunol. 158, 246–256. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ADAM-8–batimastat, 4dd8