Abstract
Unlike humans, some vertebrate animals are able to completely regenerate damaged appendages and other organs. For example, adult zebrafish will regenerate the complex structure of an amputated caudal fin to a degree that the original and replacement fins are indistinguishable. The blastema, a mass of cells that uniquely forms following appendage amputation in regenerating animals, is the major source of regenerated tissue. However, the cell lineage(s) that contribute to the blastema and their ultimate contribution(s) to the regenerated fin have not been definitively characterized. It has been suggested that cells near the amputation site dedifferentiate forming multipotent progenitors that populate the blastema and then give rise to multiple cell types of the regenerated fin. Other studies propose that blastema cells are non-uniform populations that remain restricted in their potential to contribute to different cell lineages. We tested these models by using inducible Cre-lox technology to generate adult zebrafish with distinct, isolated groups of genetically labeled cells within the caudal fin. We then tracked populations of several cell types over the entire course of fin regeneration in individual animals. We found no evidence for the existence of multipotent progenitors. Instead, multiple cell types, including epidermal cells, intra-ray fibroblasts, and osteoblasts, contribute to the newly regenerated tissue while remaining highly restricted with respect to their developmental identity. Our studies further demonstrate that the regenerating fin consists of many repeating blastema “units” dedicated to each fin ray. These blastema each have an organized structure of lineage restricted, dedifferentiated cells that cooperate to regenerate the caudal fin.
Keywords: regeneration, dedifferentiation, zebrafish, blastema, caudal fin
INTRODUCTION
Most mammalian organs, including those of humans, respond to severe tissue damage by the formation of scar tissue. In contrast, other vertebrates, such as salamanders and zebrafish, possess the incredible innate ability to fully regenerate damaged or lost body parts. This process, termed epimorphic regeneration, forms a near perfect and fully functional copy of the lost organ (Nye et al., 2003; Brockes and Kumar, 2008; Nacu and Tanaka, 2010). A common feature of epimorphic regeneration is the appearance of a population of mesenchymal cells at the wound site, termed the blastema. It is thought that the blastema is a source of progenitor-like cells that divide, differentiate, and re-organize into restored tissue. These cells are highly proliferative and express numerous developmental regulatory genes, such as growth factors, transcription factors and morphogens (Schebesta et al., 2006; Yoshinari et al., 2009).
Details of the formation of a regenerative blastema, the nature of its constituent cells, and their fate during regeneration are not well understood (Bryant et al., 2002; Slack, 2006). Early investigations into the cellular identity of the blastema in the salamander limb suggested that the major components were mesenchymal cells derived from mixed cell types of the limb stump (Hay and Fischman, 1961; Kintner and Brockes, 1984; Bryant et al., 2002; Nye et al., 2003). Dermal intra-ray fibroblasts (Muneoka et al., 1986) and Schwann cells (Kintner and Brockes, 1985) have also been proposed as sources of blastema mesenchymal cells. Studies aimed at definitively testing these possibilities have been confounded by the inability to label specific cell populations prior to amputation and then track their fates throughout regeneration.
One preferred model to explain the origin of the blastema is based on the concept of cell dedifferentiation (Hay and Fischman, 1961; Namenwirth, 1974; Slack, 2006). Here, adult cells lose key features of a fully differentiated state, such as being highly organized into tissues, expressing key functional proteins or subcellular structures that enable a defined physiological role, and frequently being cell cycle arrested. For example, non-proliferating, multinucleated skeletal muscle cells with robust myosin expression would dedifferentiate into dividing, mononucleated cells lacking myosin. In the dedifferentiation model of blastema formation, fully differentiated cells respond to amputation by transforming into “progenitor” cells that populate the blastema, proliferate, and then re-differentiate while becoming organized into a complex organ. Many otherwise silenced developmental regulatory genes are re-expressed in blastema cells, consistent with this model and leading the blastema to be considered as “dedifferentiated” (Nye et al., 2003; Schebesta et al., 2006; Brockes and Kumar, 2008; Nacu and Tanaka, 2010). A recent study suggests that several cell types of the blastema of the regenerating salamander limb maintain a restricted cell fate after dedifferentiating. These restricted cells, in conjunction with multipotent dermal cells and muscle satellite cells, participate in the regeneration of the lost tissue (Kragl et al., 2009). Transplantation studies in the regenerating tails of tadpoles also support this model (Gargioli and Slack, 2004).
Transdifferentiation, the direct conversion of one differentiated cell type into another, is an alternative proposed source of cell diversification during regeneration. The best example is lens regeneration in the newt. Following lentectomy, differentiated cells of the dorsal iris begin to proliferate, lose their pigment and then re-differentiate into transparent lens cells (Tsonis et al., 1995; Slack, 2007). Another report has proposed ectoderm to mesoderm switching during tail regeneration in the salamander whereby glia cells contribute to regenerated muscle and cartilage (Echeverri and Tanaka, 2002).
The zebrafish exhibits an outstanding ability to regenerate fins, heart ventricle and spinal cord (Akimenko et al., 2003; Poss et al., 2003). The caudal fin is a favored model of regeneration since it is easy to amputate, is not required for viability, regenerates all parts of its anatomy, and completely regenerates in a short time frame (2 weeks). The zebrafish caudal fin is composed of bony ray segments, known as lepidotrichia that are joined together by ligaments (Mari-Beffa et al., 2007; Mari-Beffa and Murciano, 2010). These rays form by direct mineralization coordinated by osteoblasts, specialized cells that deposit bone matrix (Karsenty et al., 2009). Osteoblasts develop through a tightly regulated, hierarchical process defined by the expression of runx2 and sp7 (osterix; osx), in early and intermediate stages, respectively (Brown et al., 2009; Karsenty et al., 2009). An individual ray is composed of two separate hemirays, each containing blood vessels, nerves, intra-ray fibroblasts, and osteoblasts, all of which are surrounded by an epidermis. This epidermis is composed of three layers of cells: superficial, intermediate, and basal. During the course of life, superficial epidermal cells appear to be replaced by intermediate layer cells, which are capable of robust proliferation (Le Guellec et al., 2004). In contrast, basal layer cells function to attach the epidermis to the basement membrane (Le Guellec et al., 2004). All of the fin cell types are restored in a pattern identical to that of the original fin following regeneration.
When amputated, the caudal fin responds by rapidly sealing the wound with migrating epidermal cells, forming what is known as a wound epidermis or wound epithelia. Approximately 1 day post amputation (dpa), a blastema forms beneath the wound epidermis. By 2 dpa, outgrowth is seen in cells of the blastema and it is thought that proximal blastema cells differentiate to form lost tissue. This process of outgrowth and re-differentiation continues until the tissue is completely restored in approximately 2 weeks (Akimenko et al., 2003; Poss et al., 2003). Much remains to be learned on what cell types contribute to the blastema and whether they are equivalent, multipotent cells or are a mixed population of cells restricted within a given cell lineage. Insight into blastema composition and function may ultimately allow experimental generation of a blastema in a mammalian model of limb regeneration.
Recently, several studies have addressed the nature of blastema cell identity and cell lineage relationships in the regenerating zebrafish caudal fin (Knopf et al., 2011; Sousa et al., 2011; Tu and Johnson, 2011). Knopf and colleagues observed osteoblast dedifferentiation and lineage restriction during regeneration in the fin. They demonstrated that, after fin amputation, osteoblasts in the fin down-regulate genes associated with a mature osteoblast phenotype while concomitantly expressing genes associated with immature osteoblasts. By tracking single cells during early stages of fin regeneration, they suggest that osteoblasts in the fin dedifferentiate to lineage-restricted progenitors and do not cross lineage boundaries during regeneration (Knopf et al., 2011). Tu and Johnson used a transposable element-based technique to randomly label cells during development and demonstrated that the caudal fin is composed of nine distinct cell lineages of cells. After fin amputation, none of these lineages gives rise to any other cell lineages (Tu and Johnson, 2011). Sousa and colleagues also examined osteoblasts during fin regeneration, demonstrating that osteoblasts become proliferative and undergo changes in marker gene expression consistent with the dedifferentiation model. Further, they showed genetically labeled osteoblasts only give rise to additional osteoblasts after amputation, suggesting osteoblasts remain lineage restricted (Sousa et al., 2011). As a whole, these reports did not uncover multipotent progenitors, consistent with the idea that cells in the regenerating fin are largely unipotent. However, the extent to which the progeny of such dedifferentiated cells actually contribute to fully regenerated fin remains to be determined.
We used Cre-lox technology in developing zebrafish to permanently label cells whose progeny would become various tissues of the adult caudal fin. This allowed us to follow specific populations of cells over the entire course of fin regeneration in individual animals. By monitoring several cell lineages, we did not observe any cells exhibiting multipotency. Rather, we found that cell fates were highly restricted with respect to both spatial and developmental identity during the entire course of regeneration. Our results demonstrate that the fully regenerated fin is indeed the product of lineage-restricted progenitors derived from preexisting differentiated cells that then contribute to the entire length of the regenerated fin.
MATERIALS AND METHODS
Zebrafish lines and cell labeling
Zebrafish were maintained according to University of Oregon institutional guidelines. To generate transgenic fish expressing a Cre-ERT2 fusion protein under the control of the dusp6 promoter, we first cloned a dusp6 promoter fragment sufficient to drive FGF dependent expression (Molina et al., 2007) into a 5’ element vector compatible with the Tol2Kit system (Kwan et al., 2007). Next, a Cre-ERT2 polyA (Metzger et al., 2005) cassette was cloned into a middle element vector. Finally, we used Gateway Clonase LRII enzyme (Invitrogen, Carlsbad, CA) to recombine the 5E dusp6 promoter, ME Cre-ERT2, 3E polyA (Kwan et al., 2007) and a modified Tol2 destination vector containing a cmlc2:ecfp cassette as a marker for transgenesis. The resulting construct (GW dusp6:Cre-ERT2 polyA) was co-injected with capped RNA for the Tc transposase into one cell stage AB embryos (Kawakami, 2007), which were then reared to adulthood. Founders, known heretofore as Tg(dusp6:Cre-ERT2), were selected by ECFP expression in cardiac muscle and crossed to the reporter line Tg(EAB:EGFP-FlEx-mCherry) (Boniface et al., 2009). Embryos from this cross were treated with 1 µM tamoxifen (Sigma-Aldrich, St. Louis, MO) starting at 30–50% epiboly until 48 hours post fertilization (hpf). Animals containing mCherry+ cells were selected and grown to adulthood. These F1 animals were then backcrossed to AB fish and the progeny were treated with tamoxifen and selected for robust Cre activity. For analyses of labeled cell populations, we employed F2 and F3 Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animals first selected animals with caudal fins containing one to three labeled populations that were isolated and had relatively well defined boundaries, such as a mCherry+ fin ray. Fish that had robust labeling throughout the caudal fin were not analyzed further due to the likelihood that labeled regions of the fin contained multiple cell types. Tg(sp7:EGFP) animals have been described previously (DeLaurier et al., 2010).
Regeneration time course studies
Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) heterozygous animals with representative populations of mCherry+ cells were selected and followed for 14 days after fin amputation. At t=0, animals were anesthetized in Tricaine, and caudal fins were imaged on a Leica M165FC stereomicroscope with appropriate epifluorescent illumination. Caudal fins were then amputated with a razor and the animals returned to circulating fish water. At each indicated time point, animals were anesthetized with Tricaine and re-photographed as described above.
