Abstract
Over the past 20 years, directed evolution has been seen to be the most reliable approach to protein engineering. Emulating the natural selection algorithm, ad hoc enzymes with novel features can be tailor-made for practical purposes through iterative rounds of random mutagenesis, DNA recombination and screening. Of the heterologous hosts used in laboratory evolution experiments, the budding yeast Saccharomyces cerevisiae has become the best choice to express eukaryotic proteins with improved properties. S. cerevisiae not only allows mutant enzymes to be secreted but also, it permits a wide range of genetic manipulations to be employed, ranging from in vivo cloning to the creation of greater molecular diversity, thanks to its efficient DNA recombination apparatus. Here, we summarize some successful examples of the use of the S. cerevisiae machinery to accelerate artificial evolution, complementing the traditional in vitro methods to generate tailor-made enzymes.
Keywords: directed evolution, DNA recombination, IvAM, IVOE, random mutagenesis, Saccharomyces cerevisiae
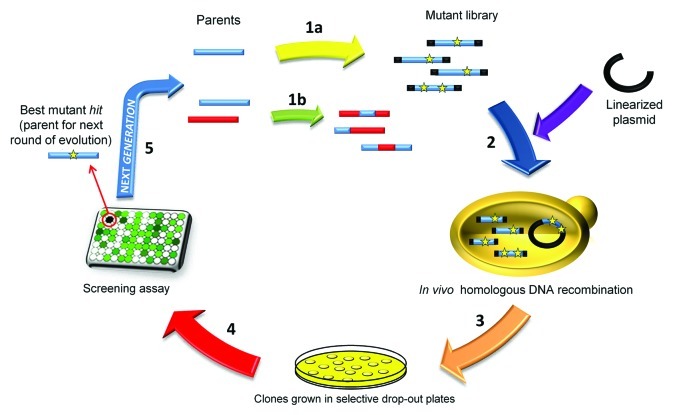
Throughout evolution, natural selection promotes the survival of specific organisms at the expense of thousands with trait/s that are not optimal to live in a given environment. Alterations to genes and enzymes are generated by processes such as random mutagenesis, DNA recombination, deletion and/or insertion, augmenting the diversity in this pool. These molecular modifications are then subjected to rigorous and constant testing by environmental factors, selection processes that drive the survival or disappearance of genes and enzymes. Typically, beneficial mutations (or neutral mutations that may become beneficial) accumulate and are recombined in the offspring. After successive generations of strict selective pressure, such mutations can give rise to new phenotypes. In February 2011, the Draper prize (considered the Nobel of Engineering) was awarded to Frances Arnold and Willem Stemmer for the development of Directed Molecular Evolution. This is a tool that has revolutionized the manner in which proteins are manipulated in the laboratory in order to improve their application in distinct industrial settings. By mimicking the mutation, recombination and selection processes that occur naturally in evolution, in vitro evolution provides a means of directing the evolution of genes toward specific goals in a manner that may not occur in a natural environment1,2 (Fig. 1).
Figure 1. A typical directed evolution experiment using Saccharomyces cerevisiae as a eukaryotic host. The cycle of evolution begins with the generation of diversity by epPCR (1A) or in vitro DNA-recombination (1B). The mutagenic library is transformed into S. cerevisiae (2) and the pool of templates is further recombined by in vivo DNA shuffling. Each template contains adequate overhangs (shown in black) that overlap with the linearized plasmid, facilitating in vivo cloning to generate the autonomously replicating and repaired vector. The clones are grown on selective drop-out plates (3) and transferred to 96-well plates where the expression of mutants is induced. After secretion, the supernatants are subjected to a high throughput assay (4) to select the best enzyme variants. Generally, consecutive re-screenings are incorporated to rule out the presence of false positives. Finally, the best hits are recovered, characterized and their genes subjected to a further generation of directed evolution (5). Yellow stars indicate single mutations.
