Abstract
Iron is an essential growth factor for virtually all organisms. However, iron is not readily available in most environments and microorganisms have evolved specialized mechanisms, such as the use of siderophores and high-affinity transport systems, to acquire iron when confronted with iron-limiting conditions. In general these systems are tightly regulated to prevent iron-induced toxicity and because they are quite costly to the microbe. Because of this tight regulation we chose to explore the response of Bifidobacterium breve UCC2003 to iron limitation. Through microarray and complementation analyses we identified and characterized a presumed ferrous iron uptake system, encoded by bfeUOB, from B. breve UCC2003 and exploited its regulated transcription to develop an inducible expression system for use in bifidobacteria.
Keywords: iron-regulation, plasmid, probiotic
Introduction
Under normal physiological conditions, iron exists in one of two interconvertible redox states: the oxidized ferric (Fe3+) or its more reduced ferrous (Fe2+) form.1 Iron is involved in important biological processes, where its functionality is largely dependent upon its incorporation into proteins, either as a mono- or binuclear species, or in a more complex form as part of iron-sulfur clusters or haem groups (see ref. 2 and references therein). It serves as a cofactor for several electron-transport proteins involved in vital metabolic processes such as aerobic and anaerobic ATP biosynthesis, making iron an essential element for most living organisms.1 The latter fact is exploited by certain higher organisms, which sequester nutrient iron to limit the growth of pathogenic organisms as a form of nutritional immunity.3 However, iron may also impose oxygen toxicity because it is a promoting factor of the Fenton reaction.4 Therefore, strict regulation of iron metabolism and associated defenses against oxidative damage is essential for aerobic life.4 Appropriate iron homeostasis requires an effective iron scavenging system to ensure adequate supplies, coupled to a delicate management of cellular, free iron levels in order to prevent iron-mediated toxicity.
Bacteria employ various physiological approaches to achieve iron homeostasis through high- and low-affinity iron transport systems, as well as to tightly control intracellular iron stores in case external supplies become exhausted.5 Furthermore, under iron-restricted conditions the expression of iron-requiring proteins can be downregulated.6
Several iron uptake mechanisms in Gram-negative bacteria have been described. For example, Escherichia coli can employ, depending on the strain investigated, more than 10 different iron-uptake routes.7 Bacterial iron uptake frequently involves so-called siderophores, which are low molecular-weight, extracellular, high-affinity iron-scavenging molecules capable of binding specific receptors in the bacterial outer membrane when complexed with iron.8 Gram-negative pathogens may acquire iron from host-derived, iron-containing compounds, e.g., transferrin, lacteroferrin, hemoglobin, haem and hemopexin. Often bacteria contain multiple ferric-complex uptake systems (six defined for Escherichia coli K-12) targeting a variety of iron sources, including siderophores produced by other organisms.5 Bacteria can also acquire the soluble, ferrous form of iron via the anaerobic-microaerophilic FeoAB pathway.9 In addition, ferrous iron can be taken up by NRAMP (natural resistance-associated macrophage protein)-like transporters (e.g., MntH of E. coli), metal-type ABC transporters (e.g., SitABCD of Salmonella) and ZIP transporters (e.g., ZupT of E. coli).10
The acquisition of iron from host proteins has been studied in just a handful of Gram-positive microbes. Some species employ surface receptors for proteins, such as transferrin or lactoferrin,11,12 while other Gram-positive bacteria, for example Bacillus subtilis, utilize siderophores to obtain ferric iron.13 The use of haem or heamo proteins mediated through dedicated surface receptors has also been reported.14,15 However, the principal machinery involved in the uptake of free or complex iron in Gram-positive bacteria involves the use of ABC-type transporters.16 A summary of known iron transporters of common Gram-positive pathogens is presented in Table S1.
Iron is essential not only for pathogenic but also for commensal bacteria. Indeed, it has been hypothesized that one of the beneficial actions of health-promoting or probiotic bacteria is to sequester iron, thus making it less available to pathogens, thus representing another form of nutritional immunity.17 A further benefit of iron sequestration is that it would abate free radical production in the gastrointestinal tract (GIT) or at sites of inflammation, where OH-free radicals are produced by the action of neutrophils or other means.18 Iron-induced free radical damage to DNA appears to promote the development of cancer, while cancer cells are also known to grow rapidly in response to iron.19
Bifidobacterium, Gram-positive, non-motile, anaerobic bacteria, first described by Tissier in 1900 are among the most dominant organisms present in the breast-fed infant gut.20 Various bifidobacterial strains are widely used as probiotic bacteria in functional foods.21 Uptake of iron by Bifidobacterium was first described by Bezkorovainy22 who demonstrated that Bifidobacterium bifidum possesses a membrane-bound ferri-reductase, which converts ferric iron to its ferrous state at the cell surface.22 Recently, B. longum DJ010A has been reported to bacteriostatically inhibit other bacteria through the production of a siderophore,23 although this characteristic seems rare among bifidobacteria and strain-dependent.24 Bifidobacteria grow under anaerobic, reducing and/or acidifying conditions, where the iron equilibrium is expected to shift from the ferric to the ferrous form, thereby allowing permeases of different protein families to take up iron without the need for a diffusible iron-sequestering compound. Iron uptake systems present in bifidobacteria have not been characterized at the genetic level and with the availability of bifidobacterial genome sequences this has now become feasible.25 Global gene expression of Bifidobacterium breve UCC2003 under conditions of iron deficiency enabled us to identify genes whose transcription is iron dependent, among which the bfeUOB gene cluster, whose encoded proteins are similar to the EfeUOB ferrous iron transporter from E. coli.26 The iron-responsive promoter region of the bfeUOB cluster was used for the construction of an inducible promoter system for bifidobacteria.
Results
Growth of bifidobacterial strains under iron-limiting conditions
In order to assess the growth inhibitory effect of the ferrous iron chelator 2′2-dipyridyl, growth was assessed for a collection of bifidobacterial strains. The strains were inoculated at 1% in CDM media and 2′2-dipyridyl was added at a final concentration that ranged from 0 to 5 µM. Following 15 h anaerobic incubation at 37°C, the OD600nm was recorded. Growth, as assessed by the OD600nm measurements, of almost all strains was inhibited by the presence of the chelator except for that of B. longum JCM7052. An inverse concentration-dependent correlation between the added amount of chelator and final optical density reached was shown following 15 h of growth (Table 1).