Immunostaining
For fluorescent immunostaining of fins, Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) or Tg(sp7:EGFP) fins were amputated and then re-amputated 48 hours later 2–4 mm proximal to the original site of amputation. Amputated fins were fixed overnight in 4% PFA/PBS, equilibrated in PBS, cryo-preserved in 30% sucrose/PBS and frozen in agarose. Frozen sections (16 µm) were prepared and stored at −20°C until use. For immunohistochemistry, frozen sections were hydrated in PBS + 0.1% Tween-20 (PBST). Sections were blocked in 10% non-fat dry milk in PBST + 0.1% Triton X-100 for 1–4 hours at room temperature. Primary antibodies were diluted in 10% milk in PBST and incubated overnight, followed by 3 × 5’ washes in PBST. For anti-dsRed and anti-PCNA antibody staining, slides were subjected to antigen retrieval for 10’ in a pressure cooker using Dako antigen retrieval buffer (Dako North America, Carpinteria, CA), and then processed as above. The anti-dsRed antibody specifically detected cells in the fin expressing mCherry (Fig. S1A–S1D). Alexa-conjugated secondary antibodies (Invitrogen) were used at 1:1000 – 1:5000 diluted in 10% non-fat dry milk in PBST and applied to sections for 1 hour at room temperature, followed by 3 × 5’ washes. Nuclei were stained with Hoechst (Invitrogen) in PBST for 10’ at room temperature followed by 2 × 5’ washes. Slides were mounted using Vectashield (Vector Labs) and immediately visualized with an Olympus confocal microscope. For confocal imaging, sections were typically analyzed at 20× magnification with 3× digital zoom. Optical sections, typically 4 µm thick, were collected and processed using ImageJ software (NIH) with maximum intensity projections generated from z-stacks. Antibodies were sourced and diluted as follows: Anti-tenascin C (United States Biologicals, Salem, MA) 1:400, anti-dsRed (Clontech, Mountain View, CA) 1:500, zns-5 (Zebrafish International Resource Center (ZIRC), Eugene, OR) 1:200, zn-3 (ZIRC) 1:200, anti-PCNA (Sigma-Aldrich) 1:5000, anti-p63 clone 4A4 (Santa Cruz Biotechnology, Santa Cruz, CA) 1:500.
Analysis of Cell Proliferation
To quantify cell proliferation in regenerating and non-regenerating tissue in fins, we performed immunohistochemistry on frozen sections of 2 dpa fins from four animals with anti-PCNA and anti-tenascin C antibodies, as described above. We used ImageJ software to count the number of PCNA+ and total nuclei in three regions of the imaged fin sections. At least three sections were scored and the average percent PCNA+ nuclei in the three regions determined for each animal.
RESULTS
A Cre/lox approach labels cells in the zebrafish caudal fin
We developed a method that would sporadically and permanently label varied cell populations in the adult caudal fin. We hypothesized that limited genetic labeling of fin tissue progenitors as they were specified during embryogenesis would produce adult fins containing different sub-populations of labeled cells. The dusp6 gene encodes a protein phosphatase that is expressed in developing fins (Kawakami et al., 2003; Tsang et al., 2004) and both the distal blastema and wound epidermis during fin regeneration (Lee et al., 2005). dusp6 expression in these settings is regulated primarily by fibroblast growth factor (FGF) signaling (Kawakami et al., 2003; Molina et al., 2007; Molina et al., 2009), which itself is essential for fin development and regeneration (Kawakami et al., 2003; Lee et al., 2005; Whitehead et al., 2005; Lee et al., 2009). This led us to speculate that the dusp6 promoter could be used in combination with Cre recombinase to selectively and irreversibly label individual cells receiving FGF signals during fin development, allowing their descendants to be identifiable in adult fins. To accomplish this, we generated a transgenic line, Tg(dusp6:Cre-ERT2), which expresses Cre recombinase fused to the estrogen receptor (Cre-ERT2) (Metzger et al., 2005) under the control of the FGF-responsive regulatory element from the dusp6 gene (Molina et al., 2007). Recombination activity of the Cre-ERT2 fusion protein requires tamoxifen (Metzger et al., 2005). Therefore the Tg(dusp6:Cre-ERT2) transgenic line provides a means to track cells in vivo during physiological process that are mediated by FGF.
We tested the Tg(dusp6:Cre-ERT2) line by crossing it to the Cre-sensitive reporter line, Tg(EAB:EGFP-FlEx-mCherry) (Boniface et al., 2009). Tg(EAB:EGFP-FlEx-mCherry) expresses EGFP robustly and ubiquitously (Boniface et al., 2009). Such EGFP expression is permanently converted to mCherry expression upon Cre-driven genetic recombination (Fig. 1A). We treated the embryos from a Tg(dusp6:Cre-ERT2) × Tg(EAB:EGFP-FlEx-mCherry) cross with a low dose of tamoxifen for 48 hours beginning at 30% epiboly. These tamoxifen treated animals exhibited expression of mCherry in several tissues, including the pectoral fin (Fig. 1B, 1C), which is consistent with reports using in situ hybridization to visualize dusp6 transcripts (Kawakami et al., 2003; Molina et al., 2007; Lee et al., 2009).
Figure 1.
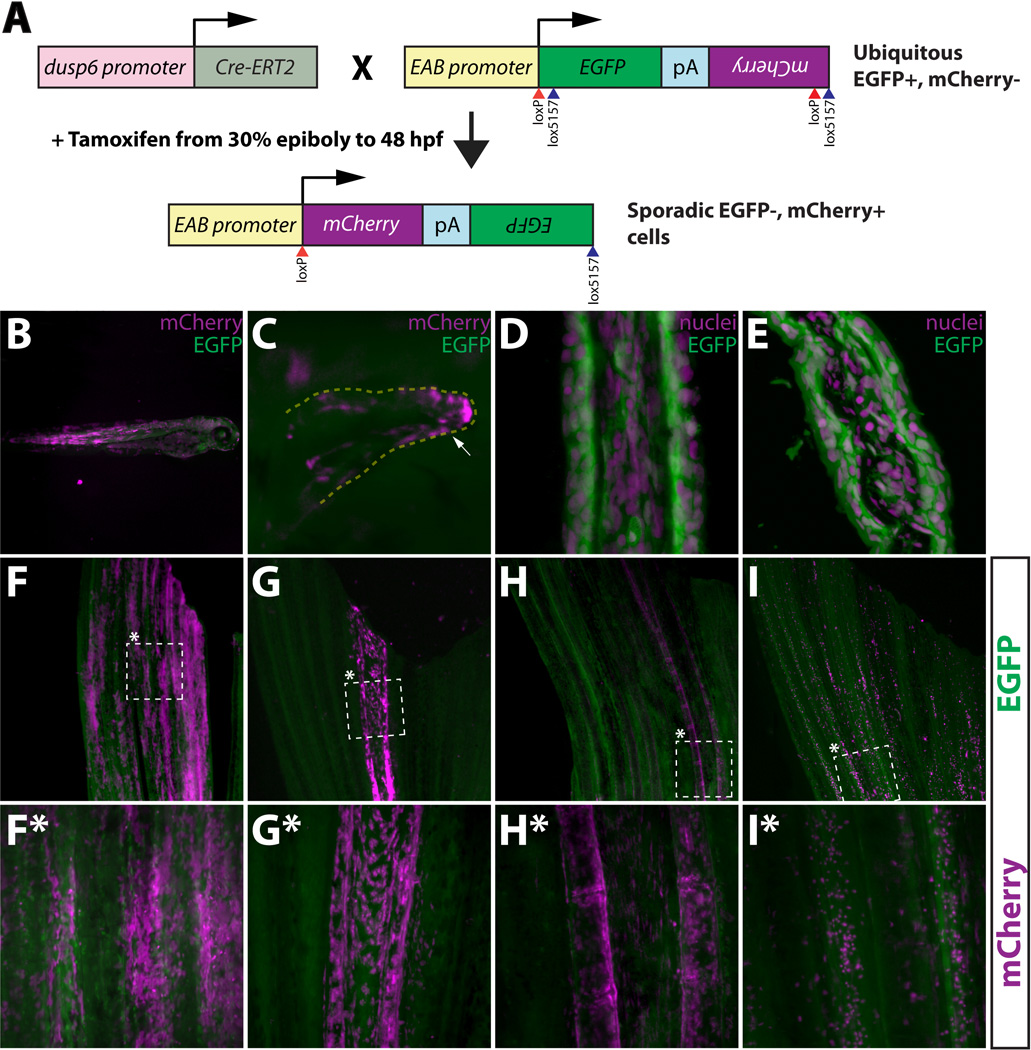
A tamoxifen-inducible Cre/lox method for mosaic cell labeling and lineage tracing in the adult zebrafish caudal fin. (A) Embryos carrying both dusp6:Cre-ERT2 and EAB:EGFP-FlEx-mCherry transgenes are briefly exposed to tamoxifen to sporadically induce rare genetic recombination events that permanently switch those cells and their descendants to mCherry from EGFP expression. (B, C) Whole mount epifluorescent images of 3 day old Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fish that were treated with tamoxifen (1 µM) at 30% epiboly for 48 hours. Mosaic mCherry+ cells (magenta) are observed in various tissues (B), including pectoral fin mesenchyme (C). The white arrow highlights mCherry+ cells. (D, E) The EAB:EGFP-FlEx-mCherry transgene is expressed in various cell lineages that make up the adult caudal fin. Longitudinal (C) and transverse (D) sections of adult caudal fins of Tg(EAB:EGFP-FlEx-mCherry) animals. EGFP expressing cells (green) were are stained with Hoechst to visualize nuclei (magenta). (F–I, F*–I*) Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) adult animals, treated as above, exhibit spatially restricted mCherry+ mosaics in four distinct classes. The dashed box marked with an asterisk represents the region shown at higher magnification in the panels directly below (F*–I*). (F and F*) Class 1 epidermal mosaics. (G and G*) Class 2 fibroblast mosaics. (H and H*) Class 3 osteoblast mosaics. (I and I*) Class 4 putative macrophage mosaics. All images show overlaid EGFP (green) and mCherry (magenta) expression. The top and bottom of each panel corresponds to the distal and proximal regions of the fin, respectively.
We examined the Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animals 1–2 months later, without any additional tamoxifen treatment, and found that many of them exhibited mosaic patterns of mCherry+ cells in an otherwise field of EGFP+ cells (Fig. 1F–1I*). Since some fins contained mosaic regions spanning the entire length of the fin and in single fin rays, we reasoned that such cells likely were derived from the same lineage. With the ultimate goal of using these animals for fin regeneration studies, we sought to characterize and identify the labeled cell populations. We first selected fish with caudal fins containing relatively isolated mCherry+ cell populations with well-defined boundaries of EGFP+ cells. We chose animals that displayed low degrees of mCherry+ mosaicism to simplify data interpretation and to minimize confusion arising from having multiple labeled cell types in overlapping regions of the fin. We failed to observe any mCherry expression without adding tamoxifen, indicating that any mCherry+ cells in adults were derived from cells labeled by tamoxifen-dependent genetic recombination events occurring months earlier during embryonic development.
We found that the populations of mCherry+ cells in these mosaic fins fell into four distinguishable, distinct classes (Fig 1F–1I*) in spite of the fact that the reporter is capable of expressing EGFP in most, if not all, cells in the adult caudal fin (Fig. 1D–1E). Class 1 cell populations were observed in 53 out of the 145 mosaic animals we examined (37%) and were characterized by broad patches of mCherry+ cells that appeared to be confined to the fin surface and exhibited pronounced cuboidal morphology (Fig. 1F and 1F*). Based on these observations, we speculated that Class 1 mosaics were comprised of labeled epidermal cells. To test this, we sectioned and stained the same fin photographed in Fig. 1F with anti-p63 antibodies. p63 is an epithelial marker that is specifically expressed in the zebrafish epidermis (Bakkers et al., 2002). We observed that the mCherry+ cells in this representative Class 1 mosaic were both p63+ and localized to all layers of the fin epidermis (Fig. S2A–2D). We concluded that Class 1 mosaics are comprised of labeled epidermal cells and are heretofore referred to as such. Also localized to the epidermis, Class 4 mosaics (Fig. 1I and I*) were found at the lowest frequency (13 out of 145; 9%) and were readily recognized by a speckled appearance under low magnification. Like Class 1 cells, Class 4 mCherry+ cells were sparsely distributed in epidermal layers, exhibited a large nucleus:cytoplasmic ratio compared to Class 1 cells (compare Fig. S2A–2D to Fig. S6), and did not express the epithelial marker p63 (Fig. S6). Based on these observations, we concluded that Class 4 mosaics likely represent labeled cells of a distinct lineage from those in Class 1 mosaics. Consistent with these observations, we noted a substantial superficial resemblance between mCherry+ Class 4 cells and tissue macrophages (Yoshinari et al., 2009; Tu and Johnson, 2011). We tentatively refer to the mCherry+ cells in Class 4 mosaic fins as “putative macrophages”.
