Abstract
Mammals employ large numbers of odorant receptors to sample and identify volatile chemicals in the environment. These receptors are thought to vary not only in specificity for particular odorants, but also in breadth of tuning. That is, some odorant receptors are narrowly focused on a few closely related structures, while other odorant receptors may be “broadly tuned”, responding to a wide variety of odorant structures. Here we have performed a detailed examination the mouse odorant receptor MOR256-17, demonstrating that this receptor is broadly tuned. This receptor responds to odorant structures that span a significant portion of a multi-dimensional odor space. However, we found that broad tuning was not a defining characteristic of other members the MOR256 subfamily. Two additional members of this odorant receptor subfamily (MOR256-8 and MOR256-22) were more narrowly focused on small sets of odorant structures. Interestingly, the receptive range of MOR256-17 encompassed a variety of nitrotoluenes, including various TNT synthesis intermediates, degradation products and TNT itself, suggesting the potential utility of odorant receptors in the development of sensing technologies for the detection of explosives and other forms of contraband.
Keywords: Olfactory receptors, molecular receptive range, ligand specificity, explosives detection
Introduction
Olfactory sensory systems allow organisms to surveil their surrounding environment for the presence of volatile chemicals. A critical component of any olfactory system is the array of receptors that bind and report the presence of the various odorants. Mammalian olfactory receptors (ORs) are members of the G protein-coupled receptor (GPCR) superfamily (Buck & Axel 1991). Each olfactory sensory neuron expresses a single type of OR (Serizawa et al. 2003) and activation of the OR is linked through a signal transduction pathway to neuronal depolarization and action potential initiation (Bakalyar & Reed 1990, Jones & Reed 1989, Lowe & Gold 1993, Nakamura & Gold 1987). In the mouse genome, there are ~ 1000 intact OR genes (Godfrey et al. 2004, Young et al. 2002, Zhang & Firestein 2002) and most or all of these genes appear to be expressed (Zhang et al. 2004), indicating the deployment of a massive receptor array to solve olfactory recognition problems.
As large as the mammalian OR array is, the number of odorant structures in the environment that may need to be recognized is far larger: the components of this “odor space” may number in the millions (Mombaerts 2004). It is generally believed that olfactory systems solve this problem by using ORs in a combinatorial manner to detect odorants and encode their identities. In this combinatorial scheme, individual ORs can recognize multiple odorants and individual odorants are detected by multiple ORs (Malnic et al. 1999). This combinatorial coding also appears to be reflected at the glomerular level (Rubin & Katz 1999).
The particular set of odorant structures recognized by an OR is termed the molecular receptive range (MRR) of the OR (Araneda et al. 2000, Mori et al. 1999) and a range of MRR breadth, with some ORs narrowly tuned and others broadly tuned, has been proposed (Reed 2004). The most extensively characterized mammalian ORs appear to be relatively narrowly tuned, with each OR responding to a set of closely related odorant structures: I7 responds to closely related aliphatic aldehydes (Araneda et al. 2000, Zhao et al. 1998), M71 (MOR171-2) responds to acetophenone and benzaldehyde (Bozza et al. 2002), S6 (MOR42-3) and S50 (MOR42-1) respond to closely related linear aliphatic carboxylic acids (Abaffy et al. 2006, Malnic et al. 1999). However, our knowledge about the MRRs of these receptors is limited by the particular set of odorants that were screened in each case and the MRRs may be more extensive than currently appreciated. For example, MOR-EG (MOR174-9) was originally thought to respond to eugenol and closely-related structures (Kajiya et al. 2001, Katada et al. 2005), but additional screening has revealed an expanded MRR, with responsiveness to several polycyclic structures (Baud et al., 2011). Earlier work examining the responsiveness of individual amphibian OSNs suggested the existence of much more broadly tuned ORs (Firestein et al. 1993, Sicard & Holley 1984) and broadly tuned mammalian ORs have recently been identified. SR1 (MOR256-3) and Olfr42 (MOR256-31) were shown to respond to a wide variety of linear aliphatic, cyclic and aromatic structures (Grosmaitre et al. 2009, Nara et al. 2011) and several mouse and human ORs have been shown to respond to a wide variety of structures (Saito et al. 2009). Thus, among the myriad ORs in the mammalian repertoire, there appears to be wide variation both in specificity for particular odorants and in the dimensions of the MRR of individual ORs.
As part of the DARPA RealNose project, we previously sought to identify and characterize canine and murine ORs that respond to and distinguish among a small panel of odorants (assigned by the RealNose project): acetophenone, amyl acetate, cyclohexanone, 2,4-dinitrotoluene (2,4-DNT), ethyl vanillin, eugenol, heptanal, 2-heptanone, limonene and methyl benzoate. We included MOR256-17 as part of this project due to a recent report that this receptor responds to cyclohexanone (Saito et al. 2009) and MOR256-17 was one of several ORs that were successfully incorporated into a carbon nanotube transistor based sensing device (Goldsmith et al. 2011). Here we examine MOR256-17 in more detail, showing that this mouse OR responds to an exceptionally wide variety of odorant structures. MOR256-17 is grouped in the same mouse OR sub-family (Zhang & Firestein 2002) as the SR1 (MOR256-3) and Olfr42 (MOR256-31) receptors, which have also been shown to be broadly tuned (Grosmaitre et al. 2009, Nara et al. 2011). The MOR256 sub-family is one of the larger groupings of mouse ORs, with at least 37 members (Godfrey et al. 2004, Zhang & Firestein 2002). However, our examination of the molecular receptive range of additional members of this OR sub-family does not support broad tuning as a characteristic feature of all members of the MOR256 sub-family.
Experimental Procedures
Materials
Xenopus laevis frogs were purchased from Nasco (Fort Atkinson, WI, USA). The care and use of X. laevis frogs in this study was approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. RNA transcription kits were from Ambion (Austin, TX, USA). Collagenase B was from Boehringer-Mannheim (Indianapolis, IN, USA). All other compounds and all odorants were from Sigma–Aldrich (St. Louis, MO, USA). CAS numbers for the odorants used in this study are listed in Supplementary Tables 1 and 3.