Table 1. Growth of bifidobacterial strains in response to increasing concentrations of 2′2 dipyridyl*.
| Species | Strain | 0 µM Dipyridyl | 0.5 µM Dipyridyl | 1 µM Dipyridyl | 5 µM Dipyridyl |
|---|---|---|---|---|---|
|
B. adolescentis |
CIP 64.61 |
++ |
+ |
- |
- |
|
B. adolescentis |
NCFB 2229 |
++ |
+ |
- |
- |
|
B. animalis |
JCM 20097 |
++ |
+ |
- |
- |
|
B. animalis |
DSM 20105 |
++ |
+ |
- |
- |
|
B. bifidum |
NCIMB8810 |
++ |
+ |
- |
- |
|
B. bifidum |
LMG 11041 |
++ |
+ |
- |
- |
|
B. breve |
UCC2003 |
++ |
+ |
- |
- |
|
B. breve |
JCM 7017 |
++ |
+ |
- |
- |
|
B. dentium |
NCFB 2843 |
++ |
+ |
- |
- |
|
B. longum |
CIP 64.63 |
++ |
++ |
+ |
+ |
|
B. longum |
JCM 7052 |
++ |
++ |
++ |
++ |
|
B. infantis |
NCFB 2205 |
++ |
++ |
+ |
- |
|
B.pseudocatenulatum |
NCIMB 8811 |
++ |
+ |
- |
- |
|
B. pseudolongum |
NCIMB 2244 |
++ |
+ |
+ |
- |
|
B. globosum |
JCM 7092 |
++ |
+ |
- |
- |
| B. thermophilum | JCM 7027 | ++ | + | - | - |
Note: *Following 24 h growth in CDM medium, each strain of Bifidobacterium was subcultured to CDM containing a range of 2′2 dipyridyl from 0 to 5 µM and incubated for a further 15 h anaerobically at 37°C. The OD600nm was established and is represented as (++) when OD600nm > 1.0; (+) when OD600nm > 0.05 and < 0.5; and (-) when OD600nm < 0.05.
Genome response of B. breve UCC2003 to iron limitation
To investigate differences in global gene expression of B. breve UCC2003 when grown at limiting concentrations of iron, global transcriptional changes were analyzed using DNA microarrays probed with cDNA from B. breve UCC2003 grown under iron-limiting conditions (see Materials and Methods). Because iron omission is likely to cause a significant impact on cell physiology, which would make it difficult to differentiate between genes directly influenced by iron-limitation from those affected by reduced metabolism, we exposed B. breve UCC2003 to 3 µM of the ferrous-iron specific chelator 2′2-dipyridyl. This resulted in the upregulation of 24 genes and the downregulation of 18 genes (fold change > 3.0 or < 0.25, p < 0.001) (Table 2). The upregulated genes were presumed to be involved in (high affinity) iron transport in B. breve UCC2003 and were subjected to further scrutiny.
Table 2. Microarray fold alteration in gene expression following exposure to 3 µM of dipyridyl for 180 min. Genes significantly up or downregulated (fold change > 3.0, p < 0.001).
| Locus | Gene | Description | Dipyridyl Array | qRT-PCR | Regulator/Motif | |
|---|---|---|---|---|---|---|
|
Bbr_0221 |
BfeU |
OFeT family high affinity iron permease |
9.6 |
64.33 |
Unknown: AAAATCAAGACTGTTGTT |
|
|
Bbr_0222 |
BfeO |
Secreted protein with iron binding domain |
9.6 |
30.9 |
|
|
|
Bbr_0223 |
BfeB |
Membrane protein |
4.3 |
69.76 |
|
|
| Bbr_0224 |
|
ABC transporter |
4.4 |
3.33 |
|
|
| Bbr_0225 |
|
ABC transporter |
4.3 |
ND |
|
|
| Bbr_0226 |
|
ABC transporter |
3.7 |
ND |
|
|
| Bbr_0227 |
|
Lipoprotein |
3.1 |
ND |
|
|
| |
|
|||||
|
Bbr_0573 |
FUR |
Ferric uptake regulation protein |
- 1.7 |
-1.0 |
Fur: GATAATGAATATCATTTGT |
|
| Bbr_0574 |
|
Conserved hypothetical |
1.2 |
ND |
|
|
| Bbr_0575 |
|
ABC transporter |
1.0 |
ND |
|
|
| Bbr_0576 |
|
Hypothetical protein |
3.3 |
ND |
|
|
| Bbr_0577 |
RpmE1 |
LSU ribosomal protein |
2.6 |
ND |
|
|
| |
|
|
|
|
|
|
| Bbr_0885 |
|
Membrane protein |
5.7 |
36.3 |
Unknown: AAAATCAAGACTGTTGTT |
|
| Bbr_0886 |
|
ABC transport permease |
6.1 |
52.5 |
|
|
| Bbr_0887 |
|
ABC transport binding protein (similar Bbr0226) |
5.4 |
32.12 |
|
|
| |
|
|
|
|
|
|
| Bbr_1364 |
GroEL |
Chaperone |
3.0 |
146.2 |
|
|
| |
|
|
|
|
|
|
| Bbr_1523 |
MerR |
MerR regulatory protein |
4.5 |
27.8 |
|
|
| |
|
|
|
|
|
|
|
Bbr_1898 |
NrdF |
Ribonucleoside diphosphatereductase-β |
3.1 |
127.2 |
|
|
| Bbr_1899 | NrdE | Ribonucleoside diphosphatereductase-α | 2.4 | 16.4 | ||
Note: Underlined is the first member of the operon. Significantly regulated genes are shown in bold. ND is not determined.
Genes differentially regulated by iron-chelation
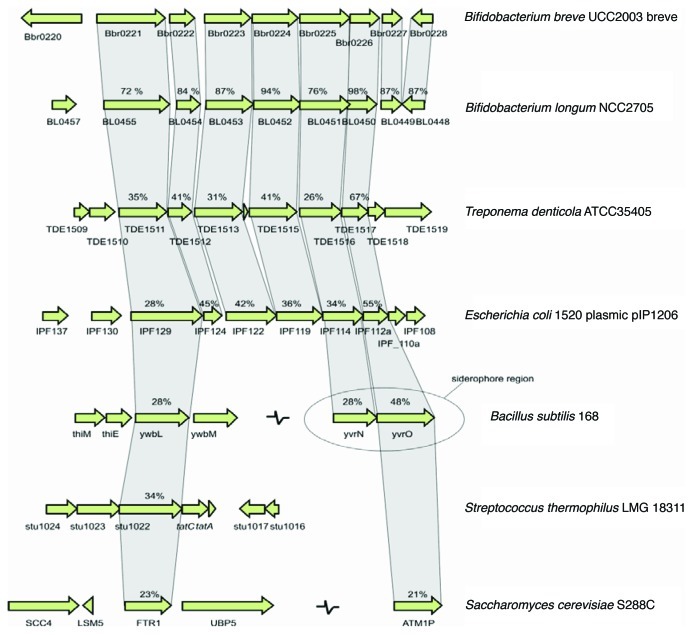
Two clusters of genes upregulated following exposure to dipyridyl were identified: the Bbr_0885–0887 gene cluster, whose products are annotated as an uncharacterized ABC transport system, and Bbr_0221–0227 (Table 3), encoding conserved hypothetical proteins with homology to the FTR1 system from Saccharomyces cerevisiae and to the EfeUOB transport system from E. coli, both ABC transporter systems responsible for iron uptake.26,27 Comparative analysis shows that the latter FTR1/EfeU-like transport system is present in virtually all kingdoms of life (Fig. 1) and form a subgroup of the iron/lead transporter (ILT) superfamily28 renamed as oxidase-dependent iron transporters (OFeT) (TC.9.A.10.1).28
Table 3. Sequence analysis of the currently available bifidobacterial genomes for the motif identified upstream of Bbr_0221 and Bbr_0885.