Class 2 (Fig. 1G and 1G*) and Class 3 (Fig. 1H and 1H*) mosaics were confined largely to the bony fin ray lepidotrichia, and were found at roughly the same frequency: 26% and 29%, respectively. One substantial difference between Class 2 and Class 3 mosaics was that the latter had a more punctate pattern of mCherry+ cells that were concentrated at lepidotrichia joints. Since intra-ray fibroblasts and osteoblasts reside within and adjacent to lepidotrichia, respectively (Akimenko et al., 2003; Poss et al., 2003; Mari-Beffa and Murciano, 2010), we sectioned the same fins photographed in Figure 1G–H* and stained them with antibodies against osteoblast specific proteins. We observed that the Class 3 cells shown in Figure 1H and H* were positive for the osteoblast marker zns-5 (Johnson and Weston, 1995; Wills et al., 2008 and Fig. S1E–S1H) and were closely associated with the fin lepidotrichia (Fig. S2M–S2P). Unlike Class 3 cells, Class 2 cells exhibited a spindle-like morphology, were found between the lepidotrichia, and were negative for the osteoblast marker zns-5 (Fig. S2I–S2L). Class 3 cells were, however, found in close contact with a tenascin-C rich extracellular matrix (Fig. S2E–S2H). Based on our results, we conclude that Class 2 mosaics comprise mCherry+ intra-ray fibroblasts and Class 3 mosaics represent labeled osteoblasts.
In addition to labeled osteoblasts, we reproducibly detected a minor adjacent mCherry+ cell population in Class 3 mosaics. These cells were observed in the tissue between adjacent lepidotrichia and therefore were not intra-ray fibroblasts (Fig. 1H, H* and 6A*–6D*, yellow arrows). Transverse sections revealed that these cells resided in a layer continuous with labeled osteoblasts but were themselves negative for expression of osteoblast markers (Fig. S3). Such cells were not identified in a previous analysis examining cell types in the fin and may be attributed to differences in methodology used to label cells or with the time of labeling itself (Tu and Johnson, 2011). The presence of these two cell types adjacent to one another could be attributed to a “mixed mosaic”, or that they are derived from a common progenitor labeled during development. The former explanation appears unlikely since we reproducibly observed both cell types in all Class 3 mosaics even if the combined mCherry+ population represented only a small fraction of the fin.
Figure 6.
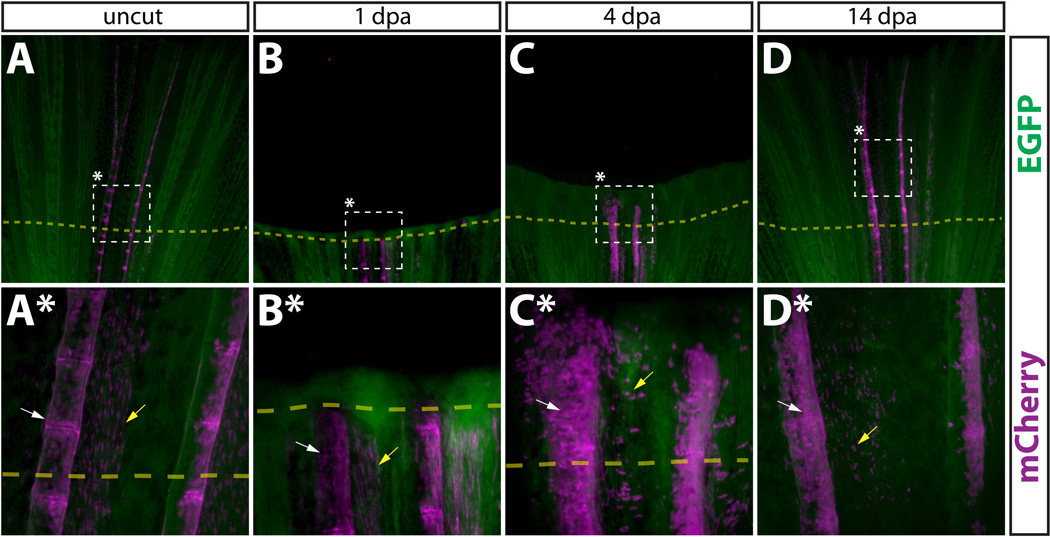
Newly regenerated bone is formed by pre-existing osteoblasts. (A–D, A*–D*) Whole mount images of a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 3 mosaic caudal fin containing mCherry labeled osteoblasts before amputation (A, A*), 1 dpa (B, B*), 4 dpa (C, C*), and 14 dpa (D, D*). The dashed box denotes the region magnified in the panel directly below. All cells are EGFP+ (green), except mCherry+ genetically recombined cells and their descendants (magenta). White arrows mark osteoblasts and yellow arrows denote cells co-labeled with mCherry+ osteoblasts.
Epidermal cells rapidly become motile and contribute only to new epidermis during fin regeneration
We collected a number of zebrafish representing each of the four mosaic classes. We then examined the behavior of each mCherry+ labeled cell type by amputating fins and periodically imaging them until the fins were fully regenerated. We hypothesized that if dedifferentiation were the primary source of replacement cells, the regenerated fin would contain mCherry+ cells in roughly the same pattern as before amputation. Further, if cells underwent only limited dedifferentiation to a lineage restricted progenitor, the same cell types would be derived from a given labeled pre-existing parental cell. In contrast, if dedifferentiation gave rise to multi- or pluripotent progenitor cells, mCherry+ cells would appear in a variety of cell types that were unlabeled before amputation. If few or no mCherry+ cells were found in the regenerated fin, we would conclude the labeled cell type is not a major contributor to replacement tissue.
We amputated Class 1 fins containing mCherry+ epidermal cells and tracked the fate of these cells and their progeny over a complete time course of regeneration by fluorescent microscopy. Even 1 day post amputation (dpa), mCherry+ cells appeared distal to the amputation site (Fig. 2A–2B*). By 4 dpa, mCherry+ cells were robustly present in the newly regenerated tissue (Fig. 2C, 2C*) and at 14 dpa, mCherry+ epidermis extended from the amputation site to the distal tip of the newly regenerated caudal fin (Fig. 2D, 2D*). Thus, pre-existing epidermal cells in the fin are a significant source of replacement epidermal cells in the regenerated fin. We did not detect any gross morphological differences between mCherry+ epidermal cells before or after regeneration. However, we noticed that labeled epidermal cells typically were widely distributed along the dorsal-ventral axis at the tip of the fin (Fig. 2A, 2B and 2A*, 2B*). By 4 dpa, this lateral population of cells was still evident (Fig. 2C, 2C*), but by 14 dpa it was no longer detected (Fig. 2D, 2D*).
Figure 2.
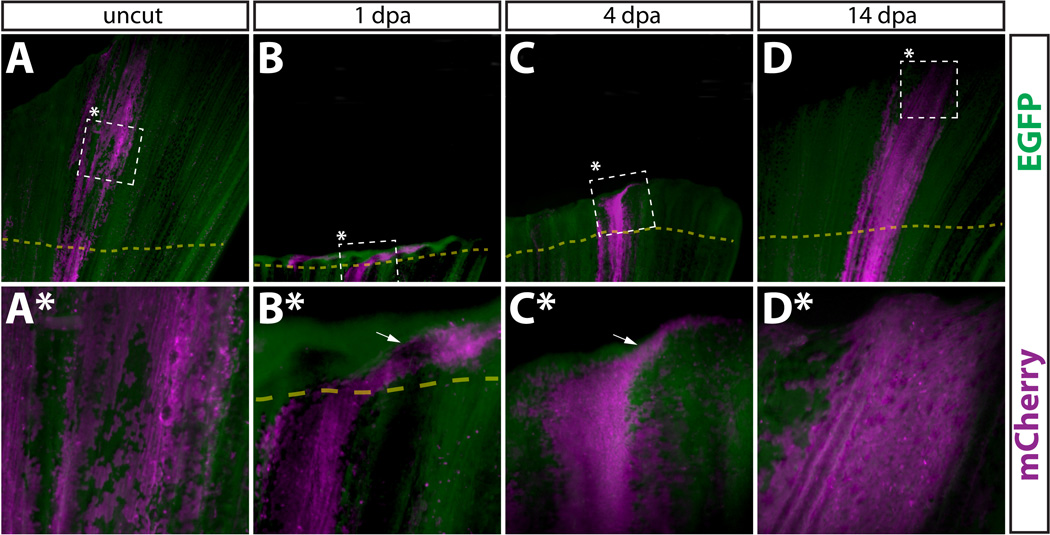
Newly regenerated epidermis is derived from pre-existing epidermal cells. (A–D, A*–D*) Whole mount epifluorescent images from the caudal fin from a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal containing Class 1 labeled epidermal cells before amputation (A, A*), 1 dpa (B, B*), 4 dpa (C, C*), and 14 dpa (D, D*). Dashed boxes marked with an asterisk represent the region shown at higher magnification in the panel directly below. (A–D) and (A*–D*) are images acquired at 25× and 120× magnification, respectively. EGFP+ cells are in green and mCherry+ cells are pseudocolored magenta. The top and bottom of each panel corresponds to the distal and proximal regions of the fin, respectively. The dashed yellow line shows the approximate amputation site and the white arrows point to epidermal cells found laterally to the starting population.
We also immunostained sectioned fins containing labeled epidermis at 2 dpa to determine whether the blastema contained any cells derived from the epidermis, including differentiated (p63+) or dedifferentiated (p63−) mCherry+ cells (Fig 3A–3D). No mCherry+ cells were found in the blastema, suggesting that the epidermal cells observed here do not contribute to this structure (Fig 3A–3D). Additionally, all mCherry+ epidermal cells remained positive for the marker p63, suggesting they retained an epithelial fate. Similarly, we found no evidence to suggest that epidermal cells gave rise to cells capable of generating replacement osteoblasts since mCherry+ cells in epidermal mosaic populations were never found localized in the blastema mesenchyme, nor did we observe any mCherry+ epidermal cell expressing osteoblast markers (Fig. S4A–S4D).
Figure 3.
