Abstract
APOBEC3G (A3G) and APOBEC3F (A3F) reduce Vif-negative HIV-1 provirus formation and cause disabling provirus G-to-A hypermutation in vitro. However, evidence conflicts about whether they negatively impact Vif-positive HIV-1, or only enhance virus genetic diversity, in vivo. We studied peripheral blood mononuclear cells (PBMC) from 19 antiretroviral-naïve, HIV-infected adults: 12 long-term non-progressors (LTNP) and 7 non-controllers (NC). Cells from LTNP had higher A3G and A3F mRNA levels, lower provirus burden, and more A3G-hypermutated positions in provirus sequence than cells from NC. A3G mRNA level was directly associated with its Hypermutation Index (HI) and inversely associated with provirus burden. Plasma HIV-1 RNA levels were inversely associated with A3G expression levels and with HI only among subjects who had HI>1. A3G HI was not associated with provirus burden. These results indicate that A3G’s deaminase-dependent activity above a threshold level, and its deaminase-independent functions, contribute to decreasing Vif-positive virus replication in vivo.
Keywords: HIV-1, APOBEC3G, APOBEC3F, G-to-A hypermutations, long-term non-progressors, cytidine deaminase
Introduction
The APOBEC3 (A3) family of cytidine deaminases defends against retrotransposition (Esnault et al, 2005; Schumacher et al., 2006; Chiu et al., 2006) and are expressed in many cell types and tissues (Koning et al., 2009; Refsland et al., 2010). Some family members, particularly APOBEC3G (A3G) and APOBEC3F (A3F), also restrict Vif-negative human immunodeficiency virus type 1 (HIV-1) replication in vitro. HIV-1 Vif counteracts their antiviral activity, however (Cullen, 2006; Goila-Gaur and Strebel, 2008; Harris and Liddament, 2004; Sheehy et al., 2002). Vif limits the cellular content of A3s, and their activity in progeny viral particles, by accelerating A3 degradation, as well as perhaps by degradation-independent mechanisms (Marin et al., 2003; Mehle et al., 2004; Opi et al., 2007; Yu et al., 2003). A3G and A3F differ in their DNA targeting and mutational signatures, levels of expression, and sensitivity to Vif. Therefore, it is important to study effects on HIV-1 of G-to-A hypermutation caused by both deaminases (called ‘hypermutation’ here).
The presence of A3-mediated hypermutation in provirus DNA from many HIV-1-infected subjects (Kieffer et al, 2005; Pace et al., 2006) indicates that Vif-mediated protection from host-mediated proviral DNA deamination is not absolute in vivo. Hypermutation in vif (Simon et al., 2005) and env genes (Knoepfel et al., 2010) of proviruses in vivo have been reported. Some evidence is consistent with hypermutation partially restricting wild-type, Vif-positive HIV-1 replication in vivo in a minority of infected subjects (Pace et al., 2006; Land et al., 2008; Vazquez-Perez et al., 2009; Amoedo et al., 2011). Physiologically higher A3G function in Th1, relative to Th2, CD4+ T cells also decreased HIV-1 replication in vitro despite the presence of Vif, whether A3G was in the virion or in the target cell cytoplasm (Vetter et al., 2009). Since A3G and A3F also have non-deaminase mediated mechanisms of antiviral activity (Luo et al., 2007; Mangeat et al., 2003; Mbisa et al., 2010), provirus hypermutation may not be the only outcome of their antiviral activity.
An alternative hypothesis is that A3 activities are not extensive enough to impair Vif-positive HIV-1 replication in vivo. Some data suggest that hypermutation in provirus facilitates HIV-1 escape from immunological and pharmacological inhibition by increasing the pool of genetic diversity available for recombination (Simon et al., 2005; Mulder et al., 2008; Jern et al., 2009; Fourati et al., 2010; Kim et al., 2010; Sadler et al., 2010(Fourati et al., 2010; Kim et al., 2010). In this formulation, A3 activity provides a favorable advantage for Vif-positive HIV-1 and is not sufficient to block its replication. This hypothesis predicts that hypermutation is either directly associated with, or not associated with, virus replication.