Heterologous Functional Expression in Saccharomyces cerevisiae
The in silico analysis of genes/enzymes by computational methods is a valuable approach to engineer “smart” libraries reducing the exploration of the vast protein sequence space. This strategy can be combined with powerful tools for HTP-screening [e.g., fluorescence activated cell sorter (FACS)], providing another twist in enzyme engineering by laboratory evolution.3-8 Still, there are 3 basic premises to carry out a laboratory evolution experiment: (1) a suitable functional expression system; (2) reliable screening assays with which to detect improvements introduced after each round of evolution; and (3) the support of in vitro or in vivo methods to create enzyme diversity. The bacteria Escherichia coli is by far the most widely used host in directed evolution as it has a well-described physiology and it reproduces rapidly, making experiments less time-consuming. Moreover, standardized protocols are available to manipulate this bacteria and to rapidly recover the screened variants. While these characteristics generally hold true for prokaryotic proteins, bacterial hosts are less appropriate when working with eukaryotic genes, often resulting in misfolded, deglycosylated, non-functional or altered proteins, and the accumulation of the desired enzyme in inclusion bodies.9-11 These shortcomings can be circumvented by using eukaryotic hosts such as Pichia pastoris or Saccharomyces cerevisiae. P. pastoris can secrete large amounts of proteins and mediate post-translational modifications. However, most vectors available for heterologous expression in P. pastoris are integrative—although there are a few exceptions12—which together with a low efficiency of integration limits their use for HTP-screening (HTPS) and laboratory evolution. Recent efforts have sought to integrate linear expression cassettes in order to express mutant libraries of hydroxynitrile lyases.13 Nevertheless, a cumbersome mutant recovery process and poor transformation rates are still big obstacles which discourage scientists to take this approach. Fortunately, S. cerevisiae provides a solution to these bottlenecks as it exhibits high transformation efficiencies (from 1 × 106 to 1 × 108 transformants/μg DNA depending on the yeast strain), it performs post-translational modifications (e.g., processing of N- and C-terminal ends, glycosylation), and it possesses a fully developed secretory machinery that directs the secretion of proteins into the culture medium (bypassing the tedious lysis steps generally required when working with E. coli and avoiding any interference of complex lysate mixtures in the screening assays14,15). S. cerevisiae may hyperglycosylate heterologous proteins (in some cases over 50% of the enzyme molecular weight) by the addition of mannose moiteties at the Golgi compartment, a side-consequence of difficulties found during the exocytosis. This effect, although generally beneficial for protein stability—at the time that protect the enzyme from proteolytic degradation—generates a pool of isoforms which makes difficult the enzyme purification and biochemical characterization. Interestingly, in recent examples tackled in our laboratory (with high redox potential peroxidases and laccases, see below), mutations discovered by directed evolution helped us to surpass this hurdle by reducing the residence time at the Golgi, which generated new variants whose glycosylation degrees were below 10% showing a noticeable improvement in secretion yields.16,17 It is also worth noting that multicopy episomal and bi-functional vectors are available to help identify and isolate the variants of interest screened from mutant libraries in S. cerevisiae. Finally, S. cerevisiae exhibits a high frequency of homologous DNA recombination with proof-reading activity, enabling in vivo recombination of the best mutant hits to occur at stages that prevent the incorporation of new mutations, as usually occurs in classical in vitro recombination protocols.18 Given these many advantages, S. cerevisiae has begun to be heavily exploited for the functional expression of evolved eukaryotic enzymes in the laboratory.
