Abstract
Selected Saccharomyces cerevisiae strains are used for wine fermentation. Based on several criteria, winemakers often use a specific yeast to improve the flavor, mouth feel, decrease the alcohol content and desired phenolic content, just to name a few properties. Scientists at the AWRI previously illustrated the potential for increased flavor release from grape must via overexpression of the Escherichia coli Tryptophanase enzyme in wine yeast. To pursue a self-cloning approach for improving the aroma production, we recently characterized the S. cerevisiae cystathionine β-lyase STR3, and investigated its flavor releasing capabilities. Here, we continue with a phylogenetic investigation of STR3 homologs from non-Saccharomyces yeasts to map the potential for using natural variation to engineer new strains.
Keywords: aromatic thiols, cystathionine β-lyase, flavor, non-Saccharomyces, self-cloning, wine, yeast
Amino acids play a central role in flavor development during alcoholic fermentation. Not only are they precursors for flavor active compounds such as higher alcohols, esters and aromatic thiols; their biosynthetic and catabolic pathways in yeast also play a key role in producing and liberating aroma.
A primary example of this are the aromatic thiols 3-mercaptohexan-1-ol (3MH) and 4-mercapto-4-methylpentan-2-one (4MMP), which are released from non-volatile cysteine-S-conjugated precursors present in grape must during fermentation with wine yeast. 3MH and 4MMP form an important part of the varietal character of Vitis vinifera L. cv Sauvignon blanc wines with grape fruit and passion fruit flavors and are found in a range of wines. 4MMP release was recently linked to a particular allele of IRC7, which encodes for an enzyme with carbon-sulfur β-lyase activity.1 IRC7 was horizontally transferred from Yersinia pestis to Saccharomyces sensu stricto species,1 and a BLAST search against all non-redundant GenBank entries of the fungi taxonomy did not identify close homologs outside this group. Efforts to identify a single gene responsible for 3MH release, on the other hand, have not proven definitive1-3 and the trait is considered to be multigenic.
We recently purified the S. cerevisiae peroxisomal cystathionine β-lyase (CBL), Str3p, and observed high catalytic efficiency against its physiological substrate, L-cystathionine, but also observed a broad substrate specificity toward other sulfur containing amino acids. We then performed reactions with the cysteine-S-conjugated thiol precursors, Cys-3MH and Cys-4MMP (Fig. 1), and quantified the thiol enzymatic release with gas chromatography-mass spectroscopy.
Figure 1.
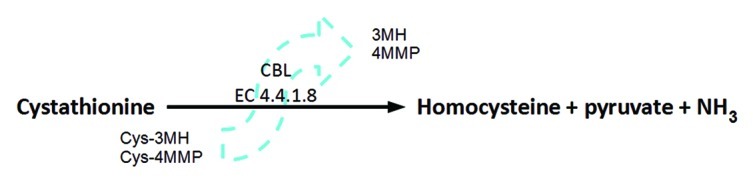
CBL enzymes (EC 4.4.1.8) cleave cystathionine in an α,β-elimination reaction to produce homocysteine and the by-products pyruvate and ammonia. The reaction is dependent of the cofactor pyridoxal-5′-phosphate. The S. cerevisiae CBL can also release the aromatic thiols 3MH and 4MMP from their respective cysteine-S-conjugated precursors present in grape must.
Str3p was able to release an amount of free 3MH and 4MMP corresponding to approximately 0.1% and 0.6%, respectively, of the specific activity against L-cystathionine. This represents a modest side activity, however, the sensory thresholds of 4MMP and 3MH are 3 and 60 ng/l, respectively,4,5 and the “best” commercial wine yeast can only release approximately 5% from the precursors.6 Therefore, even a modest increase in yeast carbon sulfur β-lyase activity could potentially alter the composition of volatile thiols in wine fermentation, and affect wine flavor.
We engineered the commercial wine yeast strain VIN13 (Anchor Yeast, South Africa) with an additional copy of the S. cerevisiae STR3 gene under the control of the PGK1 constitutive promoter, and used the wild type and engineered VIN13(STR3) to produce wines from a Sauvignon blanc grape must. The resulting wines showed an increase in 3MH concentration of 278 ng/l, and therefore illustrated the potential to engineer a self-cloned, 3MH-releasing strain using yeast CBLs.
Could we improve upon this in the future?