There is potential for A3s to both impair and benefit HIV-1 replication (Smith, et al. 2011). Indeed, most studies of associations in vivo between measures of A3G and A3F on the one hand, and Vif-positive HIV-1 replication and immunodeficiency progression on the other hand, have supported an anti-HIV effect although some have conflicted. These studies also included different subject populations and used different metrics. Measures of A3G and/or A3F have involved their RNA levels with or without cellular activation (Cho et al., 2006; Jin et al., 2005), quantitation of hypermutation in cellular HIV-1 genomes (Pace, et al., 2006; Land et al., 2008; Ulenga et al., 2008a; Piantodosi et al. 2009; Vazquez-Perez et al., 2009; Amoedo et al., 2011) or both (Gandhi et al., 2008; Vazquez-Perez et al., 2009). Parameters of HIV-1 replication have included HIV-1 plasma viral load, blood CD4+ T cell count or immunodeficiency progression classification. Subjects have been compared across immunodeficiency progression categories such as elite suppressors who have stable CD4 cells with consistently undetectable HIV-1 viremia in absence of any antiretroviral therapy (ART), long-term non-progressors (LTNP), who have stable CD4 cells with, at most, low-level HIV-1 viremia in the absence of any ART, and untreated HIV-1 infected subjects with the more typical pace of progression (non-controllers, NC). Some studies only examined untreated subjects with typical progression (Cho et al., 2006; Piantadosi et al., 2009; Ulenga et al., 2008b), or compared those who spontaneously control HIV to subjects suppressed on HAART (Gandhi et al., 2008). Genetic variations in Vif (Alexander et al., 2002; Farrow et al., 2005; Pace et al., 2006) and A3G (An et al., 2004; Pace et al., 2006) have been associated with degree of hypermutation and/or HIV immunodeficiency progression. Several groups have recently identified lower provirus burden and cell-intrinsic mechanisms that limit HIV replication in elite suppressors (Graf et al., 2011; Saez-Cirion et al., 2011; Buzon et al., 2011); however, A3s were not evaluated in those controllers. We evaluated PBMC A3G and A3F mRNA levels and provirus hypermutation, and assessed their associations with cellular provirus burden in PBMCs and plasma HIV-1 RNA levels in subjects who either did (LTNP), or did not (non-controllers, NC), spontaneously control HIV-1. The results of this study extend earlier reports and may help explain their conflicting results.
Results
A3 mRNA levels and hypermutations in PBMCs from LTNP and NC subjects
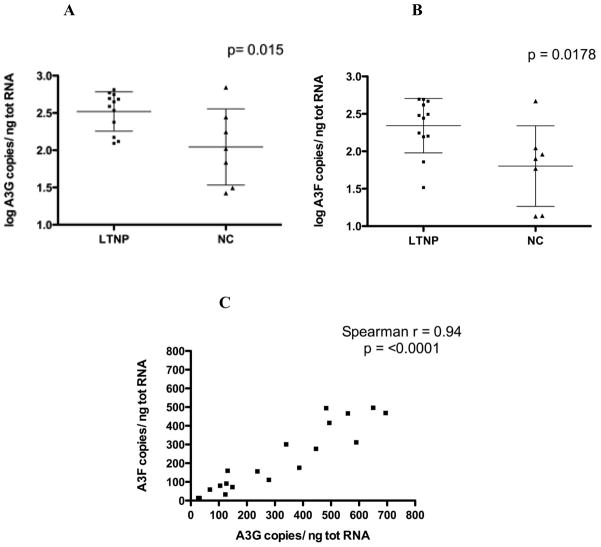
A3G and A3F mRNA levels were significantly higher in PBMC from the LTNP than in those from the NC subjects (Fig. 1A and B). Mean log transformed A3G RNA copies/ng total RNA was higher in the LTNP group than in the NC group (p=0.015, Student’s t test). Similarly, the mean A3F RNA level in LTNP subjects was higher than the mean among NC subjects (p=0.0178, Student’s t test). There was a strong and significant correlation between the expression of A3G and A3F (Spearman r = 0.94, p<0.0001), consistent with the known co-regulation of their transcription (Fig. 1C).