Expression constructs
We refer to MORs using the nomenclature of Zhang and Firestein (2002). The coding regions of MOR256-17 (AY073576.1), MOR256-6 (AY073262.1), MOR256-8 (AY073351.1) and MOR256-22 (AY073625.1) were amplified by PCR from mouse genomic DNA (Clontech, Mountain View, CA, USA) and confirmed by sequencing. MOR256-17 was subcloned into the pCI vector (Promega) with an N-terminal extension consisting of the N-terminal 20 amino acid residues of human rhodopsin (Saito et al. 2004). The coding regions of MOR256-6, MOR256-8 and MOR256-22 were each combined with a T7 RNA polymerase promoter sequence and the N-terminal rhodopsin extension in a second PCR reaction. MOR174-9 (AY73453) was also expressed from a pCI vector with the N-terminal rhodopsin extension, as previously reported (Abaffy et al. 2006). The human Gaolf construct was purchased from the UMR cDNA Resource Center. The human cystic fibrosis transmembrane regulator (CFTR) construct was kindly provided by Dr. Ian Dickerson (University of Rochester). cRNA encoding the various MORs, Gaolf and CFTR was synthesized using mMESSAGE mMACHINE Kits (Ambion).
Preparation of oocytes and cRNA injection
Oocytes were surgically removed from mature Xenopus laevis frogs (Nasco, Fort Atkinson, WI, USA). Follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem) for 2 hrs at 22–25°C. Oocytes were injected with 46 nL of water containing cRNAs: 40 ng MOR, 10 ng Gaolf, 0.5 ng CFTR. Oocytes were incubated at 16°C in Barth’s saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.5 and 12 μg/ml tetracycline) for 2 – 5 days prior to electrophysiological recording.
Electrophysiology and data analysis
Electrophysiology and data analysis were performed as described previously (Abaffy et al. 2006, Repicky & Luetje 2009). Briefly, odorant induced Cl- currents, resulting from cAMP-mediated activation of the co-expressed CFTR reporter channel (Uezono et al. 1993), were measured 2 – 5 days after cRNA injection using two-electrode voltage clamp in an automated parallel electrophysiology system (OpusExpress 6000A, Molecular Devices, Union City, CA, USA). Micropipettes were filled with 3 M KCl and had resistances of 0.2–2.0 MW. The holding potential was −70 mV. Current responses, filtered (4-pole, Bessel, low pass) at 20 Hz (−3 db) and sampled at 100 Hz, were captured and stored using OpusXpress 1.1 software (Molecular Devices). Initial analysis was performed using Clampfit 9.1 software (Molecular Devices). Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). High concentration (0.5 – 1 M) stock solutions of each odorant were prepared in dimethylsulfoxide (DMSO) or ethanol. Each odorant, diluted in ND96, was applied for 15 s, followed by a 10 min wash with ND96. IBMX was used to activate the CFTR in a receptor-independent manner. This occurs both through the inhibition of phosphodiesterase and consequent increase in cAMP concentration, and through a direct action on the CFTR (Schultz et al. 1999). Application of a high concentration of IBMX (1 mM) is needed to achieve this effect (Abaffy et al., 2006). For the dose-response analyses in Figure 1D, each odorant response was normalized to the response of the same oocyte to application of 1 μM 2-heptanone. A low concentration was used for normalization to avoid problems with receptor desensitization that might result from repeated application of a high concentration. Normalized data were fit using Prism 5 (Graphpad, San Diego, CA, USA) according to the equation: I = Imax/(1+(EC50/X)n) where I represents the current response at a given concentration of odorant, X; Imax is the maximal response; EC50 is the concentration of odorant yielding a half-maximal response; n is the apparent Hill coefficient. In Figures 2–4, each odorant response was normalized to the response of the same oocyte to application of 30 μM 2-heptanone (Figure 2), 30 μM acetophenone (Figure 3), or 100 μM 2,4-DNT (Figure 4). In Figure 6, each odorant response was normalized to the response of the same oocyte to 30 μM 2-heptanone (MOR256-17), 30 μM 2-nonanol (MOR256-8), or 30 μM trans-cinnamaldehyde (MOR256-22).
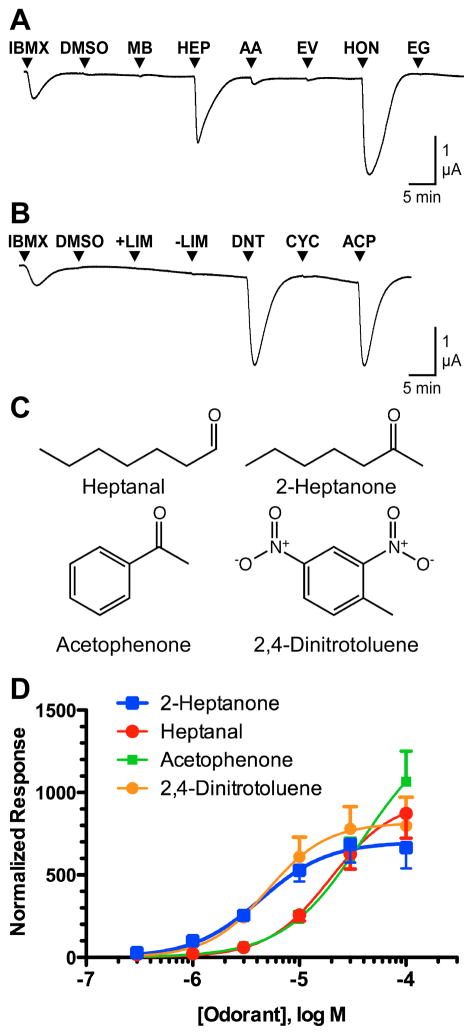
Figure 1. Screening of MOR256-17 with a small panel of odorants.
A) An oocyte expressing MOR256-17, Gαolf and CFTR was challenged with 15 s applications of 1mM IBMX, 0.006% DMSO, and 30 μM each of methyl benzoate (MB), heptanal (HEP), amyl acetate (AA), ethyl vanillin (EV), 2-heptanone (HON) and eugenol (EG). B) An oocyte expressing MOR256-17, Gαolf and CFTR was challenged with 15 sec applications of 1mM IBMX, 0.006% DMSO, and 30 μM each of (+)-limonene (+LIM), (−)-limonene (-LIM), 2,4-dinitrotoluene (DNT), cyclohexanone (CYC), and acetophenone (ACP). C) Structures of the four most active odorant ligands. D) Concentration-response analysis of MOR256-17 with 2-heptanone, heptanal, acetophenone, and 2,4-DNT. Oocytes expressing MOR256-17, Gαolf and CFTR were challenged with 15 s application of a range of concentrations from 0.3 to 100 μM. All response were normalized to the response of the same oocyte to 1 μM 2-heptanone. Data are presented as mean ± SEM (n = 5–10).