It is worth noting that the pH optimum of Str3p is 8.75, and at the S. cerevisiae cytoplasmic pH of 7.0, only 25% of CBL activity remained,7 whereas at the peroxisomal pH of 8.2, Str3p retains 80% of its maximum enzymatic activity.8 Str3p has a peroxisomal targeting signal type I (PTS1), and has been located in the peroxisome of S. cerevisiae cells grown with oleic acid as a carbon source.9,10 The other yeast enzymes involved in the sulfur amino acid biosynthetic pathway are not localized to the peroxisome but in the cytosol, therefore, the peroxisomal membrane must permit the flow of the intermediate metabolites between both compartments. Peroxisomes from S. cerevisiae contain channel (pore)-forming proteins which would allow transmembrane transfer of small solutes such as carboxylic and amino acids with a molecular mass up to 400 Da.11,12 This far exceeds the molecular sizes of either cystathionine, Cys-4MMP and Cys-3MH (the latter two substrates are 219 and 221 Da, respectively), and we can therefore infer that thiol release from the peroxisome could occur. It has been shown that the localization of peroxisomal aspartate amino transferase II, another pyridoxal-5′-phosphate dependent enzyme, fluctuates between cytosol (glucose) and peroxisomes (oleic acid) depending on the carbon source.13 The potential influence of compartmentalisation on thiol release has not been considered and represents an interesting future direction.
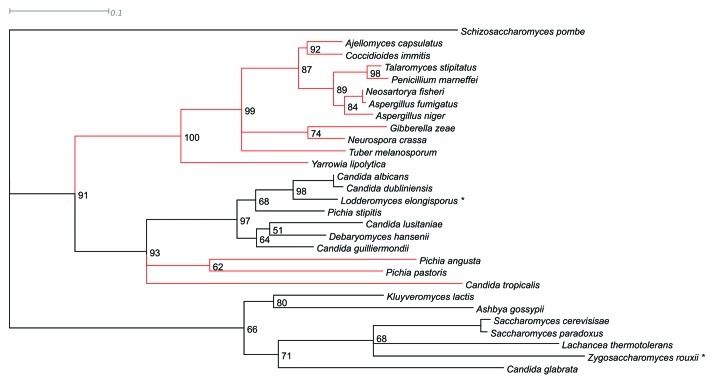
Commercial wine yeast has been selected on the basis of enhanced “fruity” characters produced during fermentation. Nonetheless, STR3 alleles from the wine yeast genomes that have been sequenced14-17 do not show significant differences in their coding regions. In order to examine the evolutionary relationship between Str3p homologs in Saccharomyces and non-Saccharomyces yeasts, and perhaps find other CBLs that may have greater capacity to release thiols, we constructed a maximum likelihood tree from fungal Str3p homologs (Fig. 2). Budding yeasts clustered together into two clades, and filamentous and other fungi, branched off with significant difference from the budding yeasts. The currently sequenced alleles of the order of Saccharomycetales fell into two clades, with S. cerevisiae, S. paradoxus, C. glabrata, A. gossypii, K. lactis, Z. rouxii and L. thermotolerans in one clade, P. pastoris, P. angusta, P. stipitis, C. guilliermondii, L. elongisporus and D. hansenii in the second clade. Members of the mitosporic Saccharomycetacae; C. albicans, C. tropicalis and C. dubliniensis that exhibit both budding and filamentous growth clustered with this second clade. Clearly the PTS1 signal was not a product of the whole genome duplication event, since both yeasts before and after have the peroxisomal signal.
Figure 2.
Phylogenetic tree constructed from alignment and maximum likelihood (quartet puzzling) analysis of 28 S. cerevisiae Str3p homologs. Each branch is indicated with confidence values (0–100%) from 100,000 puzzling steps19 and S. pombe used as an out group. Prediction of peroxisomal targeting signal type 1 (PTS1) was done with the PTS1 predictor (http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp), and sequences devoid of this signal are depicted in red. The sequences were tested for both type 1 and type 2 signals, and marked with asterisk if the prediction was indistinct.
None of the filamentous fungal Str3p homologs contained a PTS1 sequence, and had lower calculated pI values, suggesting a different evolutionary path. In contrast, the Str3p homologs of budding yeasts C. tropicalis, P. pastoris and P. angusta are not predicted for peroxisomal localization, and the branching of one clade of budding yeasts with the clade of filamentous fungi with 91% confidence, suggests that there is not a selective pressure for the PTS1 signal. The calculated pI value for the Saccharomycetacae was significantly higher (around 8, with exceptions), in the 6–7.5 range for the second clade of budding yeasts, and 6 or below for the filamentous fungi. Whether this is an evolutionary differentiation with regard to pH optimum of the STR3 gene product remains to be addressed.