Figure 1. A3G and A3F expression levels are higher in peripheral blood mononuclear cells (PBMC) of long term non-progressor (LTNP) than non-controller (NC) subjects.
(A) A3G and (B) A3F RNA levels from 12 LTNP (squares) and 7 NC (triangles). Bars represent mean ± SD of values and p value is computed by Student’s t test. (C) A3G RNA copy number strongly and significantly correlates with A3F RNA copy number. Spearman correlation coefficient and p value are listed.
Population sequences of vif amplified from high molecular weight PBMC DNA of 17 subjects were used to determine the magnitude of A3-mediated cytidine deaminase enzymatic activity in vivo, relative to each subject’s reference sequence, represented by a non-hypermutated sequence from plasma HIV-1 RNA of the same subject. Sequences from two LTNP subjects with plasma HIV-1 RNA < 50 copies/ml were not evaluable. G-to-A mutations in both the A3G and A3F signature dinucleotide contexts, GG and GA respectively, were quantified as an overall Hypermutation Index (HI) that corrected for sequence length and RT error rate (estimated as A-to-G mutation) ((Kijak et al., 2007).
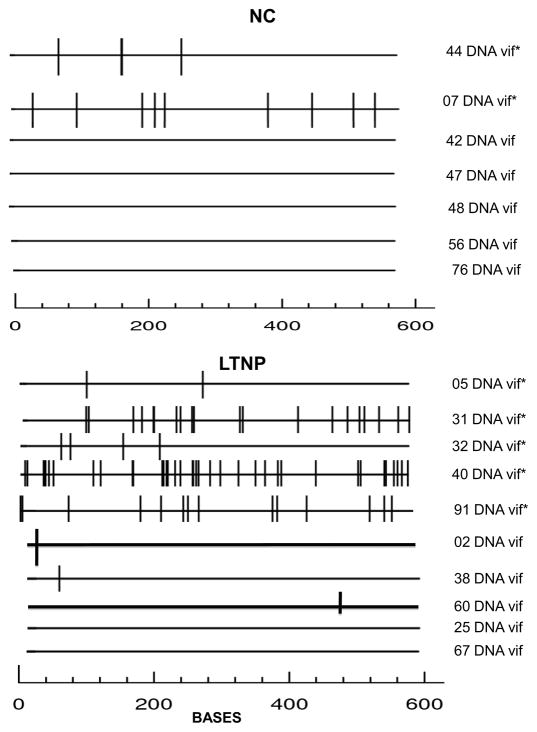
A3-mediated hypermutated positions in vif sequences were more numerous among the LTNP than the NC subjects (77 among the LTNP versus 12 among the NC subjects, p = 0.04, Mann-Whitney U) (Fig. 2). Five of 10 (50%) of the LTNP subjects had a dominant PBMC Vif sequence with a hypermutation index (HI >1) and premature stop codons, while 2 of 7 (29%) of NC had HI > 1 and premature stop codons (not significant, Fisher’s exact test) (Supplemental Fig. 1A–B) (Kijak et al, 2007; Kijak, et al. 2008). Analyses using Hypermut 2.0 software were consistent with the HI analyses in identifying the same 7 subjects (5 LTNP, 2 NC) with hypermutation relative to their own reference sequence. Population sequences of proviral PR and RT in a subset of these same subjects also showed a higher percentage of subjects with predominant G-to-A hypermutated provirus among LTNPs than NCs.
Figure 2. Alignments of vif proviral sequences from LTN and NC subjects indicate more APOBEC3-mediated hypermutations in some LTNP.