| Strain | Bbr_0221 Homolog | Motif | Bbr_0885 Homolog | Motif |
|---|---|---|---|---|
|
B. breve DSM 20213 |
BIFBRE_01998 |
AAAATCAAGACTGTTGTT |
BIFBRE_01208 |
AAAAACAAGACTTGTGTT |
|
B. infantisATCC15697 |
Blon_0196 |
AAAATCAAGACTGTTGTT |
Blon_1615 |
AAAAACAAGACTTGTGTT |
|
B. longum DJO10A |
BLD_1255 |
AAAATCAAGACTGTTCCT |
|
|
|
B. longum infantis ATCC 55813 |
HMPREF0175_0087 |
AAAATCAAGACTGTTCCT |
|
|
|
B. longum NCC2705 |
BL0455 |
AAAATCAAGACTGTTCCT |
BL0889 |
AAAAACAAGACTTGTGTT |
|
B. longum infantis CCUG 52486 |
BLIG_00474 |
AAAATCAAGACTGTTCCT |
|
|
|
B. bifidum NCIMB 41171 |
BbifN4_010100006928 |
AAAGTCAAGACTGTTCTT |
BbifN4_010100003655 |
AAAGTCAAGGCTTGTGTT |
|
B. dentium ATCC 27678 |
BIFDEN_00651 |
AAAATCAAGACTGTTCTT |
BIFDEN_02200 |
AAAGTCAAGACTTTTCTT |
|
B. angulatum DSM 20098 |
BIFANG_02052 |
AAAATCAAGACTGTTGTT |
BIFANG_02981 |
AAGTCAAGTCTTGTTTTT |
|
B.pseudocatenulatum DSM 20438 |
BIFPSEUDO_04117 |
AAAATCAAGACTGTTCTT |
|
|
|
B. catenulatum DSM 16992 |
BIFCAT_00140 |
AAAATCAAGACTGTTCTT |
BIFCAT_00981 |
AAAAACAAGACTATCTTT |
|
B. adolescentis ATCC 15703 |
BAD_0097 |
AAAATCAAGACTGTTATT |
|
|
| B. adolescentis L2–32 | BIFADO_00280 | AAAATCAAGACTGTTATT | ||
Figure 1. Comparison of the bfeU (Bbr_0221) locus in B. breve UCC 2003 with the corresponding loci in various bacteria. Each arrow indicates an ORF. Corresponding genes are indicated by the vertical links between the arrows. The levels of amino acid identity to the B. breve UCC 2003 sequence, expressed as percentages, are indicated. The siderophore locus in B. subtilis 29 is indicated.
The EfeUOB operon is conserved in E. coli and the dental pathogen Treponema denticola. Furthermore, a homolog of EfeU and part of the transport system are found in the surfactin siderophore cluster of the Gram-positive organism B. subtilis, which is responsible for high-affinity ferric iron uptake.29 A feature of all OFeT proteins is the occurrence of two REXXE amino acid residue motifs.27,30 On examination of the BfeU amino acid sequence two corresponding REGLE motifs were identified (Fig. S1).
Because of the high level of similarity to EfeUOB, we designated the Bbr_0221-Bbr_0223-encoded proteins BfeUOB. The iron-dependent transcriptional induction of bfeUOB in B. breve UCC2003, as observed using microarray analysis, was confirmed by QRT-PCR experiments, where bfeU, bfeO and bfeB were induced 64-, 30- and 69-fold, respectively, under iron-chelating conditions as compared with iron-replete conditions. This convergent expression pattern suggests that bfeUOB is controlled by a single, iron-dependent promoter.
Putative iron-dependent regulatory sequences
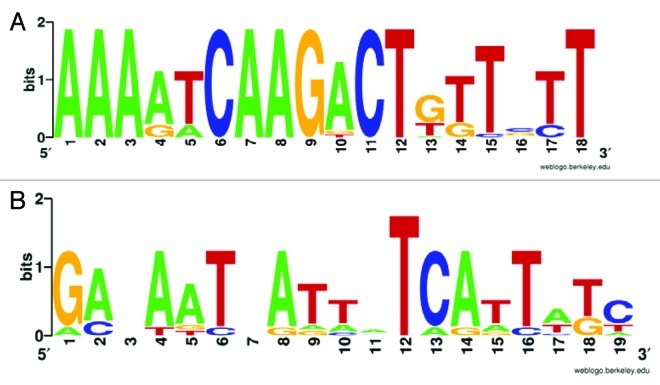
An in silico sequence analysis was performed to determine whether the genes upregulated by exposure to dipyridyl in B. breve UCC2003 contain conserved regulatory sequences in their putative promoter regions. We assumed that these genes were under direct control of one or more iron-responsive regulators, which elicit this control through binding to specific sequences in their target promoter regions. Data sets of sequences containing 400 bp upstream of the putative translation start sites of the upregulated genes were generated. In this way, a data set of five sequences was examined for the occurrence of common elements with a length between 10 bp and 30 bp using the MEME algorithm.31 Nearly identical imperfect inverted repeat (IR) sequences, AAAATCAAGACTGTTGTT and AAAAACAAGACTTGTGTT, respectively, were present in the putative promoter region of Bbr_0221 and Bbr_0885, which are both strongly upregulated (9.6 and 5.7 fold by micro array analysis, respectively, see Table 2) when B. breve UCC2003 is exposed to dipyridyl.
A screen of putative promoter regions of all currently available bifidobacterial genomes for the sequence AAAAWCAAGACTNNTGTT (Fig. 2A) revealed that the putative promoter regions of bifidobacterial homologs of Bbr_0221 and Bbr_0885 are the only sequence regions to contain this motif (Table 3). These findings suggest that this inverted repeat is involved in mediating the regulatory response to iron limitation, for instance as a binding site for a regulatory protein, although further studies need to be performed to verify this possibility and to identify the regulator involved.