Despite the advantages offered by S. cerevisiae, there are cases where the initial secretion levels of the target protein are not sufficiently high to perform artificial evolution. However, it has proved possible to adopt different strategies to considerably augment the secretion of such proteins in S. cerevisiae. One approach involves the introduction of random mutations in processing regions of the native gene to adjust the nascent polypeptide to the specific attributes of the proteases found in the secretory route. This approach has been successfully applied in the evolution of the laccase from the ascomycete Myceliophthora thermophila (MtL) in S. cerevisiae for functional expression.19 The single most beneficial mutation (producing a 10-fold enhancement in total activity) was found at the C-terminal tail of MtL, and it involved the introduction of a cleavage site for the KEX2 Golgi protease. An alternative strategy involves replacement of the signal peptide of the native protein with other signal leaders that are recognized better by S. cerevisiae. In particular, the construction of fusion genes with the α-factor prepro-leader from S. cerevisiae can drive protein secretion.20,21 Indeed, S. cerevisiae can process the native signal peptide of foreign proteins in some cases, as seen with the aspartic proteinase from Mucor pusillus and the glucoamylase from Aspergillus awamorii, among other examples.22,23 However, by replacing the native signal leader with the α-factor prepro-leader, expression can be significantly enhanced.24 As ligninolytic enzymes are remarkably difficult to express in non-fungal systems,18,25 our group has used this approach to enhance the expression of these interesting oxidoreductases in S. cerevisiae. In recent studies performed in our laboratory, the native secretion leaders of genes encoding two different high redox potential laccases (PM1L, from basidiomycete PM1, and PcL from Pycnoporus cinnabarinus) and one peroxidase (VP, the versatile peroxidase from Pleurotus eryngii) were replaced by the α-factor prepro-leader.16,17,26 The secretion of these fusion constructs was greater than that of these enzymes with their native leader (by at least one order of magnitude). Moreover, secretion could be further augmented by subjecting the entire gene (i.e., the α-factor prepro-leader plus the mature protein) to directed evolution. This strategy allowed us to adjust both the α-factor prepro-leader and the gene encoding the mature protein to the subtleties of the yeast secretory pathway. The canonical pre-leader is involved in the orientation and insertion of the nascent polypeptide during translocation to the endoplasmic reticulum (ER). Interestingly, mutations in the hydrophobic core of the pre-leader were discovered during the evolution of PcL and PM1L that enhanced secretion several fold (A[α9]D and V[α10]D, respectively). Positions 9 and 10 of the pre-leader were further analyzed by constructing individual and double mutants containing the corresponding substitutions: A[α9]D and V[α10]D mutations exerted a 2.2-fold improvement in secretion individually but not when they were introduced together in the same variant.16 Our results address that slightly increasing the hydrophilicity of the signal pre-leader may have beneficial effects on the interaction between the pre-leader and the signal recognition particle by improving the translocation of the polypeptide chain into the ER.27 We also detected several interesting mutations in the pro-leader during the directed evolution of PM1L and PcL that altered the affinity for sugar anchoring (N[α23]K and S[α58]G, respectively). As these positions correspond to 2 of the 3 N-glycosylation sites in the pro-leader, they may affect ER to Golgi protein transport.16 Recent studies demonstrated that mutations in the α-factor prepro-leader can enhance heterologous protein secretion in S. cerevisiae of a variety of proteins.28 In fact, some of these mutations that increase secretion were the same as those identified for laccase fusion genes in yeast in our laboratory (position and nucleotide change). Finally, our evolved α-factor prepro-leaders were fused with native (non-mutated) laccases, enhancing secretion by up to 40-fold and thereby corroborating the significance of the mutations induced by directed evolution.26 Taken together, these findings suggest that the directed evolution of the α-factor prepro-leader may give rise to a universal signal peptide for the heterologous expression of foreign proteins in yeast.
Exploiting the Machinery of S. cerevisiae for Directed Enzyme Evolution