Population sequence for each subject was aligned to its own non-hypermutated reference sequence and analyzed by Hypermut 2.0 (Rose et al. 2000) and HyperPack (Kijack et al., 2007). Bars indicate positions with G-to-A substitutions in a context recognized by APOBEC3G or APOBEC3F. * in right column marks 7 sequences with a Hypermutation Index (HI) >1 (5 from LTNP and 2 from NC subjects).
Analyses of clonal sequences of vif were also performed from PBMCs of 3 of the subjects described above, including 2 LTNP (#40 and #31) and 1 NC (#44), and compared to results of the population sequencing. All 13 vif clones from one LTNP (#40) had very high HI (6.1 to 6.5), and the other LTNP (#31) had 27% of clones with HIs from 1.53 to 2.38 (Supplemental Fig 1C). Only one vif clone out of 18 (5%) sequenced from the NC (#44) subject had an HI > 1; this was one of the NC subjects identified with HI > 1 in the population PCR product analysis (Supplemental Figure 1C). The cloned PCR product sequences thus were in excellent agreement with HIs determined from population sequences for those subjects (Supplemental Figure 1A). Numbers of stop codons were also similar in both population (Supplemental Fig. 1B) and clonal sequences from each of these 3 subjects (Supplemental Fig. 1D). Because of this validation, further cloned PCR product analyses were not performed here.
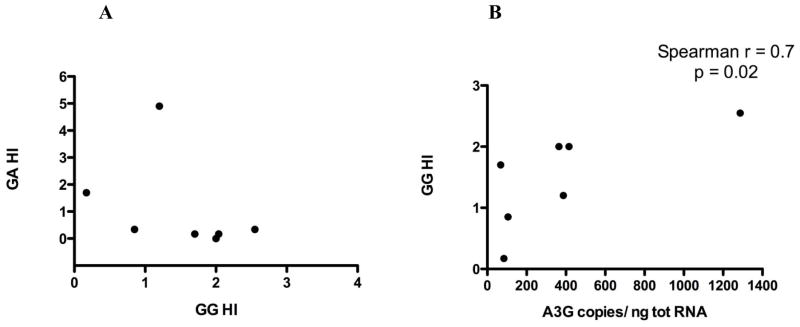
Hypermutations in the 7 subjects with an overall HI > 1 were also classified as either within GG dinucleotides targeted by A3G (GG HI) or within GA dinucleotides targeted by A3F (GA HI) (Fig. 3A). All 7 of these hypermutated sequences had A3G-specific mutations, with GG HI ranging from 0.85 to 2.5. A3G’s GG HI strongly correlated with A3G RNA expression in PBMCs (Spearman r=0.7, p=0.02) (Fig 3B). Thus, higher A3G expression was strongly and significantly associated with more hypermutation in provirus DNA, among those subjects in whom provirus hypermutation was detected. This analysis was not possible for A3F as A3F-specific mutations were additionally present in only 2 of these subjects who were both LTNP (Fig. 3A). It is of note that the highest magnitude of hypermutation observed here was a vif sequence with a GA HI of 5 from a LTNP subject.
Figure 3. Dinucleotide context of hypermutations in subjects with vif HI > 1.
(A) The 7 sequences with HI > 1 in proviral vif population sequences were further categorized as having hypermutations in the A3G dinucleotide context (GG HI) or the A3F dinucleotide context (GA HI). All 7 had some magnitude of GG HI (A3G-mediated hypermutation), and 2 also had some degree of GA HI (A3F-mediated hypermutation). (B) GG HI correlates with A3G RNA expression. Spearman correlation coefficient and p value are listed.
Among the subjects who had sequences of PR, RT, and vif genes analyzed, HIs generally increased as follows: PR<RT<vif sequences (Table 1). This is consistent with the previously described gradient in the HIV genome related to duration of a single-stranded DNA intermediate during reverse transcription (Yu et al., 2004; Suspene et al., 2006).
Table 1.