Figure 2. (A) Weblogo depiction of a motif based on the alignment of bifidobacterial sequences from the promoter regions of Bbr_0221 and Bbr_0885 homologs. (B) Weblogo depiction of FUR motif as detected in the B. breve UCC2003 genome.
The influence of the ferric uptake regulation (FUR) protein in the response by B. breve UCC2003 to iron chelation by dipyridyl is less likely. Although a putative FUR protein (Bbr_0573) has been annotated on the genome of UCC2003, screening with a model based on the FUR motif of this gene (Fig. 2B) using HMMSEARCH32 detected no FUR motifs in any of the genes upregulated by dipyridyl-mediated iron chelation (results not shown).
Expression of UCC2003 bfeUOB improves growth of an iron-uptake mutant of E. coli K12 (W3110)
In order to establish the functionality of the bfeUOB operon in iron-uptake we cloned bfeU, bfeUO and bfeUOB separately in the low-copy vector pWSK29, creating pBfeU, pBfeUO and pBfeUOB, respectively, as described in Materials and Methods. These plasmids were introduced into the E. coli K12 (W3110)-derived mutant, designated GR536 (DfecABCDE::kanDzupT::cat DmntHDfeoABCDentC),7 in order to assess plasmid-mediated effects on growth under iron limiting conditions. The GR536 mutant grows well in Luria–Bertani (LB) medium, but poorly in Tris-mineral salts medium even when iron is supplied (Fig. 3). As a positive control the E. coli Nissle strain which contains a functional EfeUOB operon33 was included in our analysis.
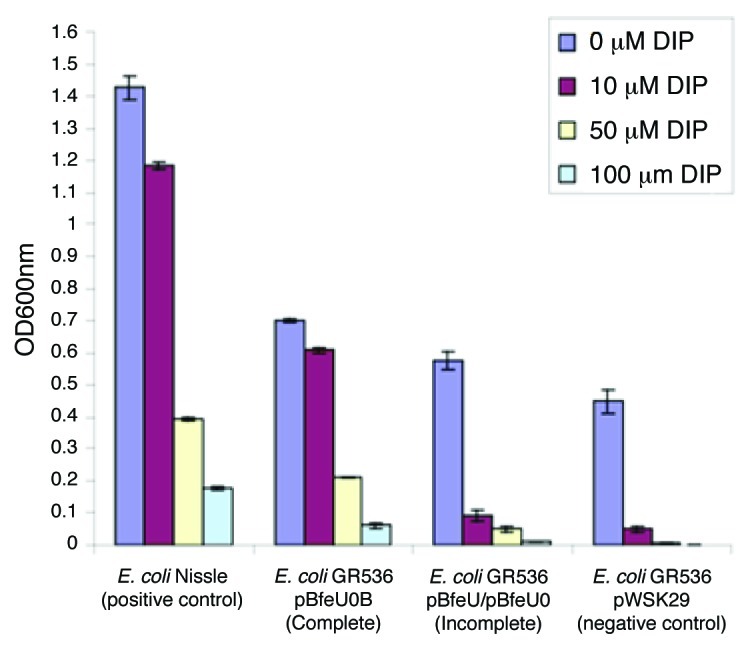
Figure 3. Complementation analysis of E. coli GR536 with the complete bfeUOB operon (pBfeUOB) in comparison to empty vector (pWSK29) or plasmids containing just bfeU (pBfeU) or bfeUO (pBfeUO) (illustrated jointly as incomplete fragments of the operon) monitored as OD600nm following 12 h of growth under increasing iron chelating conditions, dipyridyl (DIP) range 0–100 µM. E. coli Nissle containing a functional EfeUOB operon serves as a positive control.
Heterologous expression of bfeU or bfeUO in the pWSK29 plasmid in E. coli GR536 provided a modest increase in cellular growth yield in the presence of dipyridyl (ranging from 0–100 µM) when compared with a vector-only control (Fig. 3). However, strain GR536 carrying pBfeUOB showed a significant increase in cellular growth (Fig. 3) and was essentially unaffected by the presence of low concentrations of dipyridyl and only ceased growing at concentrations above 100 µM. These results clearly demonstrate that expression of functional bfeUOB is advantageous for E. coli under conditions of iron depletion.
Expression of a p272bfeU-gusA fusion is regulated by iron in multicopy plasmids
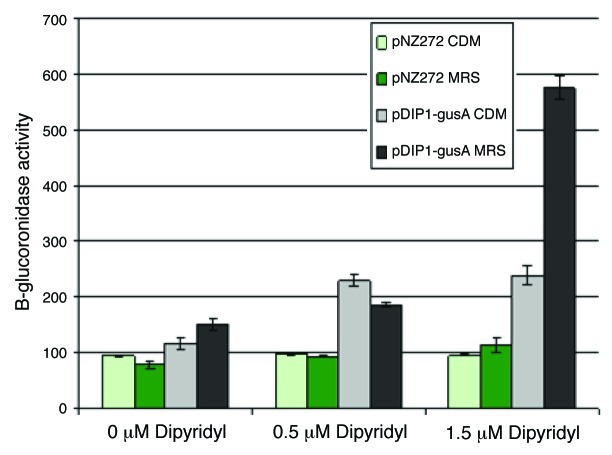
Plasmid pDIP1-gusA was constructed to investigate the putative promoter region upstream of bfeU and to establish if this promoter could be employed as an iron-dependent inducible gene expression system in B. breve UCC2003. Initial assays in CDM broth showed a 2-fold increase in GUS activity of UCC2003 harboring pDIP1-gusA when exposed to 1.5 µM dipyridyl in comparison to either the control strain (UCC2003 with plasmid pNZ272) or UCC2003 containing plasmid pDIP1-gusA in CDM (no chelator) as illustrated in Figure 4.
Figure 4. β-glucuronidase assay of B. breve UCC2003 containing pNZ272 or pDIP1-gusA in defined (CDM) or complex (MRS) media with increasing concentrations of 2′2-dipyridyl from 0 to 1.5 µM.
However, a marked improvement was observed when the media was changed to MRS broth (Fig. 4), with an at least 5-fold increase in the GUS activity of UCC2003 pDIP1-gusA on exposure to 1.5 µM dipyridyl in comparison to either UCC2003 pNZ272 or UCC2003 pDIP1-gusA in MRS (no chelator).
