Abstract
We have recently reported the first partially synthetic eukaryotic genome. Saccharomyces cerevisiae chromosomes synIXR and semi-synVIL are fully synthetic versions of the right arm of chromosome IX and the telomeric segment of the left arm of chromosome VI, respectively, and represent the beginning of the synthetic yeast genome project, Sc2.0, that progressively replaces native yeast DNA with synthetic sequences. We have designed synthetic chromosome sequences according to principles specifying a wild-type phenotype, highly stable genome, and maintenance of genetic flexibility. Although other synthetic genome projects exist, the Sc2.0 approach is unique in that we have implemented design specifications predicted to generate a wild-type phenotype until induction of “SCRaMbLE,” an inducible evolution system that generates significant genetic diversity. Here we further explore the significance of Sc2.0 and show how SCRaMbLE can serve as a genome minimization tool.
Keywords: genome minimization, Sc2.0, syn VIL, Synthetic genome, yeast
Genome-scale analyses have provided great insight into systems level phenomenon. However, while our current understanding of genomics is solidly within the experimental phase, genome engineering is in its infancy. DNA synthesis techniques have been widely used to manufacture genes or genetic elements. These efforts began with the success of Khorana and colleagues in the synthesis of a 77 bp tRNA gene in 1970 and a 207 bp tRNA suppressor in the 1976.1,2 As the costs of DNA synthesis continued to fall and new technologies were developed, synthesis at progressively larger scales became possible, ultimately enabling genome-scale construction. The first genomes to be synthesized belonged to viruses, specifically the 7.5 kb polio virus3 and the 5.4 kb φX174 genome.4 Recently, these efforts graduated in scope yet again: the synthesis of the 583 kb Mycoplasma genitalium genome was completed in 2008 and the 1.08 Mb Mycobacterium mycoides genome in 2010.5,6 We have previously described another advance in the synthetic genomics arena: the synthetic yeast genome project, Sc2.0, systematically replaces large segments of the 12 Mb Saccharomyces cerevisiae genome with synthetic designer DNA.7 Sc2.0 is one of the first genome projects of its kind and is the first effort to engineer a eukaryotic genome. Further, Sc2.0 incorporates SCRaMbLE (Synthetic Chromosome Rearrangement and Modification by LoxPsym-mediated Evolution), an inducible evolution system that restructures the synthetic yeast genome at will. Thus, Sc2.0 represents not only an increase in scale of genome projects but the progression from descriptive synthetic genomics, wherein native genomes are synthesized without significant modification, to experimental synthetic genomics, wherein new genome variants are constructed. Here we explore the relevance of this project and introduce Sc2.0 as genome minimization system.
A major challenge in the synthesis of large DNAs is the assembly step; contiguous DNA segments (contigs) are generally assembled from smaller, more easily synthesized DNAs. Small DNA fragments of approximately 500–3000 bp are now routinely synthesized from short oligos.8,9 Higher order assembly of such fragments is often performed by modified enzymatic methods, such as that performed to construct the Saccharomyces cerevisiae chromosome SynIXR,10 or by exploiting homologous recombination in vivo,5,11,12 one method we employ to assemble fragments of the synthetic yeast genome and the method employed to construct the synthetic Mycoplasma genome.