Hypermutation Index (HI) in PR, RT and vif population sequences.
| Subject | PR HI | RT HI | vif HI |
|---|---|---|---|
| LTNP 5 | 0.34 | 0.39 | 2.2 |
| LTNP31 | 1.7 | 0.39 | 3.4 |
| LTNP32 | 0 | 0.52 | 2.7 |
| LTNP40 | 2 | 6.46 | |
| LTNP91 | 0.67 | 1.18 | 2.2 |
| NC 7 | 1.35 | 1.44 | 1.36 |
| NC42 | 0.67 | 0.39 | 0.51 |
| NC44 | 1 | 0.39 | 2.04 |
A gradient with increasing HI across these genome segments is seen in most subjects, consistent with earlier reports that hypermutation increases, as follows: PR < RT < vif. HI >1 is indicated in bold.
Provirus burden in PBMCs of LTNP and NC subjects
We assessed HIV-1 provirus copy numbers in vivo by an alu-PCR assay modified from that described by O’Doherty et al. (O’Doherty et al., 2002). High molecular weight PBMC DNA was separated from nonintegrated viral DNA and used as the input template to enhance specificity for provirus. A detection limit of 1 copy per million cells allowed quantification of provirus in PBMCs from all 17 subjects who also had vif population sequence obtained. The amount of provirus in PBMC was quite stable over up to 15 months in 11 of the 13 subjects with specimens available from two time points, consistent with the known stability of provirus level (Supplemental Fig. 2).
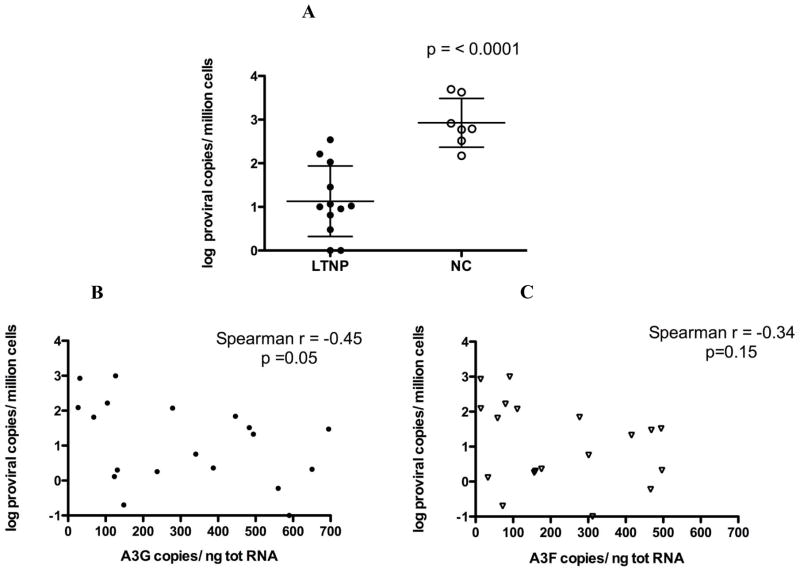
There was significantly less integrated provirus in PBMC of the LTNP than the NC subjects (Fig 4A, median = 10.2 versus 616 copies per million PBMC respectively, p<0.0001). Higher A3G mRNA levels in PBMCs correlated with fewer integration events (r= −0.45, p=0.05) (Fig. 4B). A3F mRNA levels did not correlate with provirus burden (r= −0.34, p=0.15) (Fig. 4C).
Figure 4. Alu-PCR quantitation of HIV provirus from high molecular weight PBMC DNA of LTNP and NC subjects.
(A) PBMC of the12 LTNP have significantly fewer copies of integrated HIV provirus than do PBMC of the 7 NC. Shown are mean ± SD for each group and p value computed by Student’s t test. (B) A3G RNA copy number correlates inversely with provirus copy number in PBMCs. (C) A3F RNA copy number does not correlate with provirus copy number in PBMCs. Spearman correlation coefficient and p value are listed in B and C.