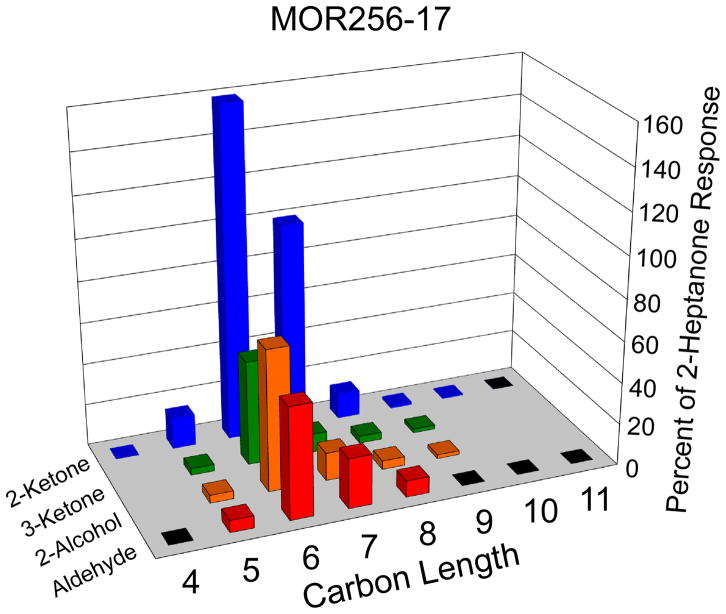
Figure 2. Responsiveness of MOR256-17 to a range of aliphatic odorants.
Oocytes expressing MOR256-17, Gαolf and CFTR were screened with a panel of aldehydes, 2-ketones, 3-ketones, and 2-alcohols, ranging in length from 4 to 11 carbons. Each compound was applied for 15 s at a concentration of 30 μM. Responses were normalized to the response of each oocyte to 30 μM 2-heptanone (mean values are displayed; error and n values may be found in Supplementary Table 2). Black squares at the base of the graph indicate tested compounds that did not yield a response. Blank areas at the base of the graph indicate compounds that were not tested.
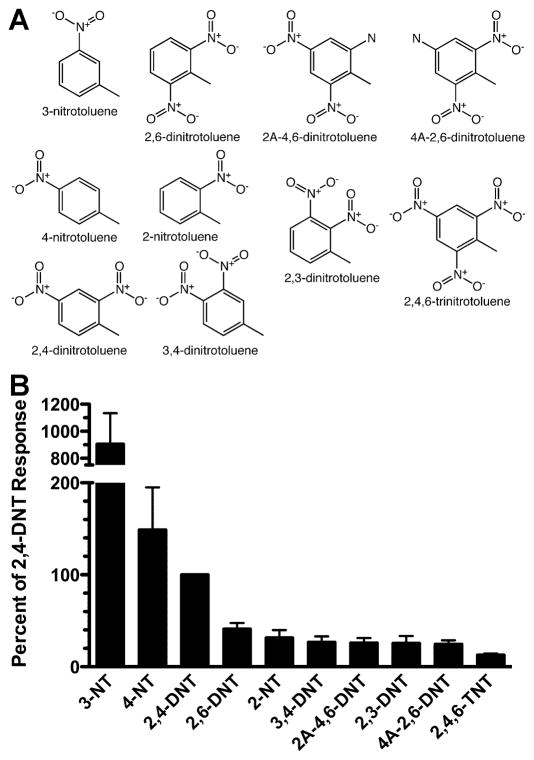
Figure 4. Screening of MOR256-17 with odorants related to 2,4-DNT.
A) Structures of odorants related to 2,4-DNT. B) Oocytes expressing MOR256-17, Gαolf and CFTR were screened with 15 s application of 100 μM of each of the compounds shown in panel A. Response were normalized to 100 μM 2,4-DNT. Values are the mean ± SEM (n = 4-7).
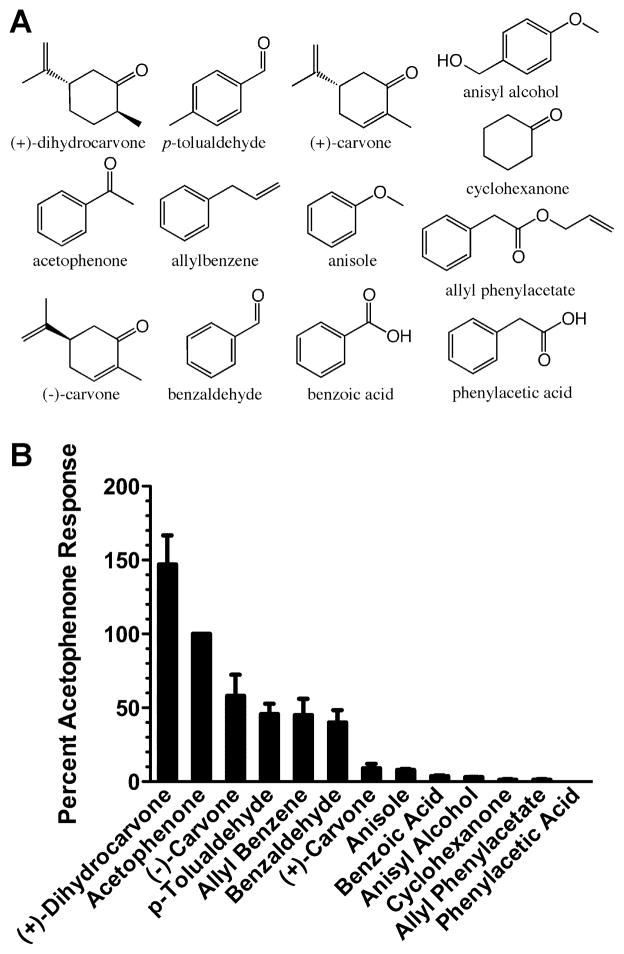
Figure 3. Screening of MOR256-17 with odorants related to acetophenone.
A) Structures of odorants related to acetophenone. B) Oocytes expressing MOR256-17, Gαolf and CFTR were screened with 15 s applications of 30 μM of each of the compounds shown in panel A. Responses were normalized to 30 μM acetophenone. Values are the mean ± SEM (n=5–9).