Discussion
In order to assess the inhibitory effect of iron limitation on bifidobacterial strains we studied the growth of a collection of bifidobacterial strains under these conditions. In addition we imposed iron limitation through the use of the ferrous-iron chelator,34 2′2-dipirydyl, and performed global transcriptional analysis on B. breve UCC2003. A chelator-dependent variation in the expression of iron-regulated outer membrane proteins in E. coli has previously been reported.35 The authors concluded that the nature of the chelator used to restrict iron supply influences the synthesis of iron-regulated proteins. Lim et al.36 have exploited this strategy for the development of an expression vector based on the PentC promoter for E. coli that is induced by iron chelation. Similarly for the actinomycetes the iron-regulated desA promoter serves as an inducible system for expression of genes in Streptomyces pilosus following iron removal with dipyridyl.37 Apart from B. longum JCM7052, growth of all other Bifidobacterium strains tested showed an inverse concentration-dependent correlation between the added amount of chelator and final optical density reached following 15 h of growth. The lack of growth inhibition of B. longum JCM7052 when exposed to dipyridyl may indicate, similar to the findings reported for B. longum DJO10A,23 that this strain produces a siderophore that endows it with a very efficient iron scavenging ability.
Following chelation of ferrous iron, B. breve UCC2003 evokes a stress-like response demonstrated by the upregulation of the chaperone GroEL (Bbr_1364).38,39 Iron-chelation was also shown to upregulate transcription of nrdF (Bbr_1898), encoding a predicted ribonucleotide reductase. The latter enzyme is known in E. coli to catalyze the reduction of ribonucleotides to deoxyribonucleotides as a first step in the pathway for DNA replication, where synthesis of nrd mRNA increases when DNA synthesis is inhibited.40 The expression of nrdF in E. coli was found to be growth-phase dependent, and growth conditions that result in a decreased DNA/mass ratio result in increased levels of ribonucleotide reductase.41 We propose that this is the reason for the upregulation of nrdF (Bbr_1898) in B. breve UCC2003 in response to iron-chelation as an obvious reduction in growth rate is observed on inclusion of dipyridyl in the growth media.
Among the genes, whose expression was repressed by iron-chelation, are those that encode transport systems, as well as a number of hypothetical proteins with predicted membrane association. Of interest is the downregulation of genes annotated to play a role in pyroxidine biosynthesis (Bbr_0549) and aspartate carbamoyltransferase (Bbr_0973–0975). Aspartate carbamoyltransferase (ACTase) catalyzes the first committed step in de novo pyrimidine biosynthesis in bacteria, and the acquisition of nucleotides is a vital process of all living cells.42 Previous work has shown that ACTase activity in H. pylori is highly regulated,43 and it is probably a major point of control of de novo pyrimidine biosynthesis pathway in this bacterium. In E. coli a transient repression of ACTase has previously been used as an early indicator of a stress response,44 and a similar effect may thus be applicable in B. breve UCC2003.
The most significantly upregulated gene-cluster in B. breve UCC2003 following iron-chelation is bfeUOB. Homologs of bfeUOB are present in many sequenced bacteria and their protein products form a subgroup of the ILT superfamily28 renamed as oxidase-dependent iron transporters (OFeT) (TC.9.A.10.1).28 The first functional report on a bacterial OFeT transporter, YwbL, came from Bacillus subtilis,45 which was shown to be necessary for growth in defined medium under conditions of low iron availability and when citrate as a siderophore is absent. The identification of the homologous system in E. coli was complicated by the fact that the commonly studied laboratory strains such as K12 contain a truncated and therefore non-functional permease (YcdN). In E. coli strains other than K12, full-size ycdN genes can be identified, which are therefore presumed to contain a functional YcdN-dependent iron-uptake system. Grosse et al. (2006) reported that the iron permease YcdN from the probiotic E. coli strain Nissle 1917 is encoded as part of a Fur-regulated tricistronic operon and renamed YcdN to EfeU (elemental ferrous iron uptake), in contrast to siderophore-chelated ferric iron-uptake systems.7 A striking feature of the OFeT family is the occurrence of two REXXE amino acid residue motifs that were found to be essential for Ftr1p function in yeast.27,30 Two corresponding REGLE motifs were identified in BfeU. In E. coli the importance of this motif was highlighted by mutation of the first glutamate residue of each motif into an alanine, which failed to improve growth of strain GR536, an E. coli K12(W3110)-derived mutant, (DfecABCDE::kanDzupT::cat DmntHDfeoABCDentC), which carries deletions and/or insertions in all known iron acquisition systems,46 suggesting that each motif is necessary for its biological function.
The involvement of B. breve bfeUOB in iron metabolism was demonstrated through complementation analysis: when bfeUOB was present on a low copy-number plasmid (pBfeUOB) it allowed increased growth of the iron transport-debilitated E. coli strain GR536 grown in the presence of the iron-chelating agent dipyridyl but provided no major advantage under iron proficient conditions, and no advantage to a “wild-type” strain. Interestingly, progressive removal of bfeO and bfeB from the complementation-vector negated the growth advantage, further confirming the functionality of bfeUOB to specify a multi-component iron-uptake system. Based on its sequence, the presence of the functional motifs (REGLE) and its conferred growth advantage when heterologously expressed in the iron-uptake mutant GR536, bfeU can be classified as encoding an OFeT-family ferrous iron permease. Although the precise functions of BfeU and BfeO are unknown,47 we speculate that they are involved in the presentation of Fe2+ to the transporter.
A primary goal of this work was to identify a promoter which is directly affected by the presence of the chelator and which can therefore be exploited to create an inducible protein expression system in B. breve UCC2003. The putative promoter region upstream of bfeU was selected and cloned into pNZ8048, thereby replacing the nisin-inducible promoter, followed by the insertion of a β-glucuronidase reporter gene (pDIP1-gusA). The actual induction level measured using this dipyridyl-inducible promoter is likely to be higher than the observed 5-fold, although it is comparable to the (2–7 fold) increase observed with the desA-amylase dipyridyl-inducible system developed for Streptomyces pilosus.37 However, the control plasmid produces a significant level of background GUS activity, which is probably due to transcription originating from elsewhere on the pNZ-derived plasmid. This inducible system could therefore be improved by the inclusion of a transcriptional “silencer” just upstream of the bfeU-derived promoter. Significantly, these results do represent a dose-dependent response to the presence of an inducer, in this case the chelator 2′2-dipyridyl. Regulation of the response to ferrous iron chelation by B. breve UCC2003 is likely to be FUR-independent and presumably involves a novel but as yet unidentified regulator. Unraveling the precise mechanism of control should allow us to improve our inducible system with the goal of creating an improved genetic tool for use in bifidobacteria for controlled expression of proteins that become toxic or insoluble when induced at high levels.