Associations of plasma HIV-1 RNA levels with provirus, A3 expression, and hypermutation
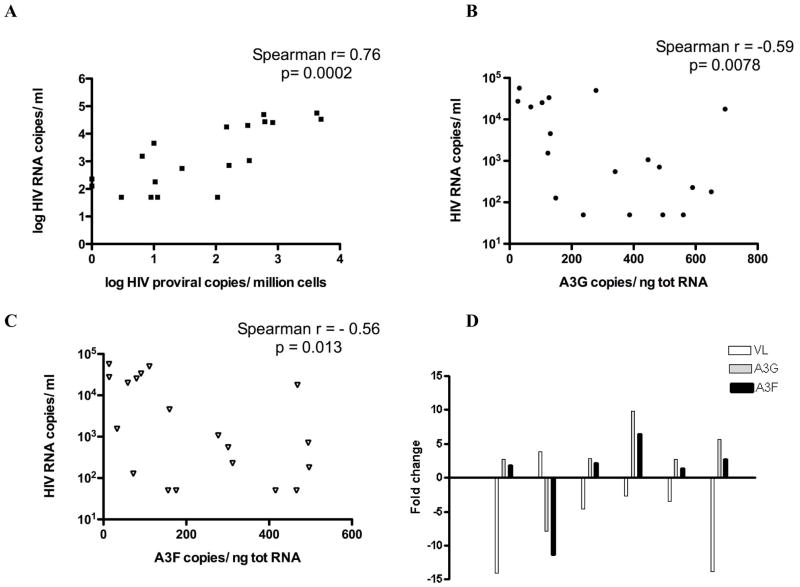
We also evaluated associations of plasma viremia level with provirus burden and A3 parameters. Provirus burden strongly and significantly correlated with the viral load in plasma (r=0.76, Spearman’s rank=0.0002) (Fig. 5A). A3G and A3F mRNA levels at the initial time point studied for each subject were inversely correlated with plasma HIV-1 RNA levels (A3G: r = −0.59; p= 0.0078; A3F: r = −0.56; p=0.013) (Fig 5B and C). Longitudinal changes in A3 transcript levels were assessed over a maximum of 15 months in 12 of the subjects who had specimens available at two time points. A3G and A3F levels did not vary over time in 6 out of 12 individuals’ cells. Changes in A3G RNA levels were found in the other 6, with substantial changes only in 3 (defined as more than a 5-fold increase or decrease mRNA level at the later time-point). These longitudinal variations were inversely associated with changes in plasma viral load in each subject (Fig 5D). This is consistent with A3 expression levels being inversely associated with viral load and supporting an A3 antiviral effect.
Figure 5. Associations of plasma HIV-1 RNA levels with provirus copies and A3G/F expression levels.
(A) HIV plasma viral load is directly associated with integrated proviral copy number. Values are log-transformed because of the wide range. (B) Plasma HIV RNA copy number is inversely correlated with A3G RNA copy number in PBMCs. (C) Plasma HIV RNA copy number is inversely correlated with A3F RNA copy number in PBMCs. APOBEC3G/F RNA expression was determined by qRT-PCR and expressed as A3G/F RNA copies per ng RNA in B and C. Spearman correlation coefficient and p value are listed for A, B and C. (D) Longitudinal changes in A3G and F RNA and plasma viral load were studied in 6 subjects who had specimens available at 2 time-points within 15 months. In each case, changes in A3G/F RNA were in the direction opposite to changes in viral load.