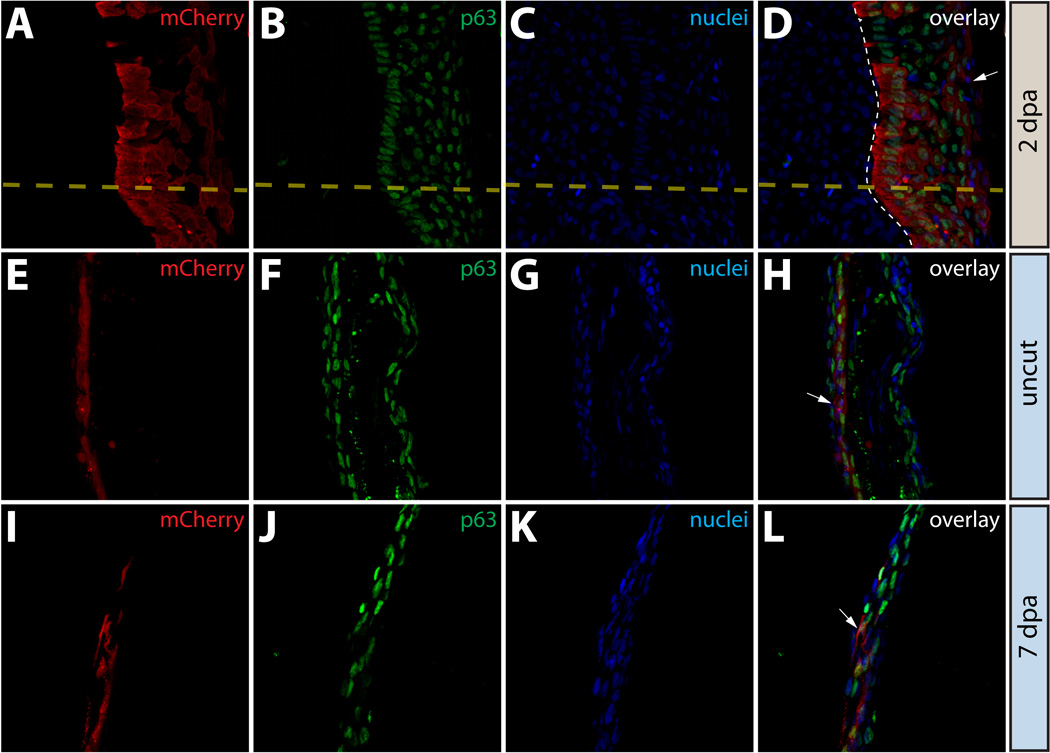
Epidermal cells do not contribute to the blastema or change fate during regeneration. (A–L) Longitudinal sections of the caudal fin of Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 1 mosaic animals demonstrating mCherry expression (red, A, E, I) and stained with anti-p63 antibodies to mark epidermal cells (green, B, F, J) and with Hoechst to mark nuclei (blue, C, G, K). The three-color overlays are shown in (D, H, L). (A–D) mCherry+ epidermal cells in Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 1 mosaic animals remain positive for the epidermal maker p63 at 2 dpa. The yellow dashed line marks the approximate amputation site and the white dashed line marks the boundary between the epidermis and the blastema. (E–L) Stained fin sections from the same Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 1 mosaic animal prior to amputation (E–H) and 7 dpa (I–L). White arrows indicate mCherry+/p63+ epidermis.
We expanded on these observations by staining fin tissue from the same animal with mCherry+ epidermis before (Fig. 3E–3H) and 7 dpa (Fig. 3I–3L), at which point regeneration was largely complete. All mCherry+ cells expressed p63 both before and after amputation, indicating that epidermal cells did not undergo any net change in cell fate. These results were consistent with our observations on whole fins in live animals and confirmed that the cells in the fin epidermis do not change fate after fin amputation or contribute to other lineages during regeneration. Collectively, these observations supported the idea that epidermal cells in the caudal fin acquire a highly motile state during regeneration (Santos-Ruiz et al., 2002; Lee et al., 2009) but maintain epidermal cell characteristics.
After amputation of fins with labeled Class 4 cells, we observed that these cells re-established their diffuse localization throughout in the epidermis by 2–4 dpa (Fig. S5). By 14 dpa, the distribution of the putative macrophages was indistinguishable from their pattern in an uncut fin (Fig. S5). We did not detect any plasticity of these cells at any point during regeneration, suggesting they remained lineage-committed. Consistent with this observation, the putative macrophages in Class 4 mosaics were uniformly p63− both before and after regeneration (Fig. S6).
Intra-ray fibroblasts contribute to blastema cells but do not become multipotent progenitors of regenerated fin tissue
To test whether any tissue of the regenerated fin was derived from intra-ray fibroblasts, we amputated Class 2 fins containing mCherry+ cells and followed them over time. At 1 dpa, we detected few mCherry+ cells beyond the amputation site (Fig. 4B, 4B*) in marked contrast to what we observed with cells residing in the epidermis (Fig. 2B, 2B*). By 4 dpa and 14 dpa, mCherry+ intra-ray fibroblasts-derived cells were widespread components of the newly regenerated fin ray (Fig. 4C–4D*). This indicates that the labeled intra-ray fibroblasts responded robustly after fin amputation to populate the intra-ray space in the newly regenerated fin. There was a notable similarity in the pattern assumed by the mCherry+ cells before amputation and 14 dpa, with intra-ray fibroblasts remaining spatially restricted within rays and not mixing with adjacent segments during regeneration. We noted significant differences in the behaviors of Class 1 and Class 2 mosaics during regeneration by observing a single fin that contained both mCherry+ epidermis and mCherry+ intra-ray fibroblasts in non-overlapping but nearby fin segments (Fig. S11). At 2 dpa, we observed the mCherry+ epidermal cells throughout the outer layer of the regenerated fin in contrast to mCherry+ intra-ray fibroblasts, which were found only immediately next to the amputation site. This observation supports the conclusion that epidermal cells uniquely become highly motile immediately after fin amputation (Santos-Ruiz et al., 2002; Lee et al., 2009).
Figure 4.
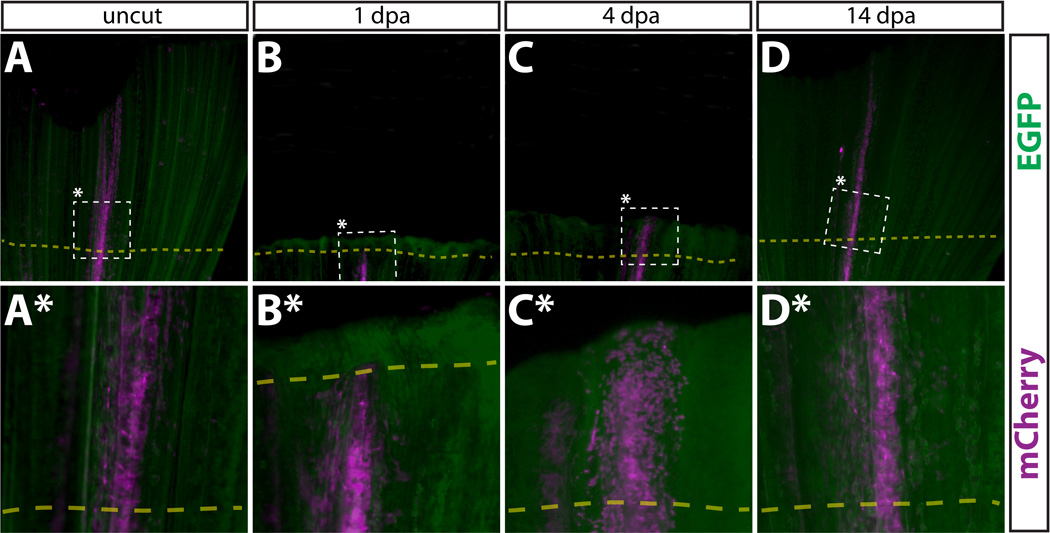
Intra-ray fibroblasts in regenerated fins are derived from pre-existing intra-ray fibroblasts. (A–D, A*–D*) Whole mount epifluorescent images of a Class 2 intra-ray fibroblast mosaic labeled caudal fin of the same Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal before amputation (A, A*), 1 dpa (B, B*), 4 dpa (C, C*), and 14 dpa (D, D*). The dashed box marks the zoomed region in the panel directly below. mCherry+ cells are shown in magenta. All other cells are EGFP+ (green). The amputation plane is shown with a dashed yellow line.
To further monitor the localization and proliferation status of intra-ray fibroblasts-derived cells during regeneration, we immunostained sections of regenerating Class 2 mosaic fins. 2 dpa, these fins contained mCherry+ cells embedded in a tenascin C matrix found throughout the blastema (Fig. 5A–5D). Although we detected labeled intra-ray fibroblasts adjacent to osteoblasts in agreement with an earlier observation (Tu and Johnson, 2011), we never observed mCherry+ intra-ray fibroblasts becoming zns-5+ osteoblasts or other non-fibroblast cells (Fig. 5E–5H). Anti-PCNA antibody staining to mark cells in M or S phase of the cell cycle confirmed a high rate of proliferation of mCherry+ intra-ray fibroblasts-derived cells within the blastema compared to labeled cells proximal to the amputation site (Fig. 5I–5L). We quantitatively analyzed proliferation of cells in 2 dpa fins stained with anti-PCNA and anti-tenascin C antibodies. (Fig. S7). We determined the percentage of PCNA+ nuclei in three regions (regions I–III) based on their positions relative to the amputation site as well as the presence of tenascin C. Region I cells were defined as blastema cells found distal to the amputation site that were localized within tenascin C+ extracellular matrix, region II was made up of cells also localized in tenascin C+ extracellular matrix but proximal to the amputation site, and region III cells consisted of cells further proximal to the amputation site in a field lacking detectable tenascin C expression. We observed significantly higher percentages of PCNA+ nuclei in both regions I and regions II relative to region III (Fig. S7), indicating a robust induction of cell proliferation in regions nearest the amputation site.
Figure 5.
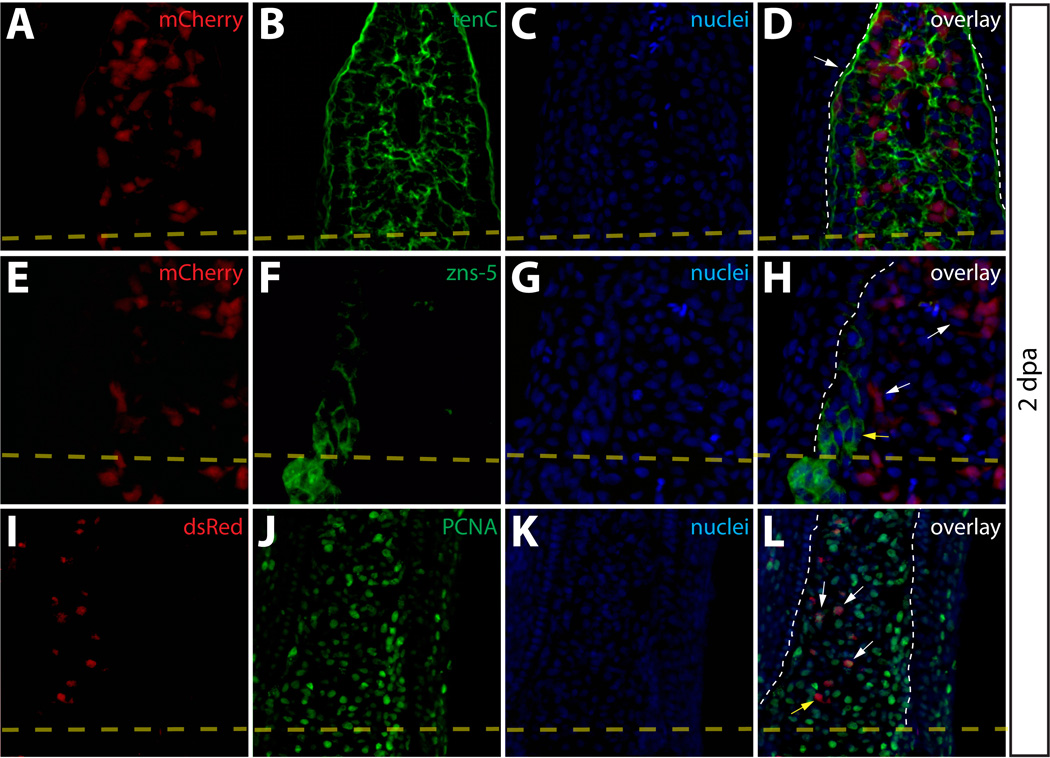
Intra-ray fibroblasts are a proliferating component of the blastema. (A–L) Longitudinal sections of Class 2 mosaic fins from Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) harvested 2 dpa and monitored for mCherry expression (A, E, I, red) and immunostained with indicated antibodies (B, F, J, green). Nuclei are stained with Hoechst (C, G, K, blue), and the three-color overlay is shown in each case (D, H, L). (A–D) Cells derived from intra-ray fibroblasts populate the blastema at 2 dpa. The section shows mCherry expression (A, red) and is immunostained with anti-tenascin-C (tenC) antibodies (B, green). The white arrow indicates mCherry+ intra-ray fibroblasts and the white dashed line denotes the boundary between epidermis and blastema. (E–H) mCherry+ intra-ray fibroblasts do not express markers for osteoblasts in regenerating tissue. mCherry+ cells are shown in red and zns-5 antibodies detect osteoblasts (F, green). White arrows indicate mCherry+ cells in the blastema that do not co-localize with zns-5+ osteoblasts (yellow arrow). The white dashed line indicates the boundary between epidermis and blastema. (I–L) Intra-ray fibroblasts are a source of proliferating cells in the blastema. A 2 dpa section co-stained with anti-dsRed antibodies to detect mCherry+ cells (I, red) and anti-PCNA antibodies to detect proliferating cells (J, green). The white arrows indicate mCherry+/PCNA+ intra-ray fibroblasts in the blastema. The yellow arrow marks a mcherry+/PCNA− intra-ray fibroblast, and the border between epidermis and blastema is marked with a white dashed line.