Methods
Bacterial strains and culture conditions
The various bacterial strains and plasmids used in this study are listed in Table 4. E. coli strain DH10B (Invitrogen) was used as a cloning host and was grown aerobically at 37°C in LB medium (Sigma). For iron limitation experiments E. coli cultures were grown overnight in LB at 37°C, diluted 1:400 in Tris-buffered mineral salts medium containing 2 ml of glycerol and 3 g/L of casamino acids and grown overnight. Cultures were inoculated (as a 1:400 dilution) in fresh Tris-buffered mineral salts medium,48 and after 2 h early log cultures were diluted 1:400 in fresh medium to which a varying amount of 2′2-dipyridyl was added, and growth ability was recorded spectrophotometrically at OD600nm. B. breve UCC2003 was routinely grown at 37°C in reinforced clostridial medium (Oxoid). For iron-limitation tests involving B. breve UCC2003 we used a chemically defined medium, which was developed from first principles (designated as chemically defined medium or CDM)49 and which was supplemented with 0.05% (w/v) cysteine-HCl. Anaerobic conditions were established and maintained using an anaerobic chamber [Mac500, Don Whitley Scientific, (atmosphere 10% H2, 10% CO2, 80% N2)]. Varying amounts of 2′2-dipyridyl and/or various antibiotics [chloramphenicol (15–20 μg ml−1), kanamycin (25 μgml−1), ampicillin (100 μg ml−1)] were added to growth media where appropriate.
Table 4. Bacterial strains and plasmids used in this study.
| Bacterial species | Strain Details | Reference |
|---|---|---|
|
B. breve |
UCC2003 Source of bfeUOB,50 |
University College Cork Culture Collection |
|
B. longum |
JCM 7052 Encodes a potential siderophore |
Japan Collection of Microorganisms |
|
B. adolescentis |
CIP 64.61 |
Pasteur Institute Collection, France |
|
B. adolescentis |
NCFB 2229 |
National Collection of Food Bacteria, Reading |
|
B. animalis |
JCM 20097 |
Japan Collection of Microorganisms |
|
B. animalis |
DSM 20105 |
Deutsche Sammlung von Mikroorganismen GmbH, Germany |
|
B. bifidum |
NCIMB 8810 |
National Collection of Industrial, Food and Marine Bacteria, Aberdeen |
|
B. bifidum |
LMG 11041 |
Laboratorium voor Microbiologie, Universiteit Gent, Gent, Belgium |
|
B. breve |
JCM 7017 |
Japan Collection of Microorganisms |
|
B. dentium |
NCFB 2843 |
National Collection of Food Bacteria, Reading |
|
B. longum |
CIP 64.63 |
Pasteur Institute Collection, France |
|
B. infantis |
NCFB 2205 |
National Collection of Food Bacteria, Reading |
|
B. pseudocatenulatum |
NCIMB 8811 |
National Collection of Industrial, Food and Marine Bacteria, Aberdeen |
|
B. pseudolongum |
NCIMB 2244 |
National Collection of Industrial, Food and Marine Bacteria, Aberdeen |
|
B. globosum |
JCM 7092 |
Japan Collection of Microorganisms |
|
B. thermophilum |
JCM 7027 |
Japan Collection of Microorganisms |
|
E. coli |
DH10B, Cloning host |
Invitrogen, Paisley, United Kingdom |
|
E. coli Nissle 1917 |
DSM 6601 |
Deutsche Sammlung von Mikroorganismen, GmbH, Germany |
| E. coli | GR536 | 7 |
| Plasmid Name | Description | Source |
|---|---|---|
| pWSK29 |
Ampr low copy vector |
59 |
| pNZ272 |
Promoterless gusA,Cmr, 4.6 kb |
61 |
| pDIP1-gusA |
Transcriptional fusion between bfeU promoter region and gusA reporter. |
This study |
| pBfeU |
pWSK29 containing bfeU only |
This study |
| pBfeUOB |
pWSK29 containing bfeUOB entire operon |
This study |
| pBfeUO | pWSK29 containing bfeUO and a truncated non-functional bfeB. | This study |
DNA techniques
Plasmid DNA was isolated from E. coli using a QIAprep Spin Miniprep kit according to the manufacturer's instructions (QIAGEN). Genomic DNA isolation from B. breve UCC2003 was performed as described previously.50 Standard procedures were used for DNA manipulation in E. coli. Plasmid DNA was introduced into B. breve by electrotransformation as previously described.51 Restriction endonucleases (Roche Diagnostics), T4 DNA ligase (Roche), and 2 × PCR mixture (Promega) were used as recommended by the manufacturers. Primers were purchased from MWG and are listed in Table S1. PCR products that needed to be cloned were generated using KOD hot-start high-fidelity DNA polymerase (Merck) and subsequently sequenced to verify sequence integrity.
DNA-microarray and QRT-PCR experimental procedures
DNA-microarrays of B. breve UCC2003 were obtained from Agilent Technologies.52 An overnight culture of B. breve UCC2003 in CDM was inoculated at 1% into fresh CDM and incubated at 37°C until an OD at 600 nm of 0.5 was reached and then divided into individual subcultures. Two subsequent subcultures were exposed to 3 µM of the iron-chelator 2′2-dipyridyl for 180 min (DIP+) while a further two acted as controls, i.e., growth at 37°C for 180 min (DIP-). In all cases, cells were harvested by centrifugation at 8,000 × g for 1 min at room temperature and immediately frozen in liquid nitrogen prior to RNA isolation.
Methods for cell disruption, RNA isolation, RNA quality control, cDNA synthesis and indirect labeling were performed as described previously.53 Labeled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part number 5188–5242) as described in the Agilent Two-Color Microarray-Based Gene Expression Analysis v4.0 manual (G4140–90050). Following hybridization, all microarrays were washed as described in the manual and scanned using Agilent's DNA microarray scanner G2565A. The scans were converted to data files with Agilent's Feature Extraction software (Version 9.5). DNA-microarray data were processed as previously described.53-55 Differential expression tests were performed with the Cyber-T implementation of a variant of the t-test.56 A gene was considered to exhibit a significantly different expression level relative to the control when p < 0.001 and an expression ratio of > 3 or < 0.25. Final data represents the average of at least two independent array experiments.