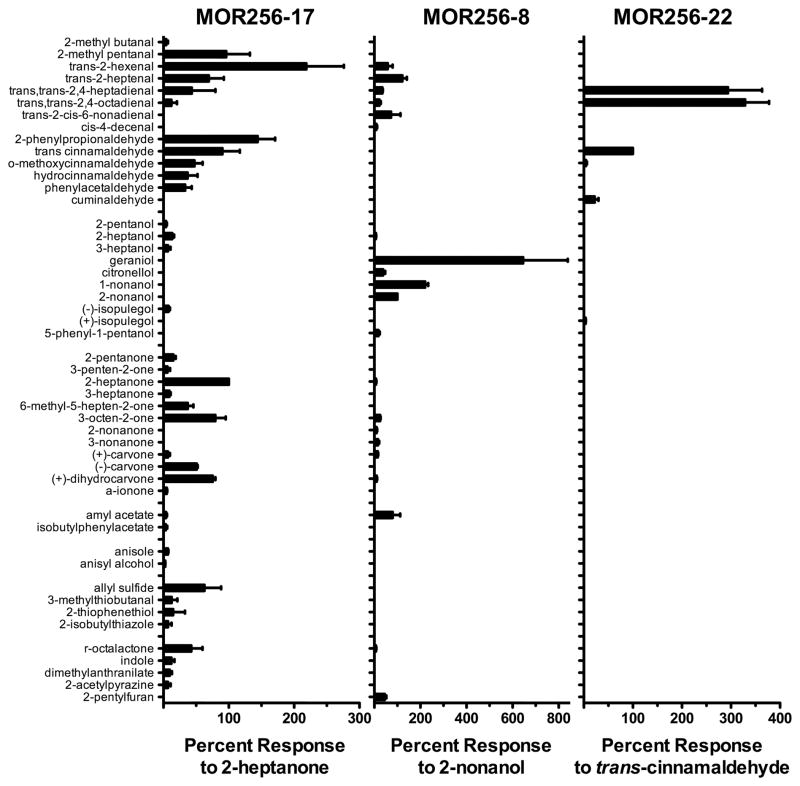
Figure 6. Responsiveness of MOR256-17, MOR256-8 and MOR256-22 to a large panel of odorants.
Oocytes expressing MOR256-17, MOR256-8 or MOR256-22 (as well as Gαolf and CFTR) were screened as shown in Figure 5. All odorants, whether applied individually or as part of mixtures, were applied for 15 s at a concentration of 30 μM. Responses were normalized to the response of the same oocyte to 30 μM 2-heptanone (MOR256-17), 30 μM 2-nonanol (MOR256-8), or 30 μM trans-cinnamaldehyde (MOR256-22). Data are presented as mean ± SEM (n = 3 – 20). All odorants yielding a response that was at least 1% of the response to the most active odorant (trans-2-hexenal for MOR256-17, geraniol for MOR256-8, trans, trans-2,4-octadienal for MOR256-22) are displayed.
The CFTR can be directly activated by a wide variety of structures (Ma et al. 2002). Thus, all odorants and odorant mixtures used in our studies were tested with oocytes expressing the human M1 muscarinic receptor, Gaolf and CFTR, but no odorant receptors, to guard against false positives. Odorants were screened at a concentration of 30 μM. Most of the odorants did not yield a response under these conditions. For the few odorants that did yield small responses in control oocytes not expressing odorant receptors, the responses seen in odorant receptor expressing oocytes were judged to be authentic receptor responses only if those responses were significantly greater than any responses seen in the control oocytes (one-way ANOVA followed by Dunnett’s post-test).
The results we obtained with MOR256-17 in Figure 1 are at variance with the results of Saito et al. (2009), who reported that MOR256-17 responds to cyclohexanone, but not to heptanal or 2-heptanone. We obtained the MOR256-17 expression construct used by the Matsunami laboratory in their study (Saito et al. 2009) and found that, in our hands, this construct yields results in agreement with what we show in Figure 1. Similarly, when the Matsunami laboratory retested their MOR256-17 expression construct in their assay system, they obtained results in agreement with our results (Dr. Hiroaki Matsunami, personal communication). Thus, the results we present here regarding the odorant specificity of MOR256-17 accurately reflect the function of this receptor.
We estimated the breadth of tuning of MOR256-17, MOR256-8 and MOR256-22 by calculating the radius of a hypersphere centered on the center of mass of all the odorants that activate each receptor (Saito et al., 2009). We also used this approach to estimate the distribution in odor space of the odorant sets used for screening in Figures 1, 5–6. We estimated odor space using 1035 odorants that are commonly used in olfaction studies (Haddad et al., 2008; Haddad et al., 2010). We obtained the molecular structure files from PubChem (http://pubchem.ncbi.nlm.nih.gov/) and used Dragon software (Talete) to compute physicochemical descriptors. This multi-dimensional odor space was based on 32 optimized physicochemical descriptors (Haddad et al., 2008). Principal component analysis was used to depict the odorants in two dimensions.
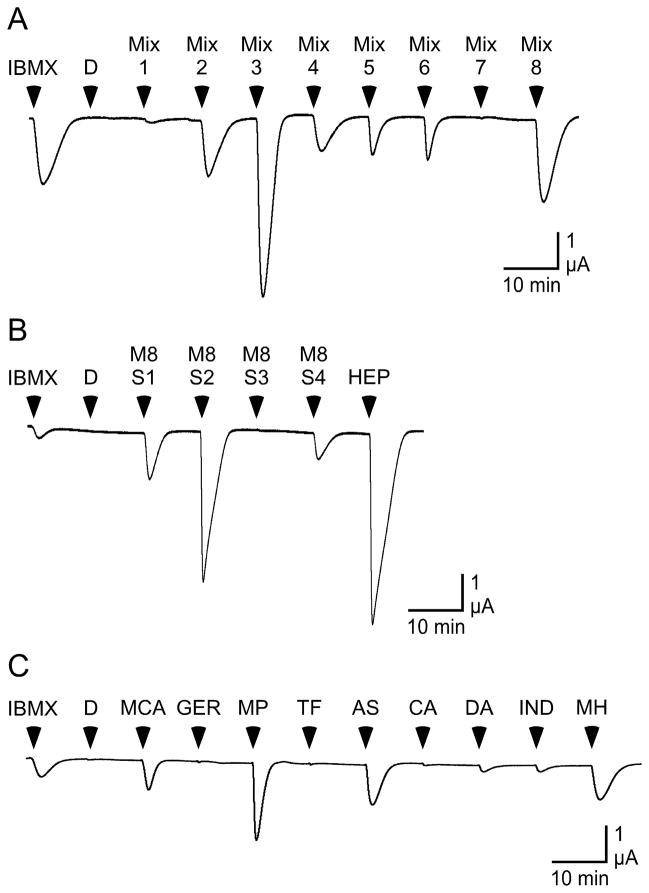
Figure 5. Screening of MOR256-17 with an expanded panel of odorants.