Several non-Saccharomyces yeast have been shown to release significant concentrations of 3MH, even where their apparent role in grape must fermentation may be minor.18,20 Indeed, P. kluyveri was recently commercialised (Frootzen, Chr. Hansen, Denmark) and used in winemaking for the enhancement of “fruity” flavors, through a 2-fold elevation of 3MH concentration.18 Among the divergent Str3p homologs detected across non-Saccharomyces yeast species, will there be any that are more active toward Cys-3MH than S. cerevisiae Str3p—alleles that could be harnessed to engineer further enhancements in flavor release? In vitro characterization of non-Saccharomyces CBLs represents a path forward. Combined with enzyme crystallization and bioinformatic analysis of the active sites residues, we can improve our understanding of volatile thiol release and engineer optimal yeast strains for enhanced wine flavor.
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/19566
References
- 1.Roncoroni M, Santiago M, Hooks DO, Moroney S, Harsch MJ, Lee SA, et al. The yeast IRC7 gene encodes a β-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol. 2011;28:926–35. doi: 10.1016/j.fm.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Thibon C, Marullo P, Claisse O, Cullin C, Dubourdieu D, Tominaga T. Nitrogen catabolic repression controls the release of volatile thiols by Saccharomyces cerevisiae during wine fermentation. FEMS Yeast Res. 2008;8:1076–86. doi: 10.1111/j.1567-1364.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- 3.Harsch M. Identification of yeast genes involved in Sauvignon Blanc aroma development. Auckland, New Zealand: University of Auckland, 2009. [Google Scholar]
- 4.Darriet P, Lavigne V. Boidron, J.-ndl, & Dubourdieu, D. Identification of a Powerful Aromatic Component of Vitis vinifera L. var. Sauvignon Wines: 4-Mercapto-4-methylpentan-2-one. Flavour Fragrance J. 1995;10:385–92. doi: 10.1002/ffj.2730100610. [DOI] [Google Scholar]
- 5.Tominaga T, Furrer A, Henry R, Dubourdieu D. Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Fragrance J. 1998;13:159–62. doi: 10.1002/(SICI)1099-1026(199805/06)13:3<159::AID-FFJ709>3.0.CO;2-7. [DOI] [Google Scholar]
- 6.Murat ML, Masneuf I, Darriet P, Lavigne V, Tominaga T, Dubourdieu D. Effect of Saccharomyces cerevisiae yeast strains on the liberation of volatile thiols in Sauvignon blanc wine. Am J Enol Vitic. 2001;52:136–9. [Google Scholar]
- 7.Holt S, Cordente AG, Williams SJ, Capone DL, Jitjaroen W, Menz IR, et al. Engineering Saccharomyces cerevisiae to release 3-Mercaptohexan-1-ol during fermentation through overexpression of an S. cerevisiae Gene, STR3, for improvement of wine aroma. Appl Environ Microbiol. 2011;77:3626–32. doi: 10.1128/AEM.03009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Roermund CWT, de Jong M, IJlst L, van Marle J, Dansen TB, Wanders RJA, et al. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J Cell Sci. 2004;117:4231–7. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- 9.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, et al. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–16. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Schäfer H, Nau K, Sickmann A, Erdmann R, Meyer HE. Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis. 2001;22:2955–68. doi: 10.1002/1522-2683(200108)22:14<2955::AID-ELPS2955>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 11.Grunau S, Mindthoff S, Rottensteiner H, Sormunen RT, Hiltunen JK, Erdmann R, et al. Channel-forming activities of peroxisomal membrane proteins from the yeast Saccharomyces cerevisiae. FEBS J. 2009;276:1698–708. doi: 10.1111/j.1742-4658.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- 12.Antonenkov VD, Mindthoff S, Grunau S, Erdmann R, Hiltunen JK. An involvement of yeast peroxisomal channels in transmembrane transfer of glyoxylate cycle intermediates. Int J Biochem Cell Biol. 2009;41:2546–54. doi: 10.1016/j.biocel.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Verleur N, Elgersma Y, Van Roermund CWT, Tabak HF, Wanders RJA. Cytosolic aspartate aminotransferase encoded by the AAT2 gene is targeted to the peroxisomes in oleate-grown Saccharomyces cerevisiae. Eur J Biochem. 1997;247:972–80. doi: 10.1111/j.1432-1033.1997.00972.x. [DOI] [PubMed] [Google Scholar]
- 14.Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–95. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 16.Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci U S A. 2009;106:16333–8. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anfang N, Brajkovich M, Goddard MR. Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust J Grape Wine Res. 2009;15:1–8. doi: 10.1111/j.1755-0238.2008.00031.x. [DOI] [Google Scholar]
- 19.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–4. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 20.Zott K, Thibon C, Bely M, Lonvaud-Funel A, Dubourdieu D, Masneuf-Pomarede I. The grape must non-Saccharomyces microbial community: impact on volatile thiol release. Int J Food Microbiol. 2011;151:210–5. doi: 10.1016/j.ijfoodmicro.2011.08.026. [DOI] [PubMed] [Google Scholar]