Osteoblasts undergo partial dedifferentiation, populate the blastema periphery, and remain committed to the osteoblast lineage during fin regeneration
We also used lineage tracing of Class 3 mosaic fins to examine the fate of osteoblasts during regeneration. As described, prior to amputation, mCherry+ osteoblasts were directly adjacent to lepidotrichia (Fig. 6A, 6A*) and expressed both zn-3 and zns-5 antigens (Fig. 7 and Fig. S8). To assess whether the osteoblast lineage contributed to newly regenerated tissue, we amputated fins containing mCherry+ osteoblasts and followed the progeny of the labeled cells over the course of regeneration. By 1 dpa, we did not detect labeled osteoblasts or derived cells in regenerated tissue (Fig. 6B, 6B*). This contrasted with both epidermal cells and intra-ray fibroblasts (compare Fig. 6B, 6B* to Fig. 2B, 2B* and to Fig. 4B, 4B*). However, by 4 dpa we observed significant numbers of labeled cells in the newly regenerated tissue (Fig 6C, 6C*) and by 14 dpa mCherry+ osteoblasts were found along the entire length of the regenerated fin ray (Fig. 6D, 6D*). Therefore, mature osteoblasts existing prior to fin amputation are the primary source of new bone forming cells in the regenerated fin. Further, as with fibroblast-derived cells, we observed no “mixing” of labeled osteoblast-derived cells from one lepidotrichia into neighboring rays. These observations suggest that the regenerating fin is composed of numerous self-contained blastema found at the distal tip of each regenerating lepidotrichia.
Figure 7.
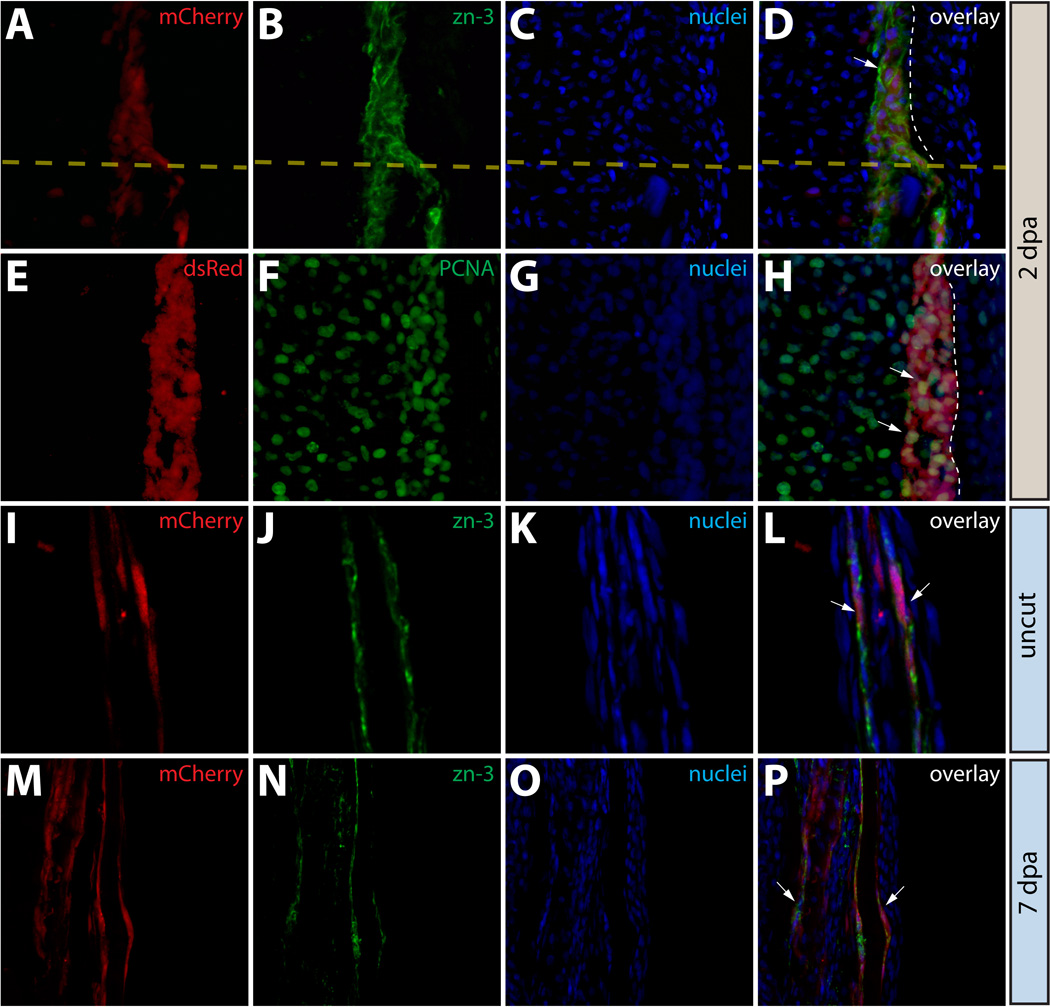
Osteoblasts in the caudal fin populate the blastema but remain fate restricted. (A–L) Longitudinal sections of the caudal fin of Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 3 mosaic animals showing mCherry expression in osteoblasts (A, E, I, M, red) and stained with indicated antibodies (B, F, J, N, green) and with Hoechst to mark nuclei (C, G, K, O, blue). The three-color overlays are shown in (D, H, L, P). (A–D) Osteoblasts contribute to the blastema. A fin section observed for mCherry expression and immunostained with zn-3 antibodies to mark osteoblasts. White arrows indicate mCherry+ cells (red) in the blastema that co-localize with zn-3+ osteoblasts (green). The dashed yellow line marks the amputation site and the dashed white line indicates the epidermis-blastema boundary. (E–H) Osteoblast-derived blastema cells proliferate. A fin section from the same animal in (A–D) stained with anti-dsRed antibodies to detect mCherry expression (red) and with anti-PCNA antibodies (green) to mark proliferating cells. White arrows indicate mCherry+/PCNA+ intra-ray fibroblasts in the blastema. The border between epidermis and blastema is marked with a white dashed line. (I–P) Osteoblasts do not change fate during regeneration. Stained fin sections from the same Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 3 mosaic animal prior to amputation (I–L) and 7 dpa (M–P). mCherry expression is red and anti-zn-3 antibody staining marks osteoblasts in green. The white arrows mark cells that are both mCherry and zn-3 positive.
We examined the behavior of mCherry+ osteoblasts and their progeny during regeneration by immunofluorescence of sectioned Class 3 fins. First, we used zn-3 and zns-5 antibodies to determine whether expression of these markers was lost in blastema cells derived from osteoblasts. We reasoned that if osteoblasts in the fin dedifferentiate to a multipotent progenitor after fin amputation, they would lose expression of mature osteoblast markers. However, at 2 dpa, mCherry+ osteoblast-derived cells localized near the amputation site maintained expression of zn-3 (Fig. 7A–7D) or zns-5 (Fig. S8A–S8D). Therefore, the mCherry+ cells near the stump at 2 dpa either remained mature osteoblasts or underwent limited dedifferentiation to a cellular state that retained expression of both of these osteoblast-specific antigens. These results also underscore that cells derived from mature osteoblasts are bona fide components of the blastema. Additionally, mCherry+ osteoblast-derived blastema cells were robustly positive for PCNA (Fig. 7E–7H), confirming that osteoblast-derived blastema cells were proliferating and actively participating in regeneration of the lost tissue. Osteoblast-derived blastema cells were also consistently localized to the periphery of the blastema adjacent to the basal epidermis, consistent with published observations (Tu and Johnson, 2011). Since we failed to observe any other lineage derived from mCherry+ cells in this class of fin after regeneration (Fig. 7I–7P), osteoblasts likely remain committed and contribute only replacement osteoblasts to regenerated tissue.
We hypothesized that even limited dedifferentiation of osteoblasts would require down-regulation of transcription factors whose function is required for osteoblast differentiation, such as sp7 (Nakashima et al., 2002). To investigate this, we amputated fins from a transgenic zebrafish line, Tg(sp7:EGFP). The Tg(sp7:EGFP) transgene includes regions surrounding the zebrafish sp7 promoter and drives expression of EGFP in mature caudal fin osteoblasts and in cells near the site of amputation in amputated fins (DeLaurier et al., 2010). EGFP expressing cells in the Tg(sp7:EGFP) animals were uniformly zn-3+ and zns-5+ (Fig. S1E–S1L). At 2 dpa, we observed robust EGFP expression in cells immediately adjacent to the amputation site of Tg(sp7:EGFP) animals (Fig. 8A, 8H, and Figs. S9 and S10). These EGFP+ cells were uniformly positive for both zn-3+ and zns-5+ antigens (Fig. 8B, 8I and Figs. S9 and S10), reinforcing the notion that these cells were osteoblasts. In these same samples, we also observed a noticeable reduction in sp7-driven EGFP+ intensity in osteoblast-derived blastema cells distal to the amputation site (Fig. 8A, 8E, and 8H, 8L, and Figs. S9 and S10). This result suggested that dedifferentiated osteoblasts downregulate sp7 promoter activity as they populate the blastema. To visualize this data graphically, 3-dimensional surface plots were generated with EGFP signal intensities in areas extending from the amputation site through the blastema plotted on the z-axis relative to the x–y positions of the cells (Fig. 8E–8G, 8L–8N and Figs. S9 and S10). Cells with low or undetectable EGFP levels still maintained high levels of both zn-3 and zns-5 (Fig. 8B, 8F, and 8I, 8M), showing they retained a partial molecular signature characteristic of caudal fin osteoblasts. These results are consistent with another report (Knopf et al., 2011) and support the idea that dedifferentiated osteoblasts are a highly organized component of the regenerating fin blastema.
Figure 8.