A) An oocyte expressing MOR256-17, Gαolf and CFTR is screened with 15 sec applications of 1mM IBMX, 0.006% DMSO (D), and 8 mixtures containing a total of 155 odorants. Each mixture contains 17 – 20 odorants, each at a concentration of 30 μM (the odorant composition of each mixture is listed in the Supplemental Table 3). B) An oocyte expressing a MOR256-17, Gαolf and CFTR is challenged with 15 sec applications of 1mM IBMX, 0.006% DMSO, and 4 sub-mixtures derived from the mixture 8. M8S1 contains geraniol, carvacrol, (S)-(−)-perillyl alcohol, thymol and 2-methyl pentanal. M8S2 contains o-methoxycinnamaldehyde, trans-3-hexenoic acid, benzyl cinnamate, ethyl butyrate and hexyl isobutyrate. M8S3 contains p-tolyl acetate, isopropyl tiglate, terpinyl formate, 1,4-cineole and dimethyl anthranilate. M8S4 contains indole, 6-methyl-5-hepten-2-one, allyl sulfide and γ-terpinene. 2-heptanone (HEP) tested separately. Each odorant was tested at a concentration of 30 μM. C) An oocyte expressing a MOR256-17, Gαolf and CFTR is challenged with 15 sec applications of 1mM IBMX, 0.006% DMSO and 30 μM of each of some of the individual odorants from the M8S1, M8S2, and M8S4 sub-mixtures: o-methoxycinnamaldehyde (MCA), geraniol (GER), 2-methyl pentanal (MP), terpinyl formate (TF), allyl sulfide, 1,4-cineole (CA), dimethyl anthranilate (DA), indole (IND), and 6-methyl-5-hepten-2-one (MH).
Results
Broad responsiveness of MOR256-17 to a small panel of odorants
We expressed MOR256-17 in Xenopus oocytes, along with human Gαolf and human CFTR to allow electrophysiological assay of receptor function (Abaffy et al. 2006). The receptor was screened with the small odorant panel that was assigned as part of the DARPA RealNose project, with each odorant applied at a concentration of 30 μM (Figure 1A and B). While this odorant panel was quite small, the individual odorants were broadly (albeit sparsely) distributed within an estimate of odor space (Supplementary Figure 1). MOR256-17 responded well to acetophenone, 2,4-DNT, heptanal and 2-heptanone, responded poorly to amyl acetate and did not respond to cyclohexanone, ethyl vanillin, eugenol, (+)-limonene, (−)-limonene or methyl benzoate. Small responses to cyclohexanone could be observed when the concentration was increased to 1 mM (data not shown). Our results are at variance with the results of Saito et al. (2009), who reported that MOR256-17 responds to cyclohexanone, but not to heptanal or 2-heptanone. However, we have confirmed that the odorant sensitivity for MOR256-17 that we show in Figure 1 is correct (see Experimental Procedures). Structures of the four highly active odorants are shown in Figure 1C. To further evaluate the responsiveness of MOR256-17 to these four ligands, we generated concentration-response curves (Figure 1D). MOR256-17 displayed an EC50 of 4 ± 1 μM for 2-heptanone, 5 ± 2 μM for 2,4-DNT, 19 ± 6 μM for heptanal, and 38 ± 30 μM for acetophenone.
Responsiveness to structures related to 2-heptanone, heptanal and acetophenone
The responsiveness of MOR256-17 to four odorants in such a small and diverse screening panel is striking. Furthermore, while heptanal and 2-heptanone could be hypothesized to adopt a conformation similar to that of acetophenone, the sensitivity to 2,4-DNT suggested that this receptor may be broadly tuned. That is, MOR256-17 may recognize a wide range of structurally diverse odorants. To examine this possibility, we screened MOR256-17 with a series of compounds that are structurally related to each of these four odorants. We began with a panel of 26 aliphatic odorants structurally related to heptanal and 2-heptanone, including aldehydes, 2-ketones, 3-ketones, and 2-alcohols, ranging in length from 4 to 11 carbons (Figure 2, Supplemental Table 2). The odorants were applied at a concentration of 30 μM. MOR256-17 responded to odorants that were 5 to 9 carbons in length, with a preference for odorants containing 6 carbons. MOR256-17 responded best to 2-ketones (2-hexanone and 2-heptanone), but also responded well to 3-hexanone, 2-hexanol and hexanal.
MOR256-17 was also responsive to acetophenone, an aromatic ketone. To further explore this structure, we screened an additional 12 odorants that are structurally related to acetophenone (Figure 3). MOR256-17 was highly responsive to (+)-dihydrocarvone and moderately responsive to (−)-carvone, p-tolualdehyde, allylbenzene and benzaldehyde. Interestingly, the receptor displayed some enantioselectivity and was less responsive to (+)-carvone. The responsiveness to allylbenzene indicated that a ketone, aldehyde or alcohol moiety was not essential for agonist activity at this receptor and suggested that the receptive range of MOR256-17 might be unusually broad.
Responsiveness to nitrotoluenes
In our initial screen, MOR256-17 was also highly responsive to 2,4-DNT, a nitrotoluene. 2,4-DNT is a minor manufacturing impurity in TNT preparations, but is a major component of emitted vapor (Jenkins et al. 2001; Leggett et al., 1977; Leggett et al., 2001). Consequently, 2,4-DNT is an attractive target for explosive detection efforts (Pinnaduwage et al., 2004; Radhika et al. 2007). To explore this aspect of MOR256-17 sensitivity, we screened a variety of nitrotoluenes, each at a concentration of 100 μM (Figure 4). Of the dinitrotoluenes, the receptor was most responsive to 2,4-DNT and was less responsiveness to 2,6-DNT, 2,3-DNT and 3,4-DNT, which yielded response amplitudes that were 41±6%, 25±8% and 27±7% of the response to 2,4-DNT, respectively. The receptor was also responsiveness to 2,4,6-trinitrotoluene (TNT) itself, albeit at a response amplitude 13±2% of the response to 2,4-DNT. While responses to 2-nitrotoluene (2-NT) were modest (31±8% of the 2,4-DNT response), the receptor was highly responsive to 3-NT and 4-NT. Concentration-response analysis for 3-NT yielded an EC50 of 37 ± 13 μM and a maximal response that was 1400 ± 163% of the maximal response to 2,4-DNT. 4-amino-2,6-DNT (4A-2,6-DNT) and 2-amino-4,6-DNT (2A-4,6-DNT), two major biological metabolites of TNT (Esteve-Nunez et al. 2001), were also able to activate MOR256-17.