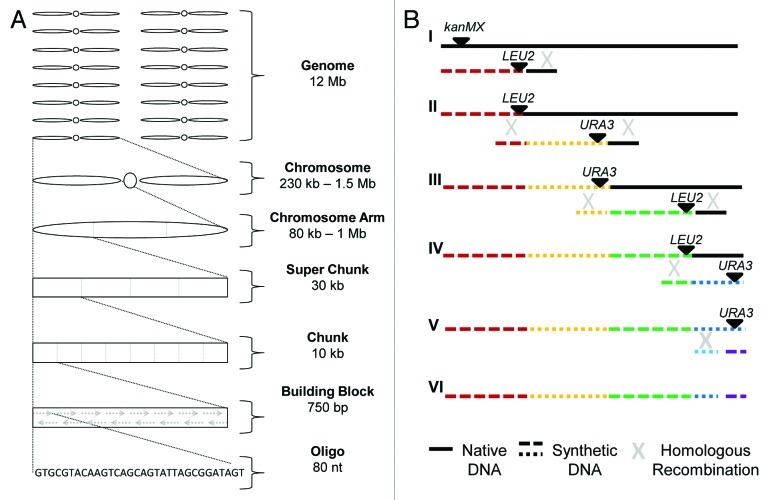
Synthesis of Sc2.0 differs from other genome projects in many ways. The first eukaryotic genome to be synthesized, the S. cerevisiae genome comprises 16 linear chromosomes totaling approximately 12 Mb, in contrast to the small (< 10 kb) viral genomes and the 583 kb and 1 Mb circular Mycoplasma genomes. The yeast genome is an order of magnitude larger than Mycoplasma mycoides, the largest genome yet assembled. As a result, new technologies must be developed, and the genome must be reduced to manageable pieces: chromosomes may be subdivided into chromosomes arms that must be then further subdivided to facilitate synthesis and construction (Fig. 1A). This artificial modularity based upon size, rather than genomic elements (e.g., genes, promoters, etc.), enables the inclusion of many collaborators, now including teams of undergraduate Build-a-Genome students9 at different institutions. This modularity also allows specific regions of the genome to be targeted for replacement, or later modified as necessary. Construction is generally performed to the level of chunks or super chunks that are then incorporated by homologous recombination, as in the case of chromosome semi-synVIL, or as circular chromosomes, as for synIXR.7 Stepwise incorporation of synthetic sequence by iterative homologous recombination is the preferred method as the native sequence is removed as the synthetic sequence is incorporated (Fig. 1B).
Figure 1. Genome modularity and integration of synthetic DNA. (A) The yeast genome is subdivided into increasingly smaller segments to facilitate construction and assembly of the synthetic Sc2.0 genome (not to scale). The assembly pipeline may be entered from multiple points. The assembly technique utilized by Build-a-Genome students begins at the bottom of the assembly pipeline, constructing building blocks from oligos.9 Commercially synthesized DNAs used to construct semi-synVIL were obtained as chunks, and assembled into a super chunk prior to integration in the yeast genome. SynIXR entered the pipeline as a chromosome arm. (B) Synthetic DNA is iteratively integrated into the yeast genome to replace native DNA (not to scale). The native chromosome, marked with kanMX, is targeted for replacement by integration of synthetic DNA, marked with LEU2. An “endcap” directs homologous recombination to the region/s flanking the synthetic sequence. The resulting Leu+ G418S semi-synthetic chromosome is then targeted for replacement by integration of a URA3-marked synthetic DNA fragment (II). Iterative transformations with synthetic DNA fragments alternately marked with LEU2 and URA3 sequentially replace the native DNA (III-IV). The final URA3 marker is replaced by transformation with synthetic sequence lacking the URA3 gene and subsequent selection on 5-FOA (V) to generate the complete unmarked synthetic chromosome.
Integration of synthetic sequence represents another difference between the synthetic yeast genome project and other efforts. In the design phase, a wrong prediction of the content of a minimal genome or change in genome structure would result in lethality using the “top down” approach employed in the Mycoplamsa construction in which the whole genome is replaced at once.6 The “bottom up” philosophy underlying the Sc2.0 genome design, incorporating 10–100 kb segments iteratively, with fitness testing at each cycle, allows maximal plasticity while preserving viability. If a modification results in a decrease in fitness, the bottom up approach allows rapid identification of the detrimental sequence change and replacement of the modified segment following correction of the offending modification.