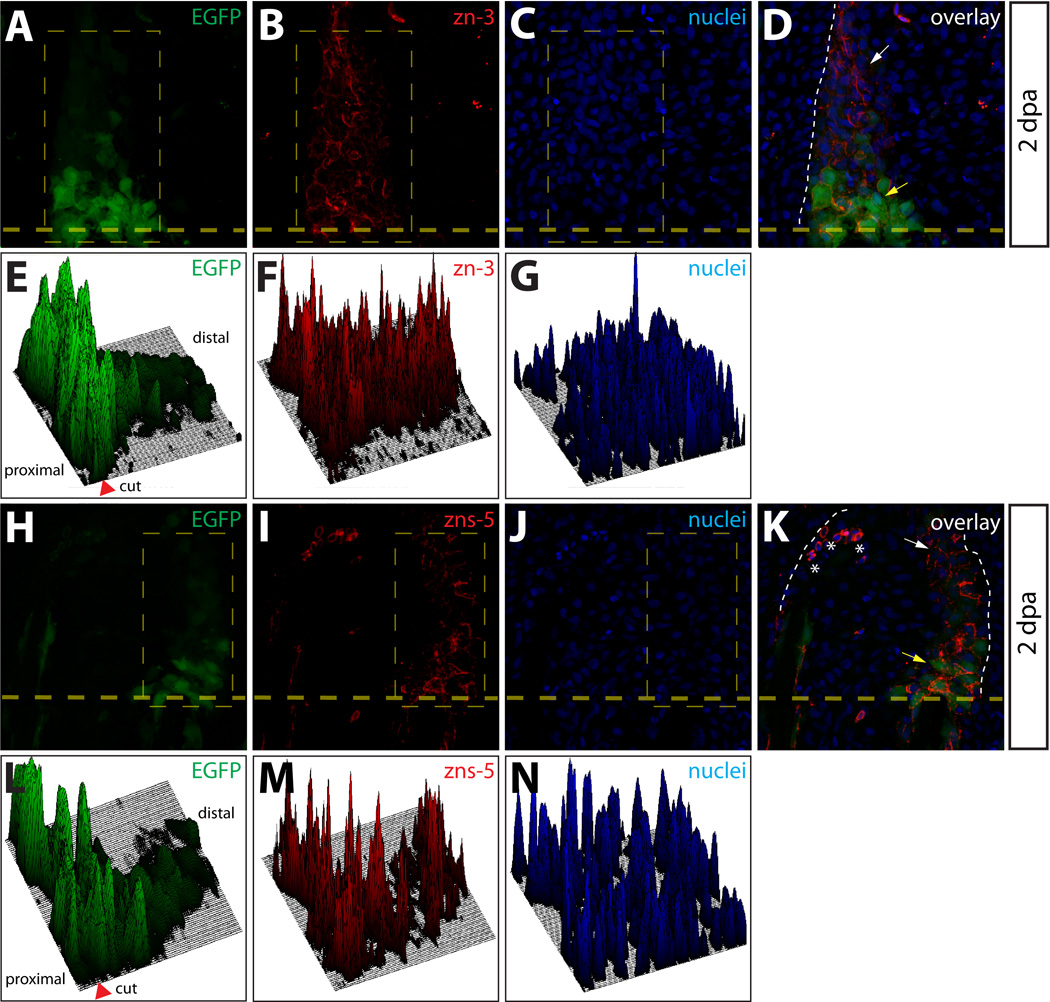
Osteoblasts dedifferentiate within the blastema. (A–D, H–K) Longitudinal sections from a Tg(sp7:EGFP) animal 2 dpa stained with zn-3 (A–D) or zns-5 antibodies (H–K). White arrows point to EGFP+/zn-3+ (or zns-5+) immature, dedifferentiated osteoblasts and yellow arrows point to EGFP+/zn-3− (or zns-5−) mature, differentiated osteoblasts. EGFP signal is shown in green, zn-3+ (B, D), or zns-5+ (I, K) cells are red, and Hoechst-stained nuclei are blue. The border between epidermis and blastema is marked with a white dashed line. (E–G, L–M) Relative signal intensity levels for EGFP (E, L), zn-3 or zns-5 antibody staining (F, M), and Hoechst (G, N). Fluorescence intensity levels (z-axis) within the indicated rectangle (thin dashed yellow line) are plotted as a 3-dimensional surface using Image J software. The amputation site is marked with an arrowhead and the proximal and distal regions relative to the amputation site are indicated. The white dashed line indicates the border between epidermis and blastema.
DISCUSSION
Using a fluorescent cell lineage tracing approach, we tracked the fate and lineage plasticity of epidermal cells, putative macrophages, intra-ray fibroblasts, and osteoblasts following caudal fin amputation in individual zebrafish. We found that each of these pre-existing cell types contributes to the regenerated fin. While showing signs of de-differentiation, each cell type remains committed to its original identity throughout the regeneration process. In the case of osteoblasts and intra-ray fibroblasts, both the axial positions and proportion of labeled cells is reestablished in the fully regenerated fin. Our results demonstrate that the progeny of lineage-restricted, dedifferentiated cells are the major source of new cells in a fully regenerated fin (Fig. 9), in strong support of recently proposed models (Knopf et al., 2011; Sousa et al., 2011; Tu and Johnson, 2011). In further concordance with these studies, our results show that fin blastemas are composed of organized, lineage restricted cells and they are segmentally organized at the distal tip of each amputated fin ray.
Figure 9.
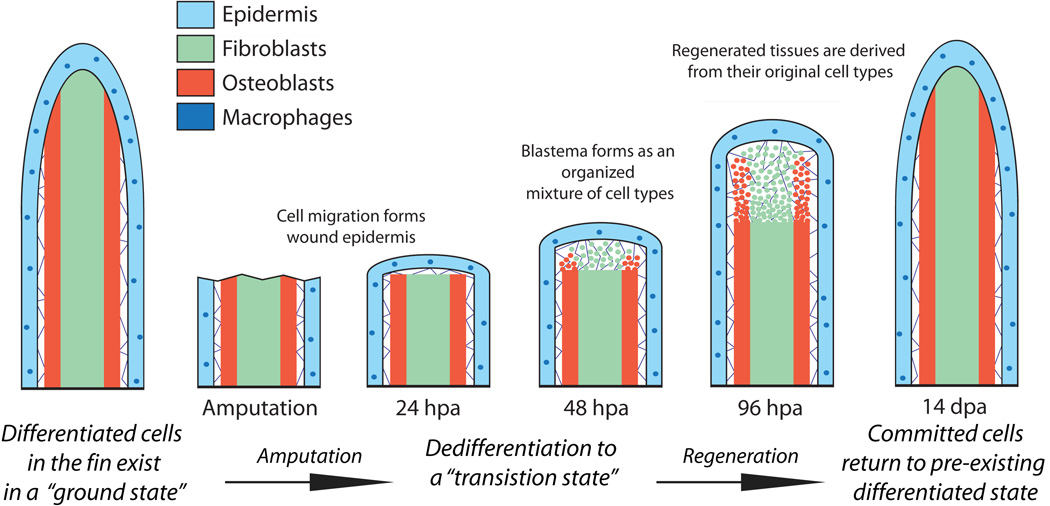
A restricted cell lineage model for fin regeneration. During homeostasis, the caudal fin is composed of various lineage restricted differentiated cells existing in a “ground state”. Fin amputation initiates wound healing by a motile epidermis and results in the conversion of ground state cells to “transition state” cells by the process of limited dedifferentiation. Transition state cells, including those derived from osteoblasts and intra-ray fibroblasts, contribute to an organized proliferating regenerative blastema capable of outgrowth and re-differentiation. The newly regenerated tissue is derived from transition state cells that differentiate into cells only of the same lineage, returning to their ground state as the fin reforms.
In fins containing labeled epidermis, we did not observe any labeled epidermal cells that were confined to a single layer, arguing that epidermal cell layers are of the same lineage. Still, some epidermal cells may switch between basal and surface epidermal fates; such a transition would not be detected by the experimental approach described here. Further analyses of the epidermal lineage(s) using a marker for the basal epidermis, such as lef1 (Poss et al., 2000), could provide insight into whether there exists switching of cell types within the epidermis itself. We were also struck by the presence of far-lateral epidermal-derived cells in the first few days following amputation. The significance of this cell population and mechanism behind this phenomenon are not clear, but it is tempting to speculate that these cells may be migrating to cues generated upon fin amputation such as hydrogen peroxide (Niethammer et al., 2009) or bioelectric signals (Zhao et al., 2006; Levin, 2007) to position them in proximity to promote regeneration.
Given the self-regenerating nature of the zebrafish epidermis (Le Guellec et al., 2004), it is not clear whether epidermal cells undergo dedifferentiation during fin regeneration. Our results demonstrate that epidermal cells do not lose expression of the marker p63, a gene which functions in epidermal development (Bakkers et al., 2002; Lee and Kimelman, 2002). In support of epidermal dedifferentiation, epidermal cells acquire the expression of developmental regulatory genes during regeneration including those of primitive epidermis, such as keratin-8, (Martorana et al., 2001) and are thought to provide developmental cues to the underlying blastema (Lee et al., 2009). Therefore, during regeneration epidermal cells remain committed to the epidermal lineage (p63+), but likely undergo partial dedifferentiation. We also tracked a non-epithelial cell population that resided in the epidermis. We identify these cells as putative tissue macrophages based on the strong resemblance of their localization to cells expressing the macrophage marker, l-plastin, in the fin under normal and regenerating conditions (Yoshinari et al., 2009). We do not know if these putative macrophages undergo dedifferentiation but our data indicate they do not switch lineages.
Our approach also allowed us to unambiguously identify the lineages of two distinct types of caudal fin blastema cells. Our results conclusively demonstrate that cells derived from mature intra-ray fibroblasts and osteoblasts take up residency in the blastema after fin amputation. Notably, we found no cells derived from the epidermal layers in the blastema, indicating they, or cells derived from them, remain spatially restricted in the epidermis throughout regeneration. It is likely additional cell lineages contribute to the blastema that were not labeled by Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) transgene, highlighting the need for additional conditional Cre transgenic lines. In addition to being developmentally restricted, intra-ray fibroblasts and osteoblasts are also spatially restricted. zn-3+ or zns-5+ cells reside only adjacent to the basal epidermal layer demonstrating an otherwise unapparent organization of resident blastema cells. This result also implies regeneration-directing signals, such as FGFs (Lee et al., 2005; Lee et al., 2009), originating from basal epidermal cells may be received by neighboring osteoblast-derived cells (Fig. 9). Finally, we failed to observe “contamination” of labeled cells into tissue of adjacent rays. This suggests that, in the case of the zebrafish caudal fin, there is a collection of individual blastemas at the distal end of each amputated ray that regenerate simultaneously but largely independent of one another.
Our lineage tracking studies of osteoblasts in the regenerating fin allowed us to observe molecular evidence for osteoblast dedifferentiation. Given that all zn-3 and zns-5 expressing cells in the blastema were derived from mature sp7+ osteoblasts, and that sub-populations of zn-3+/zns-5+ cells near the amputation site exhibited reduced sp7-driven EGFP expression (Fig. 6E, 6L), we conclude that these sub-populations represent partially dedifferentiated osteoblasts. Therefore, both zn-3 and zns-5 antigens are expressed by dedifferentiated (immature) and differentiated (mature) osteoblasts, whereas sp7 is only expressed by fully differentiated osteoblasts. It remains to be determined if zn-3 or zns-5 expression is maintained in all osteoblast-derived cells of the blastema as some may become further dedifferentiated and lose zn-3/zns-5 expression. Overall, these results argue that osteoblasts dedifferentiate to an osteoblast-like transition state, proliferate within the blastema, and eventually redifferentiate only to replacement bone cells (Fig. 9). The transcription regulators runx2 and sp7 coordinate osteogenesis in a sequential manner in mice (Karsenty et al., 2009) and dynamic patterns of expression have been observed for both runx2 and sp7 in the regenerating fin (Brown et al., 2008, Knopf et al., 2011, Sousa et al., 2011). Therefore, it will be of interest to determine whether dedifferentiation and redifferentiation of osteoblast-derived cells in the blastema is also a hierarchical process with respect to runx2 and sp7 and other regulators of the osteoblast lineage.
Our studies agree with those of others (Knopf et al., 2011; Sousa et al., 2011; Tu and Johnson, 2011) and support a model where limited dedifferentiation of cells in damaged tissue is sufficient to provide a source of cells for the regenerated zebrafish caudal fin. Conceivably, dedifferentiated progenitor-like cells generated after injury are more receptive to signals from neighboring cells and their environment, endowing them with robust proliferative and migratory properties compared to their differentiated precursors. Regeneration of damaged myocardium in the zebrafish also is mediated by limited dedifferentiation (Jopling et al., 2010; Kikuchi et al., 2010). Thus, the constituent cells of all regenerating zebrafish organs may share uncharacterized molecular mechanisms that direct dedifferentiation. Given that robust epigenetic changes occur during regeneration (Yakushiji et al., 2007; Stewart et al., 2009; Katsuyama and Paro, 2011) and upon derivation of fully dedifferentiated induced pluripotent stem (iPS) cells (Hochedlinger and Plath, 2009), studies of how chromatin mechanisms facilitate regenerative dedifferentiation are particularly attractive.