MOR256-17 responds to numerous odorants in an expanded screening panel
The data that we have presented in Figures 1–4 suggests that MOR256-17 may be broadly tuned. In particular, the responsiveness of this receptor to allylbenzene and various nitrotoluenes indicates that odorant structures beyond ketones and aldehydes can be accommodated by the receptor. To assess how broadly tuned MOR256-17 might be, we screened with a large and diverse set of odorants. This panel included 155 odorants chosen from different chemical categories, including aldehydes, alcohols, ketones, esters, amines, carboxylic acids, heterocycles, hydrocarbons, and sulfur-containing compounds (Supplemental Table 3). This odorant panel is distributed across a large portion of odor space (Supplementary Figure 2). The panel was initially screened as a set of 8 mixtures, each containing 17–20 compounds, and all applied at a concentration of 30 μM. Several odorants that we have already screened against MOR256-17 were included as controls, including three odorants that yield large responses (2-heptanone, (+)-dihydrocarvone, and (−)-carvone) and several odorants that yield little (amyl acetate, anisole, anisyl alcohol, (+)-carvone, 2-pentanol, 2-heptanol, 2-pentanone, 3-heptanone) or no response (2-nonanol, phenylacetic acid). Figure 5 illustrates our screening approach. In Figure 5A, we found that MOR256-17 was responsive to 6 of the 8 mixtures. In contrast, MOR174-9 (mOR-EG), a receptor known to recognize eugenol and related structures, as well as several polycyclic structures (Kajiya et al. 2001, Katada et al. 2005, Baud et al. 2011), responded well to eugenol, but failed to respond to any of the odorant mixtures (Supplementary Figure 3). This confirmed that the broad responsiveness to multiple odorant mixtures that we observed with MOR256-17 is a function of this receptor and not an artifact of our assay system. Each active mixture was then divided into 4 sub-mixtures. For example, in Figure 5B we screened 4 sub-mixtures containing the components of mixture 8. Mixture components already known to be highly active, such as 2-heptanone (a component of mixture 8) were not included in the sub-mixtures and were applied separately. The components of active sub-mixtures were then applied individually (Figure 5C). Through this screening process, we identified a large number of additional agonists for MOR256-17 (Figure 6). The wide variety of odorants that activate MOR256-17 was apparent when these compounds were plotted within in an estimate of odor space (Figure 7A, E), providing further support for this receptor being broadly-tuned.
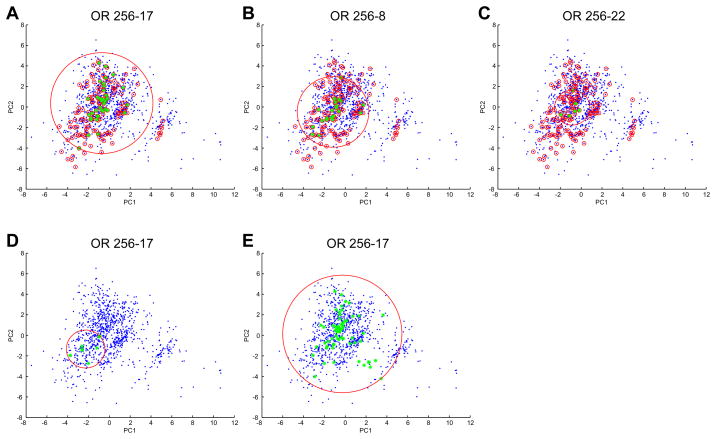
Figure 7. MOR256-17 is broadly tuned.
Odor space was estimated using 1035 odorants (blue dots) in a multi-dimensional odor space was based on 32 optimized physicochemical descriptors (Haddad et al., 2008) and first and second principal components were used to plot this odor space. A 32D hypersphere enclosing all 1035 odorants in this odor space has a radius of 18.5. The 155 odorants in our screening set are indicated by small red circles. The large red circle in each panel is a 2D representation of a 32D hypersphere that encloses the odorants that activate the receptor. A) Odorants from the screening set that activate MOR256-17 are indicated by green stars. A 32D hypersphere that encloses the odorants that activate MOR256-17 has a radius of 11.4. B) Odorants from the screening set that activate MOR256-8 are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 5.3. C) Odorants from the screening set that activate MOR256-22 are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 4. D) Odorants previously shown to activate MOR256-17 (Dahoun et al., 2011) are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 4.8. E) All odorants shown to activate MOR256-17 in this study are indicated by green stars. A 32D hypersphere that encloses these odorants has a radius of 16.4.
Other members of the MOR256 subfamily are more narrowly tuned
The broad tuning of MOR256-17, together with recent reports that MOR256-3 (SR1) and MOR256-31 (Olfr42) are broadly-tuned (Grosmaitre et al. 2009; Nara et al., 2011), suggested that other members of the MOR256 sub-family might also be broadly-tuned. To explore this possibility, we cloned and expressed several additional members of the MOR256 sub-family. MOR256-6, MOR256-8 and MOR256-22 were chosen to represent the various sub-groups (Godfrey et al. 2004) within this large (at least 37 members) MOR subfamily (Supplementary Fig. 4). Oocytes expressing each receptor were screened with the panel of 155 odorants, as in Figure 5. While MOR256-6 failed to respond to any odorant mixtures, MOR256-8 and MOR256-22 each responded to multiple mixtures. Upon screening with the sub-mixtures and individual odorants, we found that, in contrast to MOR256-17, MOR256-8 and MOR256-22 were not broadly tuned (Figure 6). MOR256-8 responds primarily to linear aliphatic aldehydes, alcohols and esters, with 2-pentylfuran being the only aromatic structure yielding even small responses. The MRR of MOR256-8 overlaps with that of MOR256-17 with responsiveness to aldehydes such as trans-2-hexenal and trans-2-heptenal, but diverges from the MOR256-17 MRR with responsiveness to alcohols such as geraniol and 1-nonanol. MOR256-22 was even more narrowly tuned, responding strongly to just 3 odorants (trans,trans-2,4-heptadienal, trans,trans-2,4-octadienal, trans-cinnamaldehyde) and displaying a small response to 1 odorant (cuminaldehyde). With the exception of cuminaldehyde, the MRR for MOR256-22 is a small subset of the much more extensive MRR of MOR256-17. Plotting the odorants that activate MOR256-8 and MOR256-22 within in an estimate of odor space (Figure 7B, C) revealed both receptors to be less broadly tuned than MOR256-17. In particular, MOR256-22 was much more narrowly tuned, suggesting that broad tuning is not a defining characteristic of the MOR256 subfamily.
Discussion
MOR256-17 has recently been characterized, to varying extents, by us and others (Dahoun et al. 2011, Goldsmith et al. 2011, Saito et al. 2009). Our interest in pursuing a more detailed examination of this receptor stemmed from indications that this receptor might be broadly tuned (Goldsmith et al. 2011, Saito et al. 2009) and that this receptor responded to 2,4-DNT (Goldsmith et al. 2011), a major component of vapor emitted from TNT based explosives (Leggett et al., 1977; Leggett et al., 2001 (Jenkins et al. 2001). There have been only a few detailed studies of broadly tuned ORs (Grosmaitre et al. 2009, Saito et al. 2009) and there is obvious utility in characterizing ORs that can detect explosives (Corcelli et al. 2010, Goldsmith et al. 2011, Radhika et al. 2007).