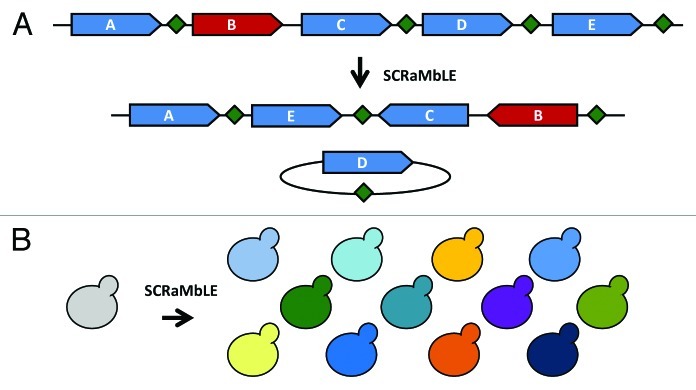
Another fundamental difference between Sc2.0 and the existing genome projects is the design philosophy. Engineers of the polio virus, ϕX174, and Mycoplasma genomes did not incorporate significant modifications in the sequence design, beyond the addition of a few “watermarks” in the Mycoplasma sequences.3,4,6 As such, these genomes are rigid synthetic models of the extant native genome and do not include provisions for more extensive genome remodeling, the ultimate long-term goal of most genome synthesis projects. Therefore, making extensive genome wide changes will require a full resynthesis in these cases, increasing costs. In contrast, Sc2.0 incorporates the inducible evolution system, SCRaMbLE, allowing unparalleled restructuring of a nearly wild-type genome to generate genomes of highly variable structures and contents. While the synthetic yeast has a near-wild type genotype with regard to gene content, induction of SCRaMbLE results in significant modifications both in genome structure and content (Fig. 2).7 The symmetry inherent in the loxPsym site should generate inversions and deletions between the same pair of sites on one chromosome with theoretical equal probability and may also allow formation of translocations and duplications.13 Following genome SCRaMbLEing, only viable configurations are recovered. Thus, Sc2.0 is highly plastic and can generate a wide variety of genome variants with little additional expenditure of time or money.
Figure 2. SCRaMbLE restructures the synthetic genome. (A) LoxPsym sites (green diamonds) are inserted in the 3′UTR of each non-essential gene (blue arrows); essential genes (red arrow) do not have an associated loxPsym site. The symmetry of loxPsym sites permits both translocations/inversions and deletions at each site. Complex rearrangements result from induction of SCRaMbLE. In the example shown, genes “B” and “C” are inverted, “E” has been excised and reintegrated, and “D” has been lost from the SCRaMbLEd chromosome. (B) Induction of SCRaMbLE in a synthetic strain (gray) results in a significant increase in genetic diversity (colors). Following selection for a desired phenotype, which can range from simple viability to increased ability to produce a desirable substance, genome content and structure of SCRaMbLEd strains can be analyzed by PCRTag analysis,7 comparative genome hybridization (CGH), molecular karyotyping and/or whole-genome sequencing.
One major goal in the development of synthetic genomes is the design and construction of a “minimal genome,” a genome containing the bare minimum of genetic elements required to support life. The synthesis of a minimal genome would be extremely valuable as a genetic answer to the question, “What is life?” Many attempts have been made to predict the contents of a minimal genome, yet the construction of a reduced genome by a top down approach would be extremely difficult. The challenge of identifying a minimal genome is further increased by interactions or redundancy between genes that may render some genes essential in the presence or absence of others, or non-laboratory conditions that may require activation of otherwise non-essential genetic pathways. The SCRaMbLE approach employed in the design of Sc2.0 provides an alternate route to genome minimization. The Sc2.0 genome is constructed to be nearly wild type, but the SCRaMbLE system allows the genome content and structure to be randomly shuffled at will.
Another goal of synthetic genome projects is the development of a “chassis” upon which synthetic pathways can be assembled. Pathway engineering, the rational design and heterologous expression of a biosynthetic pathway in a host organism, has proven extremely valuable in the production of many pharmaceutical and industrial compounds.14 One drawback to this production method is the potential interference of endogenous pathways that may draw off synthetic pathway intermediates and/or products for consumption in other systems. Elimination of competing non-essential endogenous pathways, or decreasing flux through essential endogenous pathways that could siphon off intended heterologous products would increase yields of synthetic engineered pathways. Whereas other synthetic genome projects have focused predominantly on small, easily-synthesized genomes of organisms with little to no industrial relevance, S. cerevisiae is a proven workhorse of metabolic engineering and industrial-scale production.15 Further tailoring of the content yeast genome via SCRaMbLE will provide a sleek genetic chassis from which superfluous and/or interfering metabolic pathways can be eliminated or downregulated. It is unlikely such custom tailoring of genomes will be feasible exclusively by design as it is impossible to predict all metabolic interactions, and such a “top-down” approach is expensive and labor-intensive.