The QRT-PCR amplifications were performed on an ABIPRISM 7000 using SYBR®Green I dye assay chemistry. A 12.5 μl PCR assay for each gene of interest consisted of 6.25 μl of 2X RT PCR master mix (Biogene, Kimbolton, UK), 0.625 μl 1/3000 SYBR green dye (Biogene), 4.225 μl of H20, 0.2 μl (3 μmol) of forward and 0.2 μl (3 μmol) of reverse primers, and 1 μl (10 ng) of cDNA template. All QRT-PCR’s were run in triplicate; three biological replicates were taken for both iron restricted and non-restricted cultures, resulting in nine measurements per gene for each environmental condition. No amplification was observed for the QRT-PCR controls (no reverse transcriptase and no template; data not shown). Cycling conditions used for all amplifications were one cycle of 95°C for 10 min and 40 cycles of 95°C for 15 sec and 58°C for 1 min. From the QRT-PCR data, an average cycle threshold (Ct) value was calculated from the triplicate reactions. Averaged Ct values were then normalized (to adjust for varying amounts of cDNA for each reaction) relative to the control gene, rnpA, which was selected because the average gene expression did not appear to change in previously published microarray studies performed on B. breve UCC2003.52,57 The average fold ratio differences from the biological triplicates of the genes from iron restricted and non-restricted samples were determined as described previously.58
Plasmid constructions
Plasmid pBfeU was constructed as follows; the bfeUgene (Bbr_0221) was amplified by PCR from chromosomal DNA of B. breveUCC2003 using primers bfe1 and bfe2and cloned as a KpnI-BamHI insert into the vector pWSK29.59 Plasmid pBfeUOB (Bbr_0221-0223) was constructed by cloning a DNA fragment encompassing the entire bfeUOBgene cluster including the presumptive promoter region of bfeU (500 bp immediately upstream of bfeU). This bfeUOB-containing fragment was obtained by PCR using primers bfe3 and bfe4, and chromosomal DNA of B. breve UCC2003 as a template and cloned into pWSK29 as a SacI-BamHI insert. Plasmid pBfeUO was constructed by restricting plasmid pBfeUOB with BstB1 and BamHI, which removes 282 bp from the 3′ end of bfeB. Following digestion, the larger DNA fragment, which contains the pWSK29 vector plus the bfeUOB gene cluster with a truncated bfeB gene, was isolated, treated with Klenow (New England Biolabs) to polish single strand overhangs and re-ligated. The dipyridyl-inducible plasmid pDIP1 was constructed by cloning the presumed promoter region of bfeU, which was amplified by PCR from chromosomal DNA of B. breve UCC2003 using primers PbfeUfw and PbfeUrev, as a BglII-Nco I insert into the corresponding sites of vector pNZ8048, thereby replacing the nisin-inducible promoter. Plasmid pDIP1-gusA was subsequently constructed by inserting the gusA gene from plasmid pNZ272 (66), amplified by the primers gusAfw and gusArev as an Nco I–Hind III insert in the corresponding sites in pDIP1, thereby creating a transcriptional fusion of the presumed iron-responsive promoter of bfeU and gusA.
β-glucuronidase activity
Following overnight growth B. breve UCC2003 containing plasmid pNZ272 (control) and B. breve UCC2003 containing plasmid pDIP1-gusA(test) were subcultured and grown until the OD600nm had reached approximately 0.5, at which point the cultures were split and exposed to increasing amounts of 2′2-dipyridyl, the final concentration of which ranged from 0 to 5 µM. Following 180 min incubation with this chelator the cells were harvested and the levels of β-glucuronidase (GUS) activity determined as described previously.60
Supplementary Material
Disclosure of Potential Conflicts of Interest
The authors declare no competing or financial interests.
Acknowledgments
All authors are members of The Alimentary Pharmabiotic Centre, which is a Centre for Science and Technology (CSET) at University College Cork financially supported by Science Foundation Ireland (SFI) through the National Development Plan and specifically grants 02/CE/B124 and 07/CE/B1368. Furthermore the authors wish to acknowledge the support of a DAF/HRB FHRI award to the ELDERMET project to D.V.S. and an EMBARK fellowship from the Irish Research Council for Science Engineering and Technology to A. Z.
Author Contributions
MC and AZ contributed equally to the laboratory work, MC wrote the manuscript. GF assisted with planning the experiments. DvS supervised the project and critically reviewed the manuscript. All authors read and approved the final manuscript.
Author Information
Michelle Cronin is a research fellow at Cork Cancer Research Centre, Mercy University Hospital and Leslie C. Quick Jnr. Laboratory, University College Cork; Aldert Zomer is a senior scientist at the Laboratory of Pediatric Infectious Diseases, Centre for Molecular and Biomolecular Informatics Radboud University Nijmegen Medical Centre; Gerald Fitzgerald is Professor of Food Microbiology and Head of Department of Microbiology at University College Cork, and deputy director of the Alimentary Pharmabiotic Centre (APC); Douwe van Sinderen is Associate Professor in Department of Microbiology at UCC and a Principal Investigator in the APC.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/18985
References
- 1.Guerinot ML. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–72. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC. Iron storage in bacteria. Adv Microb Physiol. 1998;40:281–351. doi: 10.1016/S0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- 3.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 5.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 6.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–9. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62:120–31. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 8.Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15:328–34. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Jeon J, Kim H, Yun J, Ryu S, Groisman EA, Shin D. RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J Bacteriol. 2008;190:7326–34. doi: 10.1128/JB.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, et al. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol. 2005;187:1604–11. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modun B, Evans RW, Joannou CL, Williams P. Receptor-mediated recognition and uptake of iron from human transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 1998;66:3591–6. doi: 10.1128/iai.66.8.3591-3596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartford T, O'Brien S, Andrew PW, Jones D, Roberts IS. Utilization of transferrin-bound iron by Listeria monocytogenes. FEMS Microbiol Lett. 1993;108:311–8. doi: 10.1111/j.1574-6968.1993.tb06121.x. [DOI] [PubMed] [Google Scholar]
- 13.Montanez GE, Neely MN, Eichenbaum Z. The streptococcal iron uptake (Siu) transporter is required for iron uptake and virulence in a zebrafish infection model. Microbiology. 2005;151:3749–57. doi: 10.1099/mic.0.28075-0. [DOI] [PubMed] [Google Scholar]
- 14.Bates CS, Montanez GE, Woods CR, Vincent RM, Eichenbaum Z. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect Immun. 2003;71:1042–55. doi: 10.1128/IAI.71.3.1042-1055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt MP. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J Bacteriol. 1999;181:5330–40. doi: 10.1128/jb.181.17.5330-5340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JS, Holden DW. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 2002;4:1149–56. doi: 10.1016/S1286-4579(02)01640-4. [DOI] [PubMed] [Google Scholar]