Our results demonstrate that MOR256-17 responds to a large number of odorant ligands that seem to fall into many distinct structural categories. Does this mean that MOR256-17 is broadly tuned? The lack of a simple metric with which to quantify odorant similarity makes answering this question difficult. The breadth of OR tuning has often been defined simply by the number of odorants in a screening panel to which the OR responds. This approach can be problematic if many of the active odorants are obviously (or non-obviously) related. Recent more quantitative approaches have used large sets of physicochemical descriptors to define odorant similarity (Haddad et al. 2008, Saito et al. 2009). For example, in Haddad et al (2008) a multidimensional metric was used, in which each odorant molecule was represented as a vector of 1,664 physicochemical descriptor values. An optimized set of 32 descriptors was then used to examine the distribution of odorants in a multi-dimensional odor space. Using this approach to estimate the extent to which odorants that activate MOR256-17 are distributed across odor space, we found that MOR256-17 was responsive to a broadly distributed set of odorant structures that span a large portion of the odor space (Figure 7E). Using similar analyses, Saito et al. (2009) proposed that the tuning breadths of mammalian ORs vary along a continuum, from narrowly tuned to broadly tuned. Our results place MOR256-17 at the broadly tuned end of the spectrum.
Characterization of the MRR of MOR256-17 has been recently reported (Dahoun et al. 2011). To the extent that we can compare our results with those of Dahoun et al. (2011), we find good agreement. For example, we observed receptor responsiveness to (+)-carvone and (−)-carvone, but not to cyclohexanone, (−)-menthone, D-camphor or P-cymene. We did not observe responses to (+)-limonene or (−)-limonene, while Dahoun et al. (2011) did observe responsiveness to these odorants. However, the responses to (+)-limonene and (−)-limonene reported by Dahoun et al. (2011) only occurred at concentrations above the 30 μM used in our screening protocol. When we examine the distribution of active odorants identified by Dahoun et al. (2011) in our estimate of odor space (Figure 7D), we find that these odorants cover a relatively small portion of odor space. In contrast, when we plot all of the active odorants that we have identified within our estimated odor space (Figure 7E), we find that the MRR of MOR256-17 encompasses a large portion of odor space.
Much of our screening of MOR256-17 has involved the use of odorants that are broadly, but sparsely, scattered across odor space (Supplementary Figure 2). However, in Figures 2–4 we examine the chemical space around particular odorants in detail. In each case, we find a group of related structures that can also activate the receptor. This suggests that each active odorant identified in our broad screen represents a “node” of activity, where multiple related active structures reside. This dense screening within very small regions of odor space also reveals that while MOR256-17 is capable of detecting a wide range of odorant structures, it is also capable of making very subtle distinctions among odorant structures. For example, MOR256-17 easily distinguishes between the two enantiomers of carvone. The receptor can also distinguish between 3-nitrotoluene and 4-nitrotoluene, as well as between 2,4-dinitrotoluene and 2,6-dinitrololuene. The receptor displays a strict length preference for linear, aliphatic odorants, preferring 6 and 7 carbon lengths, but can also make subtle distinctions between 2- and 3-ketones. Understanding how a receptor can recognize widely differing chemical structures, while also making subtle distinctions among closely related structures, is a major challenge in olfaction.
The MRRs of closely related members of several mammalian OR subfamilies have been examined at various levels of detail. For example, MOR174-9 (mOR-EG) and MOR174-4 (mOR-EV) are 78% identical at the amino acid level and both receptors responded to vanillin and ethyl vanillin (Kajiya et al. 2001). However, MOR174-9 also responded to a variety of additional compounds (such as eugenol) that did not activate MOR174-4. MOR171-2 (M71) responded to acetophenone and benzaldehyde, while MOR171-3 (M72), with 96% identity to MOR171-2 at the amino acid level, responded to acetophenone, but not benzaldehyde (Bozza et al. 2002, Feinstein et al. 2004). Human OR1A1 and OR1A2 share 83% identity and have been shown to have very similar MRRs, both responding to a variety of 7–10 carbon, aliphatic aldehydes and alcohols (Schmiedeberg et al. 2007). The one major difference between these two receptors was that while OR1A1 responds well to (S)-(−)-citronellol, OR1A2 does not. Finally, MOR42-1 and MOR42-3, with 89% amino acid identity, have highly overlapping MRRs, with both receptors responding to 8–12 carbon dicarboxylic acids (Abaffy et al. 2006). A third member of the MOR42 subfamily (MOR42-2) is much less closely related (55% and 57% identity with MOR42-1 and MOR42-3, respectively) and has a more divergent MRR (Abaffy et al. 2006). Similarly, the three MORs that we have examined here display a greater divergence at the amino acid level (54–57% identity) and a greater divergence in MRR. Interestingly, the divergence in MRR among MOR256-17, MOR256-8 and MOR256-22 is manifested not only in the particular odorants that are recognized, but also in the breadth of the MRR. These studies suggest that the degree to which MRRs overlap may positively correlate with the extent of amino acid identity of the ORs. A scheme for the functional organization of ORs, based on the side-chain properties at a subset of residue positions, has been proposed (Man et al., 2007). A systematic examination of a large receptor set, for which functional data was obtained, has demonstrated that the side chain character of residues at 16 positions can account for a substantial portion of the variance in odorant responsiveness (Saito et al., 2009).
MOR256-17 is of particular interest due to the ability to respond to a variety of nitrotoluenes, including various TNT synthesis intermediates and degradation products, as well as TNT itself. Such receptors may have value in biosensor applications. However, while MOR256-17 can detect TNT-based explosives, use of this receptor alone, with its broad tuning, would lead to many false positives. A more narrowly tuned and selective receptor would be useful, if it exists and can be identified. Alternatively, an array of receptors, ranging from broadly to narrowly tuned, may be able to achieve a useful level of specificity. If MOR256-17 were part of such an array, a signal from MOR256-17, but not from other ORs that overlap with various non-explosive portions of the MOR256-17 MRR, could constitute a positive signal for nitrotoluenes. The advent of nanoelectronic sensing devices containing functional mammalian ORs (Goldsmith et al. 2011) allows such sensing strategies to be contemplated.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health [RO1 DC008119 to CWL] and by the DARPA RealNose project.
We thank Ana Castro and Benjamin Sherman for excellent technical assistance, Tali Weiss for help with odor space calculations and Dr. Selvan Bavan for critical reading of the manuscript.