Developing successful directed evolution experiments requires an appropriate array of molecular methods to allow the user to generate diversity. In this context, the power of S. cerevisiae cannot be underestimated. S. cerevisiae constitutes a simple and efficient vehicle to create libraries for directed evolution, exhibiting a high frequency of homologous DNA recombination with multiple recombination pathways generated by double-strand breaks.29 A recent study reported that S. cerevisiae can recombine up to 38 overlapping single-stranded oligonucleotides and a linear double-stranded vector in just one transformation event.30 Crossover areas can contain as few as 20 base pairs and as many as 200 homologous nucleotides. The importance of the length of the overlapping ends in the crossover region between the DNA fragment and the linearized plasmid to achieve high recombination efficiencies has been demonstrated. Thus, a homologous region of at least 40 base pairs appears to be necessary to obtain recombination efficiencies of over 60%.31
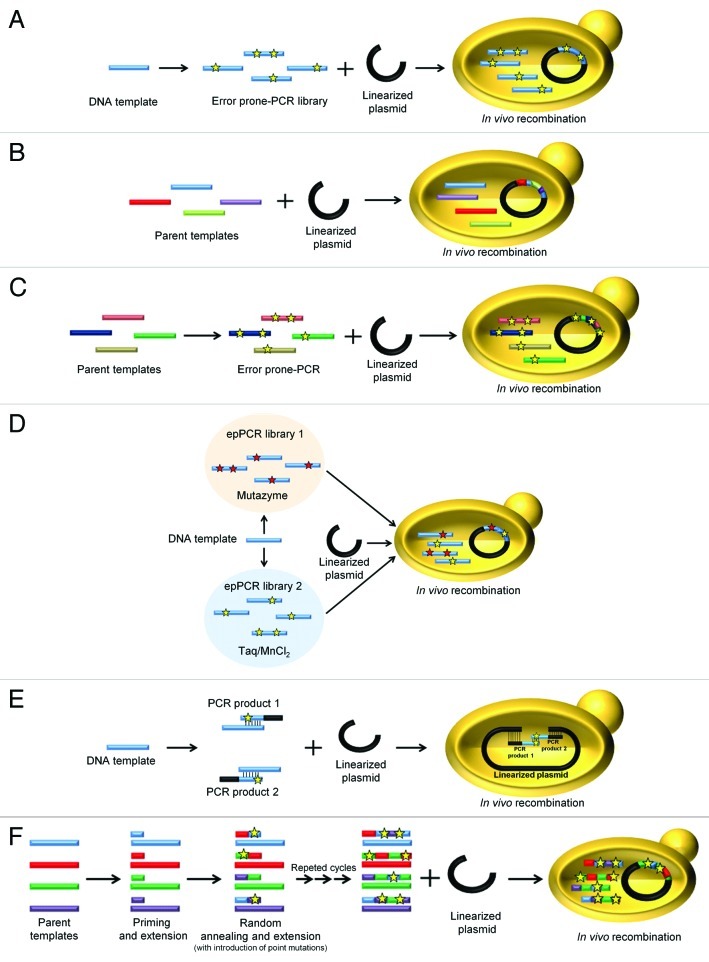
Recently, the full capacities of S. cerevisiae were challenged by a methodology known as DNA assembler, which was used to successfully assemble an entire biochemical pathway in a single step via in vivo homologous recombination.32 In directed evolution, we use the DNA recombination machinery of S. cerevisiae to in vivo clone and recombine mutant libraries with the linearized vector, avoiding tedious ligation steps (Fig. 2A). To perform this type of experiment, it is necessary to engineer overlapping areas of approximately 40 bp of homology with the ends of the linearized vector, coupling the mutants generated to the corresponding screening assay. The number of crossover events among the inserts can also be enhanced (increasing the likelihood of recombining beneficial mutations between templates) by testing different overlapping regions with less homology to the linear vector, although the transformation efficiency may be compromised. In this context, in vivo DNA shuffling based on the S. cerevisiae recombination machinery is a powerful tool, speeding up the evolution process by shuffling parental genes with sequence homologies of ~70% at one point in the process where the whole autonomously replicating vector is repaired by the yeast’s in vivo gap repair mechanism (Fig. 2B). One of the first pioneering works of in vivo DNA shuffling was reported by Cherry and coworkers to engineer oxidative stability into the low-medium redox potential peroxidase from Coprinopsis cinerea (CiP).33 Although in vivo DNA shuffling relies on the proof-reading device of S. cerevisiae, we observed better improvements in each cycle of evolution when error-prone PCR (epPCR) products of different templates were recombined in vivo in order to introduce new mutations in conjunction with recombination (Fig. 2C). In addition to our own works,16,17,26,34,35 in vivo DNA shuffling has also been applied to other studies such as the engineering of chimeric enzymes from four different templates of Trametes C30 laccase with low and high redox potentials.36
Figure 2. Different methods used to generate diversity using the S. cerevisiae toolbox. (A) epPCR from a single template followed by in vivo recombination in S. cerevisiae. (B) In vivo DNA shuffling. Several parental genes are recombined and cloned with a linearized vector into S. cerevisiae in a single step. (C) epPCR in conjunction with in vivo DNA shuffling. (D) IvAM (In vivo Assembly of Mutant libraries with different mutational spectra). Two or more distinct mutant libraries are generated by epPCR using polymerases with different biases. S. cerevisiae is transformed with the mutant libraries together with the linearized plasmid. (E) IVOE for combinatorial saturation mutagenesis or site-directed mutagenesis. The gene is amplified in two independent PCR reactions using mutagenized/degenerate primers. By engineering specific overhangs, the PCR products are then cloned into S. cerevisiae together with the linearized plasmid in a single transformation. (F) Mutagenic StEP (Staggered Extension Process). Several parental genes are used as templates during mutagenic StEP, promoting the random introduction of mutations during the short cycles of annealing and extension. The resulting mutant/recombined library is further shuffled by S. cerevisiae, together with the linearized plasmid. Stars represent single mutations.