- 17.Kot E, Bezkorovainy A. Binding of ferric iron to the cell walls and membranes of Bifidobacterium thermophilum: effect of free radicals. J Agric Food Chem. 1999;47:4606–10. doi: 10.1021/jf990474l. [DOI] [PubMed] [Google Scholar]
- 18.Ghio AJ, Piantadosi CA, Crumbliss AL. Hypothesis: iron chelation plays a vital role in neutrophilic inflammation. Biometals. 1997;10:135–42. doi: 10.1023/A:1018387308517. [DOI] [PubMed] [Google Scholar]
- 19.Ullen H, Augustsson K, Gustavsson C, Steineck G. Supplementary iron intake and risk of cancer: reversed causality? Cancer Lett. 1997;114:215–6. doi: 10.1016/S0304-3835(97)04666-1. [DOI] [PubMed] [Google Scholar]
- 20.Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie van Leeuwenhoek. 2008;94:35–50. doi: 10.1007/s10482-008-9232-4. [DOI] [PubMed] [Google Scholar]
- 21.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21:149–56. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Bezkorovainy A, Topouzian N, Miller-Catchpole R. Mechanisms of ferric and ferrous iron uptake by Bifidobacterium bifidum var. pennsylvanicus. Clin Physiol Biochem. 1986;4:150–8. [PubMed] [Google Scholar]
- 23.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Sullivan DJ. Screening of intestinal microflora for effective probiotic bacteria. J Agric Food Chem. 2001;49:1751–60. doi: 10.1021/jf0012244. [DOI] [PubMed] [Google Scholar]
- 25.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, et al. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009;5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol. 2007;65:857–75. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 27.Severance S, Chakraborty S, Kosman DJ. The Ftr1p iron permease in the yeast plasma membrane: orientation, topology and structure-function relationships. Biochem J. 2004;380:487–96. doi: 10.1042/BJ20031921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saier MH, Jr., Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman TH, Tuckman M, Ellestad S, Osburne MS. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfpo and Escherichia coli entD genes. J Bacteriol. 1993;175:6203–11. doi: 10.1128/jb.175.19.6203-6211.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–7. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 31.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–9. [PubMed] [Google Scholar]
- 32.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J Mol Biol. 1994;235:1501–31. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 33.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–41. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezkorovainy A, Topouzian N. The effect of metal chelators and other metabolic inhibitors on the growth of Bifidobacterium bifidus var. Pennsylvanicus. Clin Biochem. 1981;14:135–41. doi: 10.1016/S0009-9120(81)90281-2. [DOI] [PubMed] [Google Scholar]
- 35.Chart H, Buck M, Stevenson P, Griffiths E. Iron regulated outer membrane proteins of Escherichia coli: variations in expression due to the chelator used to restrict the availability of iron. J Gen Microbiol. 1986;132:1373–8. doi: 10.1099/00221287-132-5-1373. [DOI] [PubMed] [Google Scholar]
- 36.Lim JM, Hong MJ, Kim S, Oh DB, Kang HA, Kwon O. Iron chelator-inducible expression system for Escherichia coli. J Microbiol Biotechnol. 2008;18:1357–63. [PubMed] [Google Scholar]
- 37.Flores FJ, Rincon J, Martin JF. Characterization of the iron-regulated desA promoter of Streptomyces pilosus as a system for controlled gene expression in actinomycetes. Microb Cell Fact. 2003;2:5. doi: 10.1186/1475-2859-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura M, Canchaya C, Zink R, Fitzgerald GF, van Sinderen D. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl Environ Microbiol. 2004;70:6197–209. doi: 10.1128/AEM.70.10.6197-6209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakunst D, Larisch C, Huser AT, Tauch A, Puhler A, Kalinowski J. The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes. J Bacteriol. 2007;189:4696–707. doi: 10.1128/JB.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanke PD, Fuchs JA. Regulation of ribonucleoside diphosphate reductase mRNA synthesis in Escherichia coli. J Bacteriol. 1983;154:1040–5. doi: 10.1128/jb.154.3.1040-1045.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 42.Burns BP, Hazell SL, Mendz GL, Kolesnikow T, Tillet D, Neilan BA. The Helicobacter pylori pyrB gene encoding aspartate carbamoyltransferase is essential for bacterial survival. Arch Biochem Biophys. 2000;380:78–84. doi: 10.1006/abbi.2000.1920. [DOI] [PubMed] [Google Scholar]
- 43.Burns BP, Mendz GL, Hazell SL. In situ properties of Helicobacter pylori aspartate carbamoyltransferase. Arch Biochem Biophys. 1997;347:119–25. doi: 10.1006/abbi.1997.0328. [DOI] [PubMed] [Google Scholar]
- 44.Faber F, Egli T, Harder W. Transient repression of the synthesis of OmpF and aspartate transcarbamoylase in Escherichia coli K12 as a response to pollutant stress. FEMS Microbiol Lett. 1993;111:189–95. doi: 10.1111/j.1574-6968.1993.tb06384.x. [DOI] [PubMed] [Google Scholar]
- 45.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–73. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosse R, Lund U, Caruso V, Fischer R, Janka GE, Magnano C, et al. Non-transferrin-bound iron during blood transfusion cycles in beta-thalassemia major. Ann N Y Acad Sci. 2005;1054:429–32. doi: 10.1196/annals.1345.074. [DOI] [PubMed] [Google Scholar]
- 47.Sturm A, Schierhorn A, Lindenstrauss U, Lilie H, Bruser T. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J Biol Chem. 2006;281:13972–8. doi: 10.1074/jbc.M511891200. [DOI] [PubMed] [Google Scholar]
- 48.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–34. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petry S, Furlan S, Crepeau MJ, Cerning J, Desmazeaud M. Factors affecting exocellular polysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus grown in a chemically defined medium. Appl Environ Microbiol. 2000;66:3427–31. doi: 10.1128/AEM.66.8.3427-3431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacConaill LE, Fitzgerald GF, Van Sinderen D. Investigation of protein export in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2003;69:6994–7001. doi: 10.1128/AEM.69.12.6994-7001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maze A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–53. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191:7039–49. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, Larsen R, et al. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics. 2005;6:77. doi: 10.1186/1471-2164-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics. 2003;2:241–4. [PubMed] [Google Scholar]
- 55.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108:11217–22. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–44. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 57.Pokusaeva K, O'Connell-Motherway M, Zomer A, Fitzgerald GF, van Sinderen D. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2009;75:1135–43. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–9. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 60.Sangrador-Vegas A, Stanton C, van Sinderen D, Fitzgerald GF, Ross RP. Characterization of plasmid pASV479 from Bifidobacterium pseudolongum subsp. globosum and its use for expression vector construction. Plasmid. 2007;58:140–7. doi: 10.1016/j.plasmid.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Platteeuw C, Simons G, de Vos WM. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–93. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.