Abbreviations
- CFTR
cystic fibrosis transmembrane regulator
- DMSO
dimethylsulfoxide
- DNT
dinitrotoluene
- GPCR
G-protein coupled receptor
- MOR
mouse odorant receptor
- MRR
molecular receptive range
- NT
nitrotoluene
- OR
odorant receptor
- TNT
trinitrotoluene
Footnotes
The authors have no conflict of interest to declare.
References
- Abaffy T, Matsunami H, Luetje CW. Functional analysis of a mammalian odorant receptor subfamily. J Neurochem. 2006;97:1506–1518. doi: 10.1111/j.1471-4159.2006.03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- Bakalyar HA, Reed RR. Identification of a specialized adenylyl cyclase that may mediate odorant detection. Science. 1990;250:1403–1406. doi: 10.1126/science.2255909. [DOI] [PubMed] [Google Scholar]
- Baud O, Etter S, Spreafico M, Bordoli L, Schwede T, Vogel H, Pick H. The mouse eugenol odorant receptor: Structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry. 2011;50:843–853. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Corcelli A, Lobasso S, Lopalco P, Dibattista M, Araneda R, Peterlin Z, Firestein S. Detection of explosives by olfactory sensory neurons. J Hazard Mater. 2010;175:1096–1100. doi: 10.1016/j.jhazmat.2009.10.054. [DOI] [PubMed] [Google Scholar]
- Dahoun T, Grasso L, Vogel H, Pick H. Recombinant Expression and Functional Characterization of Mouse Olfactory Receptor mOR256-17 in Mammalian Cells. Biochemistry. 2011 doi: 10.1021/bi2008596. [DOI] [PubMed] [Google Scholar]
- Esteve-Nunez A, Caballero A, Ramos JL. Biological degradation of 2,4,6-trinitrotoluene. Microbiol Mol Biol Rev. 2001;65:335–352. doi: 10.1128/MMBR.65.3.335-352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Firestein S, Picco C, Menini A. The relation between stimulus and response in olfactory receptor cells of the tiger salamander. J Physiol. 1993;468:1–10. doi: 10.1113/jphysiol.1993.sp019756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A. 2004;101:2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith BR, Mitala JJ, Josue J, Castro A, Lerner MB, Bayburt TH, Khamis SM, Jones RA, Brand JG, Sligar SG, Luetje CW, Gelperin A, Rhodes PA, Discher BM, Johnson ATC. Biomimetic chemical sensors using nanoelectronic readout of olfactory receptor proteins. ACS Nano. 2011;5:5408–5416. doi: 10.1021/nn200489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29:14545–14552. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Khan R, Takahashi YK, Mori K, Harel D, Sobel N. A metric for odorant comparison. Nat Methods. 2008;5:425–429. doi: 10.1038/nmeth.1197. [DOI] [PubMed] [Google Scholar]
- Hadda R, Weiss T, Khan R, Nadler B, Mandairon N, Bensafi M, Schneidman E, Sobel N. Global features of neural activity in the olfactory system form a parallel code that predicts olfactory behavior and perception. J Neurosci. 2010;30:9017–9026. doi: 10.1523/JNEUROSCI.0398-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TF, Leggett DC, Miyares PH, Walsh ME, Ranney TA, Cragin JH, George V. Chemical signatures of TNT-filled land mines. Talanta. 2001;54:501–513. doi: 10.1016/s0039-9140(00)00547-6. [DOI] [PubMed] [Google Scholar]
- Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DC, Jenkins TF, Murrmann RP. Composition of vapors evolved from military TNT as influenced by temperature, solid composition, age and source. US Army CRREL Special Report 77-16 1977 [Google Scholar]
- Leggett DC, Cragin JH, Jenkins TF, Ranney T. Release of explosive related vapors from land mines. US Army ERDC/CRREL Technical Report TR-01-6 2001 [Google Scholar]
- Lowe G, Gold GH. Nonlinear amplification by calcium-dependent chloride channels in olfactory receptor cells. Nature. 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- Ma T, Vetrivel L, Yang H, Pedemonte N, Zegarra-Moran O, Galietta LJ, Verkman AS. High-affinity activators of cystic fibrosis transmembrane conductance regulator (CFTR) chloride conductance identified by high-throughput screening. J Biol Chem. 2002;277:37235–37241. doi: 10.1074/jbc.M205932200. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Man O, Willhite DC, Crasto CJ, Shepherd GM, Gilad Y. A framework for exploring functional variability in olfactory receptor genes. PLoS One. 2007;2:e682. doi: 10.1371/journal.pone.0000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Gold GH. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature. 1987;325:442–444. doi: 10.1038/325442a0. [DOI] [PubMed] [Google Scholar]
- Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31:9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnaduwage LA, Thundat T, Hawk JE, Hedden DL, Britt PF, Houser EJ, Stepnowski S, McGill RA, Bubb D. Detection of 2,4-dinitrotoluene using microcantilever sensors. Sensors and Actuators B. 2004;99:223–229. [Google Scholar]
- Radhika V, Proikas-Cezanne T, Jayaraman M, Onesime D, Ha JH, Dhanasekaran DN. Chemical sensing of DNT by engineered olfactory yeast strain. Nat Chem Biol. 2007;3:325–330. doi: 10.1038/nchembio882. [DOI] [PubMed] [Google Scholar]
- Reed RR. After the holy grail: establishing a molecular basis for Mammalian olfaction. Cell. 2004;116:329–336. doi: 10.1016/s0092-8674(04)00047-9. [DOI] [PubMed] [Google Scholar]
- Repicky SE, Luetje CW. Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids. J Neurochem. 2009;109:193–202. doi: 10.1111/j.1471-4159.2009.05925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BD, Katz LC. Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron. 1999;23:499–511. doi: 10.1016/s0896-6273(00)80803-x. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Schultz BD, Singh AK, Devor DC, Bridges RJ. Pharmacology of CFTR chloride channel activity. Physiol Rev. 1999;79:S109–144. doi: 10.1152/physrev.1999.79.1.S109. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Sicard G, Holley A. Receptor cell responses to odorants: similarities and differences among odorants. Brain Res. 1984;292:283–296. doi: 10.1016/0006-8993(84)90764-9. [DOI] [PubMed] [Google Scholar]
- Uezono Y, Bradley J, Min C, McCarty NA, Quick M, Riordan JR, Chavkin C, Zinn K, Lester HA, Davidson N. Receptors that couple to 2 classes of G proteins increase cAMP and activate CFTR expressed in Xenopus oocytes. Receptors Channels. 1993;1:233–241. [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. Functional expression of a mammalian odorant receptor. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.