Given the inherent degeneracy of the genetic code and the fact that some errors in the genetic code cause silent mutations, diversity is more fault-tolerant to point mutations. Moreover, epPCR methods (Fig. 2A) tend to introduce transitions (A↔G/T↔C) rather than transversion (A↔T/G↔C), limiting the mutagenic spectrum and introducing the intrinsic bias of each specific polymerase.37 We sought to offset this tendency by designing new molecular tools based on the physiology of S. cerevisae (IvAM, IVOE). IvAM (In vivo Assembly of Mutant libraries with different mutational spectra) permits the combination of two or more mutant libraries created by different mutagenic approaches. The mutant libraries to be in vivo recombined can be developed by epPCR using polymerases with different biases. Despite the intrinsic bias derived from the codon usage of S. cerevisiae, this technique helps to enhance the mutational spectrum34 (Fig. 2D). IvAM has been applied to the directed evolution of the MtL in order to confer organic co-solvent tolerance.35 We identified two beneficial mutations in two consecutive codons during the same cycle of evolution (G614D and E615K), probably induced as a consequence of the IvAM technique. Similarly, we employed IvAM to evolve VP toward thermal stability, raising the T50 by 8°C in the final VP mutant.17 IVOE (In Vivo Overlap Extension) is a simple protocol applied to semi-rational or rational approaches such as combinatorial saturation mutagenesis (CSM), site-directed mutagenesis, site-directed recombination, insertions and deletions. IVOE is based on conventional SOE (Splicing by Overlap Extension),38 although several of the in vitro steps in SOE are missing. Our method involves the engineering of mutagenic primers that generate PCR products with homologous regions, both with one another and with the linearized plasmid. These PCR fragments are transformed into the yeast together with the linearized plasmid, promoting in vivo DNA recombination and generating a circular plasmid with the desired mutation/s (Fig. 2E).39,40 We previously improved the properties of hot-spot residues in MtL by combining CSM with IVOE.39 Moreover, in an attempt to enhance the activity and stability of PM1L using IVOE, we performed site-directed mutagenesis studies to recover beneficial mutations discarded during the evolutionary pathway. The final PM1L mutant was readily secreted by S. cerevisiae (~8 mg/L) in an active and stable form with regards temperature, pH range and organic co-solvents.16 We also used IVOE to demonstrate how VP secretion was affected by linking an extra four amino acid N-terminal tail to the mature protein (EAEA). The truncated VP variant was engineered by deletion mutagenesis through IVOE, confirming that the STE13 protease failed to process the extra N-terminal extension in the Golgi compartment of S. cerevisiae.17 Another interesting application of IVOE developed in our laboratory involves the fusion of different enzymes with the α-factor prepro-leader of S. cerevisiae.16,17,26 We engineered primers with homologous overhangs in order to generate fragments that were spliced in vivo to produce proteins fused to the α-factor prepro-leader, replacing the native signal peptide.
It is feasible to combine in vitro and in vivo methods for DNA-recombination to perform directed enzyme evolution. Indeed, in vitro DNA recombination and in vivo DNA shuffling were combined to increase the mutagenic spectrum of a given library (a method known as CLERY; Combinatorial Libraries Enhanced by Recombination in Yeast).41 Similarly, we modified conventional StEP (Staggered Extension Process)42 to enhance the likelihood of introduction of random mutations in the process (Mutagenic StEP, Figure 2F), and we combined this strategy with in vivo DNA shuffling in the same round of evolution to create a temperature, peroxide and alkaline-pH tolerant VP that was secreted readily by yeast (~22 mg/L).17
In the past decade, S. cerevisiae has been used widely in the directed evolution of proteins. As a host, S. cerevisiae possesses all the necessary cellular machinery required to secrete active and functional eukaryotic proteins. As a biomolecular toolbox, S. cerevisiae permits new strategies to be designed that boost and direct the evolutionary process, complementing the traditional methods used to tailor enzymes ´ la carte. We hope that in the near future, S. cerevisiae will serve as a platform to support the directed evolution of artificial operons and metabolic pathways, thereby providing us with a powerful microbial cell factory for synthetic biology.
Acknowledgments
This paper is based on research funded by EU Projects (FP7–3D-Nanobiodevices: NMP4-SL-2009–229255; FP7-KBBE-2010–4-26537: Peroxicats; COST Action CM0701) and a National project (Evofacel, BIO2010–19697).