A corollary of the limited dedifferentiation model is that regeneration of complex vertebrate tissues and structures does not require pluripotent cells. Eventually, recapitulating similar mechanisms to coerce human cells to undergo limited reprogramming in vivo could prove an effective alternative to using pluripotent stem cells for regenerative medicine (Zhou et al., 2008; Graf and Enver, 2009). Continued progress understanding the cellular and molecular mechanisms underlying dedifferentiation during epimorphic regeneration will facilitate such approaches.
Highlights.
Regenerating zebrafish fin blastemas are temporally and spatially organized
Regeneration proceeds by formation of isolated blastemas at the tip of each fin ray
Osteoblasts undergo partial dedifferentiation while populating blastema peripheries
Cell lineages remain fate restricted during fin regeneration
Progeny of dedifferentiated cells are the major source of regenerated fin tissue
Supplementary Material
Figure S1. Antibody validation. (A–D) An anti-dsRed antibody detects mCherry expression in sectioned fins. A section from a Class 1 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fin immunostained with an anti-dsRed antibody (A, green) with mCherry expression shown in red (B). Nuclei are stained with Hoechst (C, blue) and the overlay is shown (D). All mCherry+ cells are positive for dsRed (white arrows). (E–L) The zns-5 and zn-3 antibodies stain osteoblasts. Sections of non-regenerating caudal fins from Tg(sp7:EGFP) fish showing EGFP expression (E, I green) and stained with zns-5 or zn-3 antibodies (F, J, red) and Hoechst (G, K, blue). The overlays (H, L) demonstrate co-localization of EGFP+ cells with zns-5+ or zn-3+ cells, respectively (cells marked with white arrows).
Figure S10. Additional evidence for dedifferentiation of osteoblast-derived cells populating the blastema. (A–D, H–K) Sections from two Tg(sp7:EGFP) animals 2 dpa imaged for EGFP expression (A, H, green) and stained with zn-3 antibodies (B, I, red). Nuclei are stained with Hoechst (C, J, blue) and the overlay is shown (D, K). A heavy dashed yellow line indicates the amputation plane and a white dashed line marks the epidermis-blastema boundary. White arrows point to mCherry+/zn-3− immature, dedifferentiated osteoblasts and the yellow arrows indicate mCherry+/zn-3+ mature, differentiated osteoblasts. Asterisks mark background-staining red blood cells. (E–G, L–N) Fluorescent signal intensities for EGFP (E, L), zn-3 staining (F, M) and Hoechst (G, N) within the indicated rectangle (thin, dashed yellow line in A–C and H–J) are plotted as 3-dimensional surfaces using Image J software. The amputation site is marked with an arrowhead and proximal and distal regions relative to the amputation site are indicated.
Figure S11. Comparison of epidermal cells and intra-ray fibroblasts during regeneration. (A–D, A*–D*) Whole mount epifluorescent images of a caudal fin from a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal containing both Class 1 epidermis and Class 2 intra-ray fibroblast mosaics before amputation (A), 2 dpa (B, B*, B**), and 14 dpa (C). Both EGFP (green) and mCherry (magenta) are shown in all panels. The dashed box marked with single asterisk outlines a region of labeled epidermis shown at higher magnification in (B*) and the dashed box marked with double asterisk indicates a region of labeled intra-ray fibroblasts shown at higher magnification (B**). The dashed yellow line shows the approximate amputation site. White arrows point at mCherry+ epidermal cells and yellow arrows indicate mCherry-labeled fibroblasts or fibroblast-derived cells.
Figure S2. Identification of mosaic cell populations in the fins shown in Figure 1. (A–D) Class 1 mosaics contain labeled epidermal cells. The fin shown in Fig. 1F and 1F* sectioned and stained with anti-p63 antibodies (B, green). mCherry+ (A, red) cells are found exclusively in epidermal layers and all are p63+ (white arrows). (E–L) Class 2 mosaics are composed of labeled intra-ray fibroblasts. The fin shown in Fig. 1G and 1G* sectioned and stained with antibodies recognizing tenascin C (F, green) or zns-5 (J, green). mCherry+ (red) cells are in a tenascin C rich extracellular matrix (white arrows in H) but do not express the osteoblast marker zns-5 (white arrows in L). (M–P) Class 3 mosaics comprise lepidotrichia-localized osteoblasts. The fin shown in Fig. 1H and 1H* sectioned and stained with zns-5 antibodies (N, green). mCherry+ (M, red) cells in lepidotrichia are also zns-5+ (white arrows). For all experiments, nuclei are stained with Hoechst (C, G, K, O, blue) and the three-color overlay is shown in the last column (D, H, L, P).
Figure S3. Osteoblasts and adjacent inter-ray cells are labeled in Class 3 mosaic fins. (A) Whole mount image of a Class 3 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fin demonstrating lepidotrichia-localized osteoblasts (white arrows) and adjacent non-osteoblast cells (yellow arrows). (B–I) Transverse sections from the same non-regenerating fin imaged for mCherry expression (B, F, red) and stained with either zn-3 or zns-5 antibodies (C, G, green) showing mCherry+/(zn-3 or zns-5)+ osteoblasts and inter-ray, non-osteoblast mCherry+/(zn-3 or zns-5)− cells. Nuclei are stained with Hoechst (D, H, blue), and the overlays are shown (E, I). White arrows indicate mCherry+/(zn-3 or zns-5)+ osteoblasts and yellow arrows indicate mCherry single positive cells.
Figure S4. Epidermal cells do not give rise to zns-5 expressing blastema-populating osteoblasts. (A–D) High magnification visualization of mCherry+ epidermis from 2 dpa Class 1 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fins imaged for mCherry expression (A, red) and stained with zns-5 antibodies (B, green). Nuclei are stained with Hoechst (blue). White arrows indicate mCherry+ epidermis that is negative for the osteoblast marker zns-5. Zns-5 positive osteoblasts are marked with a yellow arrow. The dashed yellow line indicates the site of amputation and a white dashed line indicates the border between epidermis and blastema.
Figure S5. Putative macrophages in Class 4 mosaic fins are fate restricted during fin regeneration. (A–D, A*–D*) Whole mount epifluorescent images of a Class 4 mosaic caudal fin of a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal containing mCherry+ putative macrophages before amputation (A, A*), 1 dpa (B, B*), 4 dpa (C, C*), and 14 dpa (D, D*). EGFP (green) and mCherry (magenta) signals are overlaid. The dashed box marked with an asterisk indicates the region shown at higher magnification in the panel directly below. The dashed line shows the approximate site of amputation. (A–D) and (A*–D*) are images acquired at 25× and 120× magnification, respectively.
Figure S6. Putative macrophages exhibit no net change in cell fate during regeneration. (A–H) Stained fin sections from the same Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 4 mosaic animal prior to amputation (A–D) and 7 dpa (E–H). mCherry expression is red (A, E) and p63 antibody staining marks epidermal cells in green (B, F). Nuclei are stained with Hoechst (C, G, blue) and the overlays are shown in (D, H). White arrows mark cells within the epidermis that are mCherry positive and p63 negative.
Figure S7. Cell proliferation is increased in tenascin C matrix localized blastema cells. (A–D) A representative image of a caudal fin harvested 2 dpa stained with anti-PCNA (A, red) and anti-tenascin C antibodies (B, green). Hoechst staining marks nuclei (C, blue). The amputation site is shown with a dashed light yellow line. The vertical solid yellow lines in the overlay (D) distinguish three regions in the regenerating fin marked by being distal (region I) or proximal (regions II, III) to the amputation site and by the presence of a tenascin C+ extracellular matrix (regions I, II). (E) A bar graph showing the mean percent of PCNA+ proliferating cells in each denoted region of the regenerating fin. Error bars represent the standard deviation between four animals. P values are calculated using a Student’s t-test.
Figure S8. mCherry+ osteoblast-derived blastema cells adjacent to the amputation site do not down regulate zns-5 antigen expression. (A–D) A longitudinal fin section from a Class 3 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal 2 dpa containing mCherry+ osteoblasts (A, red) and stained with zns-5 antibodies (B, green). Hoechst staining marks nuclei (C, blue). White arrows indicate mCherry+/zns-5+ cells and the border between epidermis and blastema is denoted with a white dashed line. The yellow dashed line shows the approximate site of amputation.
Figure S9. Osteoblasts dedifferentiate as they enter the blastema. (A–D, H–K) Sections from two Tg(sp7:EGFP) animals 2 dpa imaged for EGFP expression (A, H, green) and stained with zns-5 antibodies (B, I, red). Nuclei are stained with Hoechst (C, J, blue) and the overlay is shown (D, K). A heavy dashed yellow line indicates the amputation plane and a white dashed line marks the epidermis-blastema boundary. White arrows point to zns-5+/EGFP− immature, dedifferentiated osteoblasts and the yellow arrows indicate zns-5+/EGFP+ mature, differentiated osteoblasts. Asterisks mark background-staining red blood cells. (E–G, L–N) Fluorescent signal intensities for EGFP (E, L), zns-5 staining (F, M) and Hoechst (G, N) within the indicated rectangle (thin, dashed yellow line in A–C and H–J) are plotted as 3-dimensional surfaces using Image J software. The amputation site is marked with an arrowhead and proximal and distal regions relative to the amputation site are indicated.