The SCRaMbLE system is effective in generating varying genotypes and phenotypes,7 but has not been explored as a genome minimization method. Chromosome engineering utilizing site-specific recombination has previously been employed to generate structural changes in eukaryotic genomes;16 however, these systems have predominantly been used in a targeted approach. As Sc2.0 will contain many loxPsym sites scattered throughout the genome, it is possible many different types of genome structures may be generated. As the symmetry of loxPsym sites theoretically allows inversions and deletions with equal frequency, successive rounds of (low level) SCRaMbLE induction should result in progressively increasing loss of loxPsym-flanked segments, and ongoing selection against non-fit variants. Our previous work demonstrated that SCRaMbLE generates gene deletions. Thus, repeated rounds of SCRaMbLE should be an efficient method to generate minimal genomes. To explore this capability, we performed a proof of principle experiment utilizing semi-synVIL, a ~30 kilobase pair (kb) synthetic segment on the telomeric end of the left arm of chromosome VIL. Semi-synVIL contains five loxPsym sites, including one at the telomere, and one at the centromeric end of the synthetic sequence. We induced SCRaMbLE in haploid yeast carrying semi-synVIL and passaged cells under SCRaMbLEing conditions for 12 d. PCR using primers flanking the first and last loxPsym sites revealed a viable full semi-synVIL deletion, suggesting that SCRaMbLE may serve as an effective genome minimization tool.
Attempts to design Sc2.0 have necessitated the invention of new approaches to customize the genome for maximum usefulness. The SCRaMbLE system of Sc2.0 is the first example of systematic insertion and genome-wide usage of site-specific recombination to broadly restructure a genome. We have previously shown SCRaMbLE is an effective mutagenesis tool, and restructures synthetic chromosomes at will;7 here we have shown SCRaMbLE can also be used as an effective genome minimization tool. These proof of principle experiments confirm the complete Sc2.0 genome will allow unparalleled manipulation of genome structure and content, ushering in a new era of experimental genomics.
Glossary
Abbreviations:
- PCR
polymerase chain reaction
- bp
base pair
- kb
kilobase pair
- Mb
megabase pair
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/19543
References
- 1.Agarwal KL, Büchi H, Caruthers MH, Gupta N, Khorana HG, Kleppe K, et al. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature. 1970;227:27–34. doi: 10.1038/227027a0. [DOI] [PubMed] [Google Scholar]
- 2.Khorana HG, Agarwal KL, Besmer P, Büchi H, Caruthers MH, Cashion PJ, et al. Total synthesis of the structural gene for the precursor of a tyrosine suppressor transfer RNA from Escherichia coli. 1. General introduction. J Biol Chem. 1976;251:565–70. [PubMed] [Google Scholar]
- 3.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–8. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 4.Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A. 2003;100:15440–5. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–20. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 6.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–6. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 7.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–6. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 9.Dymond JS, Scheifele LZ, Richardson S, Lee P, Chandrasegaran S, Bader JS, et al. Teaching synthetic biology, bioinformatics and engineering to undergraduates: the interdisciplinary Build-a-Genome course. Genetics. 2009;181:13–21. doi: 10.1534/genetics.108.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blake WJ, Chapman BA, Zindal A, Lee ME, Lippow SM, Baynes BM. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 2010;38:2594–602. doi: 10.1093/nar/gkq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sosio M, Bossi E, Donadio S. Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:E37. doi: 10.1093/nar/29.7.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A. 2008;105:20404–9. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–8. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 15.Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2008;72:379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–90. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]