Glossary
Abbreviations:
- bp
base pair
- CiP
Coprinopsis cinerea peroxidase
- CLERY
combinatorial libraries enhanced by recombination in yeast
- CSM
combinatorial saturation mutagenesis
- epPCR
error prone-PCR
- ER
endoplasmatic reticulum
- FACS
fluorescence activated cell sorter
- HTPS
high-throughput screening
- IvAM
in vivo assembly of mutant libraries with different mutational spectra
- IVOE
in vivo overlap extension
- MtL
Myceliophthora thermophila laccase
- PcL
Pycnoporus cinnabarinus laccase
- SOE
splicing by overlap extension
- StEP
staggered extension process
- VP
versatile peroxidase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/19544
References
- 1.Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci U S A. 2009;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–76. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalby PA. Strategy and success for the directed evolution of enzymes. Curr Opin Struct Biol. 2011;21:473–80. doi: 10.1016/j.sbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Jäckel C, Hilvert D. Biocatalysts by evolution. Curr Opin Biotechnol. 2010;21:753–9. doi: 10.1016/j.copbio.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Shivange AV, Marienhagen J, Mundhada H, Schenk A, Schwaneberg U. Advances in generating functional diversity for directed protein evolution. Curr Opin Chem Biol. 2009;13:19–25. doi: 10.1016/j.cbpa.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12:151–8. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Wong TS, Roccatano D, Schwaneberg U. Steering directed protein evolution: strategies to manage combinatorial complexity of mutant libraries. Environ Microbiol. 2007;9:2645–59. doi: 10.1111/j.1462-2920.2007.01411.x. [DOI] [PubMed] [Google Scholar]
- 8.Lutz S. Beyond directed evolution--semi-rational protein engineering and design. Curr Opin Biotechnol. 2010;21:734–43. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv. 2011;2012 doi: 10.1016/j.biotechadv.2011.09.013. In press. [DOI] [PubMed] [Google Scholar]
- 10.Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115:113–28. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Khow O, Suntrarachun S. Strategies for production of active eukaryotic proteins in bacterial expression system. Asian Pacific Journal of Tropical Biomedicine 2012: 159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CC, Williams TG, Wong DW, Robertson GH. An episomal expression vector for screening mutant gene libraries in Pichia pastoris. Plasmid. 2005;54:80–5. doi: 10.1016/j.plasmid.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Pscheidt B, Avi M, Gaisberger R, Hartner FS, Schuster C, et al. Laboratory evolved biocatalysts for stereoselective syntheses of substituted benzaldehyde cyanohydrins. Chembiochem. 2008;9:58–61. doi: 10.1002/cbic.200700514. [DOI] [PubMed] [Google Scholar]
- 14.Arnold F, Georgious G. Directed enzyme evolution: screening and selection methods. Humana Press 2003, Totowa, New Jersey (USA). [Google Scholar]
- 15.Arnold F, Georgious G. Directed evolution: library creation. Methods and protocols. Humana Press 2003, Totowa, New Jersey (USA). [Google Scholar]
- 16.Mate´ D, Garci´a-Burgos C, Garci´a-Ruiz E, Ballesteros AO, Camarero S, Alcalde M. Laboratory evolution of high-redox potential laccases. Chem Biol. 2010;17:1030–41. doi: 10.1016/j.chembiol.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Ruiz E, Gonzalez-Perez D, Ruiz-Dueñas FJ, Marti´nez AT, Alcalde M. Directed evolution of a temperature-, peroxide- and alkaline pH-tolerant versatile peroxidase. Biochem J. 2012;441:487–98. doi: 10.1042/BJ20111199. [DOI] [PubMed] [Google Scholar]
- 18.Mate´ D, Garci´a-Ruiz E, Camarero S, Alcalde M. Directed evolution of fungal laccases. Curr Genomics. 2011;12:113–22. doi: 10.2174/138920211795564322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulter T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl Environ Microbiol. 2003;69:987–95. doi: 10.1128/AEM.69.2.987-995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zsebo KM, Lu HS, Fieschko JC, Goldstein L, Davis J, Duker K, et al. Protein secretion from Saccharomyces cerevisiae directed by the prepro-alpha-factor leader region. J Biol Chem. 1986;261:5858–65. [PubMed] [Google Scholar]