ACKNOWLEDGEMENTS
We are grateful to the University of Oregon Zebrafish Facility for animal care; the University of Oregon histology facility; the University Oregon zebrafish research community for discussions and support; M. Tsang for providing the dusp6 promoter fragment; W. Chen for providing Tg(EAB:EGFP-FlEx-mCherry) fish; A. Delaurier and C. Kimmel for providing Tg(sp7:EGFP) fish; H. Zong and his laboratory for use of their confocal microscope; Z.-Y. Tsun, V. Devasthali, J. Eisen, and C. Kimmel for manuscript revisions; and members of the Stankunas lab for discussions. This work was supported by funds from the NIH/NHLBI (5R00HL087598) and the University of Oregon (K.S.) and an Oregon Scientist Development Award from the Oregon Medical Research Foundation (S.S.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- Brown AM, Fisher S, Iovine MK. Osteoblast maturation occurs in overlapping proximal-distal compartments during fin regeneration in zebrafish. Dev. Dyn. 2009;238:2922–2928. doi: 10.1002/dvdy.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant SV, Endo T, Gardiner DM. Vertebrate limb regeneration and the origin of limb stem cells. Int. J. Dev. Biol. 2002;46:887–896. [PubMed] [Google Scholar]
- DeLaurier A, Eames BF, Blanco-Sanchez B, Peng G, He X, Swartz ME, Ullmann B, Westerfield M, Kimmel CB. Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis. 2010;48:505–511. doi: 10.1002/dvg.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298:1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- Gargioli C, Slack JM. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Hay ED, Fischman DA. Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev. Biol. 1961;3:26–59. doi: 10.1016/0012-1606(61)90009-4. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS Lett. 2011;585:1617–1624. doi: 10.1016/j.febslet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisúa Belmonte JC. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell. Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastemal cells derived from dedifferentiating limb regeneration. Nature. 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies to the cells of a regenerating limb. J. Embryol. Exp. Morphol. 1985;89:37–55. [PubMed] [Google Scholar]
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, Weidinger G. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Le Guellec D, Morvan-Dubois G, Sire JY. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio) Int. J. Dev. Biol. 2004;48:217–231. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA, Black BL, Weidinger G, Poss KD. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 2009;331:270–280. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 2007;17:261–270. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Mari-Beffa M, Murciano C. Dermoskeleton morphogenesis in zebrafish fins. Dev. Dyn. 2010;239:2779–2794. doi: 10.1002/dvdy.22444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari-Beffa M, Santamaria JA, Murciano C, Santos-Ruiz L, Andrades JA, Guerado E, Becerra J. Zebrafish fins as a model system for skeletal human studies. ScientificWorldJournal. 2007;7:1114–1127. doi: 10.1100/tsw.2007.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorana ML, Tawk M, Lapointe T, Barre N, Imboden M, Joulie C, Geraudie J, Vriz S. Zebrafish keratin 8 is expressed at high levels in the epidermis of regenerating caudal fin. Int. J. Dev. Biol. 2001;45:449–452. [PubMed] [Google Scholar]
- Metzger D, Li M, Chambon P. Targeted somatic mutagenesis in the mouse epidermis. Methods Mol. Biol. 2005;289:329–340. doi: 10.1385/1-59259-830-7:329. [DOI] [PubMed] [Google Scholar]
- Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat. Chem. Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev. Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Nacu E, Tanaka EM. Limb Regeneration: A New Development? Annu. Rev. Cell Dev. Biol. 2010 doi: 10.1146/annurev-cellbio-092910-154115. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Namenwirth M. The inheritance of cell differentiation during limb regeneration in the axolotl. Dev. Biol. 1974;41:42–56. doi: 10.1016/0012-1606(74)90281-4. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye HL, Cameron JA, Chernoff EA, Stocum DL. Regeneration of the urodele limb: a review. Dev. Dyn. 2003;226:280–294. doi: 10.1002/dvdy.10236. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Keating MT. Induction of lef1 during zebrafish fin regeneration. Dev Dyn. 2000;219:282–286. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1045>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev. Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Santos-Ruiz L, Santamaria JA, Ruiz-Sanchez J, Becerra J. Cell proliferation during blastema formation in the regenerating teleost fin. Dev. Dyn. 2002;223:262–272. doi: 10.1002/dvdy.10055. [DOI] [PubMed] [Google Scholar]
- Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal. 2006;6(Suppl 1):38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM. Amphibian muscle regeneration--dedifferentiation or satellite cells? Trends Cell Biol. 2006;16:273–275. doi: 10.1016/j.tcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Slack JM. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat. Rev. Mol. Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simoes M, Leon J, Roehl H, Cancela ML, Jacinto A. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- Stewart S, Tsun ZY, Izpisúa Belmonte JC. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. USA. 2009;106:19889–19894. doi: 10.1073/pnas.0904132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131:2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- Tsonis PA, Washabaugh CH, Del Rio-Tsonis K. Transdifferentiation as a basis for amphibian limb regeneration. Semin. Cell Biol. 1995;6:127–135. doi: 10.1006/scel.1995.0018. [DOI] [PubMed] [Google Scholar]
- Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell. 2011;20:725–732. doi: 10.1016/j.devcel.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji N, Suzuki M, Satoh A, Sagai T, Shiroishi T, Kobayashi H, Sasaki H, Ide H, Tamura K. Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev. Biol. 2007;312:171–182. doi: 10.1016/j.ydbio.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Yoshinari N, Ishida T, Kudo A, Kawakami A. Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev. Biol. 2009;325:71–81. doi: 10.1016/j.ydbio.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, Sasaki T, Suzuki A, Forrester JV, Bourne HR, Devreotes PN, McCaig CD, Penninger JM. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Antibody validation. (A–D) An anti-dsRed antibody detects mCherry expression in sectioned fins. A section from a Class 1 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fin immunostained with an anti-dsRed antibody (A, green) with mCherry expression shown in red (B). Nuclei are stained with Hoechst (C, blue) and the overlay is shown (D). All mCherry+ cells are positive for dsRed (white arrows). (E–L) The zns-5 and zn-3 antibodies stain osteoblasts. Sections of non-regenerating caudal fins from Tg(sp7:EGFP) fish showing EGFP expression (E, I green) and stained with zns-5 or zn-3 antibodies (F, J, red) and Hoechst (G, K, blue). The overlays (H, L) demonstrate co-localization of EGFP+ cells with zns-5+ or zn-3+ cells, respectively (cells marked with white arrows).
Figure S10. Additional evidence for dedifferentiation of osteoblast-derived cells populating the blastema. (A–D, H–K) Sections from two Tg(sp7:EGFP) animals 2 dpa imaged for EGFP expression (A, H, green) and stained with zn-3 antibodies (B, I, red). Nuclei are stained with Hoechst (C, J, blue) and the overlay is shown (D, K). A heavy dashed yellow line indicates the amputation plane and a white dashed line marks the epidermis-blastema boundary. White arrows point to mCherry+/zn-3− immature, dedifferentiated osteoblasts and the yellow arrows indicate mCherry+/zn-3+ mature, differentiated osteoblasts. Asterisks mark background-staining red blood cells. (E–G, L–N) Fluorescent signal intensities for EGFP (E, L), zn-3 staining (F, M) and Hoechst (G, N) within the indicated rectangle (thin, dashed yellow line in A–C and H–J) are plotted as 3-dimensional surfaces using Image J software. The amputation site is marked with an arrowhead and proximal and distal regions relative to the amputation site are indicated.
Figure S11. Comparison of epidermal cells and intra-ray fibroblasts during regeneration. (A–D, A*–D*) Whole mount epifluorescent images of a caudal fin from a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal containing both Class 1 epidermis and Class 2 intra-ray fibroblast mosaics before amputation (A), 2 dpa (B, B*, B**), and 14 dpa (C). Both EGFP (green) and mCherry (magenta) are shown in all panels. The dashed box marked with single asterisk outlines a region of labeled epidermis shown at higher magnification in (B*) and the dashed box marked with double asterisk indicates a region of labeled intra-ray fibroblasts shown at higher magnification (B**). The dashed yellow line shows the approximate amputation site. White arrows point at mCherry+ epidermal cells and yellow arrows indicate mCherry-labeled fibroblasts or fibroblast-derived cells.
Figure S2. Identification of mosaic cell populations in the fins shown in Figure 1. (A–D) Class 1 mosaics contain labeled epidermal cells. The fin shown in Fig. 1F and 1F* sectioned and stained with anti-p63 antibodies (B, green). mCherry+ (A, red) cells are found exclusively in epidermal layers and all are p63+ (white arrows). (E–L) Class 2 mosaics are composed of labeled intra-ray fibroblasts. The fin shown in Fig. 1G and 1G* sectioned and stained with antibodies recognizing tenascin C (F, green) or zns-5 (J, green). mCherry+ (red) cells are in a tenascin C rich extracellular matrix (white arrows in H) but do not express the osteoblast marker zns-5 (white arrows in L). (M–P) Class 3 mosaics comprise lepidotrichia-localized osteoblasts. The fin shown in Fig. 1H and 1H* sectioned and stained with zns-5 antibodies (N, green). mCherry+ (M, red) cells in lepidotrichia are also zns-5+ (white arrows). For all experiments, nuclei are stained with Hoechst (C, G, K, O, blue) and the three-color overlay is shown in the last column (D, H, L, P).
Figure S3. Osteoblasts and adjacent inter-ray cells are labeled in Class 3 mosaic fins. (A) Whole mount image of a Class 3 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fin demonstrating lepidotrichia-localized osteoblasts (white arrows) and adjacent non-osteoblast cells (yellow arrows). (B–I) Transverse sections from the same non-regenerating fin imaged for mCherry expression (B, F, red) and stained with either zn-3 or zns-5 antibodies (C, G, green) showing mCherry+/(zn-3 or zns-5)+ osteoblasts and inter-ray, non-osteoblast mCherry+/(zn-3 or zns-5)− cells. Nuclei are stained with Hoechst (D, H, blue), and the overlays are shown (E, I). White arrows indicate mCherry+/(zn-3 or zns-5)+ osteoblasts and yellow arrows indicate mCherry single positive cells.
Figure S4. Epidermal cells do not give rise to zns-5 expressing blastema-populating osteoblasts. (A–D) High magnification visualization of mCherry+ epidermis from 2 dpa Class 1 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) fins imaged for mCherry expression (A, red) and stained with zns-5 antibodies (B, green). Nuclei are stained with Hoechst (blue). White arrows indicate mCherry+ epidermis that is negative for the osteoblast marker zns-5. Zns-5 positive osteoblasts are marked with a yellow arrow. The dashed yellow line indicates the site of amputation and a white dashed line indicates the border between epidermis and blastema.
Figure S5. Putative macrophages in Class 4 mosaic fins are fate restricted during fin regeneration. (A–D, A*–D*) Whole mount epifluorescent images of a Class 4 mosaic caudal fin of a Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal containing mCherry+ putative macrophages before amputation (A, A*), 1 dpa (B, B*), 4 dpa (C, C*), and 14 dpa (D, D*). EGFP (green) and mCherry (magenta) signals are overlaid. The dashed box marked with an asterisk indicates the region shown at higher magnification in the panel directly below. The dashed line shows the approximate site of amputation. (A–D) and (A*–D*) are images acquired at 25× and 120× magnification, respectively.
Figure S6. Putative macrophages exhibit no net change in cell fate during regeneration. (A–H) Stained fin sections from the same Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) Class 4 mosaic animal prior to amputation (A–D) and 7 dpa (E–H). mCherry expression is red (A, E) and p63 antibody staining marks epidermal cells in green (B, F). Nuclei are stained with Hoechst (C, G, blue) and the overlays are shown in (D, H). White arrows mark cells within the epidermis that are mCherry positive and p63 negative.
Figure S7. Cell proliferation is increased in tenascin C matrix localized blastema cells. (A–D) A representative image of a caudal fin harvested 2 dpa stained with anti-PCNA (A, red) and anti-tenascin C antibodies (B, green). Hoechst staining marks nuclei (C, blue). The amputation site is shown with a dashed light yellow line. The vertical solid yellow lines in the overlay (D) distinguish three regions in the regenerating fin marked by being distal (region I) or proximal (regions II, III) to the amputation site and by the presence of a tenascin C+ extracellular matrix (regions I, II). (E) A bar graph showing the mean percent of PCNA+ proliferating cells in each denoted region of the regenerating fin. Error bars represent the standard deviation between four animals. P values are calculated using a Student’s t-test.
Figure S8. mCherry+ osteoblast-derived blastema cells adjacent to the amputation site do not down regulate zns-5 antigen expression. (A–D) A longitudinal fin section from a Class 3 mosaic Tg(dusp6:Cre-ERT2, EAB:EGFP-FlEx-mCherry) animal 2 dpa containing mCherry+ osteoblasts (A, red) and stained with zns-5 antibodies (B, green). Hoechst staining marks nuclei (C, blue). White arrows indicate mCherry+/zns-5+ cells and the border between epidermis and blastema is denoted with a white dashed line. The yellow dashed line shows the approximate site of amputation.
Figure S9. Osteoblasts dedifferentiate as they enter the blastema. (A–D, H–K) Sections from two Tg(sp7:EGFP) animals 2 dpa imaged for EGFP expression (A, H, green) and stained with zns-5 antibodies (B, I, red). Nuclei are stained with Hoechst (C, J, blue) and the overlay is shown (D, K). A heavy dashed yellow line indicates the amputation plane and a white dashed line marks the epidermis-blastema boundary. White arrows point to zns-5+/EGFP− immature, dedifferentiated osteoblasts and the yellow arrows indicate zns-5+/EGFP+ mature, differentiated osteoblasts. Asterisks mark background-staining red blood cells. (E–G, L–N) Fluorescent signal intensities for EGFP (E, L), zns-5 staining (F, M) and Hoechst (G, N) within the indicated rectangle (thin, dashed yellow line in A–C and H–J) are plotted as 3-dimensional surfaces using Image J software. The amputation site is marked with an arrowhead and proximal and distal regions relative to the amputation site are indicated.