- 21.Shuster JR. Gene expression in yeast: protein secretion. Curr Opin Biotechnol. 1991;2:685–90. doi: 10.1016/0958-1669(91)90035-4. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu R, Yamashita T, Aikawa J, Horinouchi S, Beppu T. The prepro-peptide of Mucor rennin directs the secretion of human growth hormone by Saccharomyces cerevisiae. Appl Environ Microbiol. 1990;56:2125–32. doi: 10.1128/aem.56.7.2125-2132.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innis MA, Holland MJ, McCabe PC, Cole GE, Wittman VP, Tal R, et al. Expression, glycosylation, and secretion of an Aspergillus glucoamylase by Saccharomyces cerevisiae. Science. 1985;228:21–6. doi: 10.1126/science.228.4695.21. [DOI] [PubMed] [Google Scholar]
- 24.Rourke IJ, Johnsen AH, Din N, Petersen JGL, Rehfeld JF. Heterologous expression of human cholecystokinin in Saccharomyces cerevisiae. Evidence for a lysine-specific endopeptidase in the yeast secretory pathway. J Biol Chem. 1997;272:9720–7. doi: 10.1074/jbc.272.15.9720. [DOI] [PubMed] [Google Scholar]
- 25.Pe´rez-Boada M, Doyle WA, Ruiz-Dueñas FJ, Martinez MJ, Martinez AT, Smith AT. Expression of Pleurotus eryngii versatile peroxidase in Escherichia coli and optimisation of in vitro folding. Enzyme Microl Tech. 2002;30:518–24. doi: 10.1016/S0141-0229(02)00008-X. [DOI] [Google Scholar]
- 26.Camarero S, Cañas AI, Pardo I, Molina P, Marti´nez AT, Marti´nez MJ, et al. Engineering platforms for the directed evolution of laccase from Pycnoporus cinnabarinus. Appl Environ Microbiol. 2012;78:1370–84. doi: 10.1128/AEM.07530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanos MA, Scorer CA, Clare JJ. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–88. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 28.Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009;37:6984–90. doi: 10.1093/nar/gkp687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–2. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry JR, Lamsa MH, Schneider P, Vind J, Svendsen A, Jones A, et al. Directed evolution of a fungal peroxidase. Nat Biotechnol. 1999;17:379–84. doi: 10.1038/7939. [DOI] [PubMed] [Google Scholar]
- 34.Zumárraga M, Camarero S, Shleev S, Marti´nez-Arias A, Ballesteros A, Plou FJ, et al. Altering the laccase functionality by in vivo assembly of mutant libraries with different mutational spectra. Proteins. 2008;71:250–60. doi: 10.1002/prot.21699. [DOI] [PubMed] [Google Scholar]
- 35.Zumárraga M, Bulter T, Shleev S, Polaina J, Marti´nez-Arias A, Plou FJ, et al. In vitro evolution of a fungal laccase in high concentrations of organic cosolvents. Chem Biol. 2007;14:1052–64. doi: 10.1016/j.chembiol.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Cusano AM, Mekmouche Y, Meglecz E, Tron T. Plasticity of laccase generated by homeologous recombination in yeast. FEBS J. 2009;276:5471–80. doi: 10.1111/j.1742-4658.2009.07231.x. [DOI] [PubMed] [Google Scholar]
- 37.Wong TS, Zhurina D, Schwaneberg U. The diversity challenge in directed protein evolution. Comb Chem High Throughput Screen. 2006;9:271–88. doi: 10.2174/138620706776843192. [DOI] [PubMed] [Google Scholar]
- 38.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–8. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 39.Alcalde M, Zumárraga M, Polaina J, Ballesteros A, Plou FJ. Combinatorial saturation mutagenesis by in vivo overlap extension for the engineering of fungal laccases. Comb Chem High Throughput Screen. 2006;9:719–27. doi: 10.2174/138620706779026079. [DOI] [PubMed] [Google Scholar]
- 40.Alcalde M. Mutagenesis protocols in Saccharomyces cerevisiae by in vivo overlap extension. In Bramman, J (Ed.), In vitro Mutagenesis Protocols, Towota, New Jersey US: Humana Press 2010; 634:3-14. [DOI] [PubMed] [Google Scholar]
- 41.Abe´cassis V, Pompon D, Truan G. High efficiency family shuffling based on multi-step PCR and in vivo DNA recombination in yeast: statistical and functional analysis of a combinatorial library between human cytochrome P450 1A1 and 1A2. Nucleic Acids Res. 2000;28:E88. doi: 10.1093/nar/28.20.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16:258–61. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]