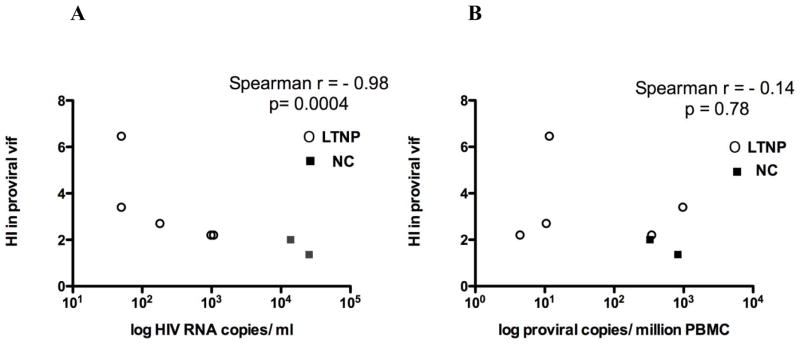
Plasma viral load was also strongly inversely correlated with the overall HI of vif population sequences, the measure of A3-specific hypermutations in proviruses (r= −0.98, Spearman p=0.0004) (Fig. 6A). This is consistent with a physiological antiviral effect of A3G cytidine deaminase-mediated editing of provirus. However, if the analysis was performed including all subjects, counting also those with GG HI<1, the correlation between HI and plasma HIV-1 RNA level was not significant (data not shown). This result support the hypothesis that A3G hypermutation is associated with viral load only if it is above a threshold level, which is present in some and not all subjects.
Figure 6. Hypermutation Index (HI) in proviral vif population sequences determined using HyperPack and measures of HIV burden among the 7 subjects with HI > 1.
(A) HI strongly correlates inversely with plasma viral load among subjects with HI > 1 in proviral vif population sequences. LTNP sequences are depicted by open circles; NC sequences by closed squares. Spearman correlation coefficient is -0.98 and p value = 0.0004. In contrast, no significant correlation was seen when all 17 subjects, with and without HI > 1 were included in the analysis. (B) Provirus burden was not significantly associated with vif HI either among subjects with HI > 1 or all subjects (latter not shown). LTNP sequences depicted by open circles, NC sequences by closed squares. Spearman correlation coefficient and p value are listed.
The overall HI of vif population sequences for subjects with HI > 1, in contrast, did not correlate with provirus burden (r= −0.14, p = 0.78) (Fig. 6B). These results suggest the hypothesis that a non-deaminase mediated activity of A3G, and not its cytidine deaminase enzyme activity, is responsible for the inverse association between A3G expression and provirus burden.
Hypermutation is not associated with amino acid polymorphisms in Vif functional motifs
Distinct motifs in Vif have been mapped that are critical for its interaction with A3G and/or A3F that leads to A3 degradation (Dang et al., 2009; Donahue et al., 2008; Goila-Gaur and Strebel, 2008; He et al., 2008; Iwabu et al., 2010; Russell and Pathak, 2007). We analyzed a putative translation of the non-hypermutated reference HIV-1 vif sequences from each subject, which represented their dominant replication-competent genome, for amino acid polymorphisms in regions critical for Vif suppression of A3G and/or A3F. This determined if Vif variation in its capacity to degrade A3s was associated with the observed magnitude of hypermutation. Vif SOCS Box (144SLQYLA149) and Zn-binding motifs that are critical for degrading A3G and A3F by bridging to the cellular E3 ubiquitin ligase complex (Yu et al., 2003) were conserved in all non-hypermutated sequences. The Vif multimerization motif 161PPLP164, which is necessary for A3G binding and degradation (Bernacchi et al., 2011; Miller et al., 2007), was also preserved in all sequences. We also did not identify amino acid polymorphisms in other motifs involved in Vif binding to A3G or A3F, including 21WxSLVK26 and 40YRHHY44, that are involved in interaction with A3G, and 11WxxDRMR17 and 74TGERxW79, that are essential for binding to A3F (Tian C, 2006). Polymorphisms in A3G binding sites were seen in 2 single clonal sequences of vif from one LTNP (LTNP 31) (K26I and Y44H), but neither of these polymorphisms in vif was present in the population-based sequence or any of the other 36 non-hypermutated clones from that subject.
Discussion
The current results add support for an antiviral effect of A3G against Vif-positive HIV-1 in vivo in certain circumstances and help explain earlier conflicting results. An association with plasma HIV-1 RNA level was not seen here when all levels of A3-mediated hypermutation were analyzed, including subjects with no detectable hypermutation in population sequences. However, A3-mediated hypermutations in provirus DNA above a threshold level (HI > 1) were inversely associated with plasma viral load. Our longitudinal analysis of patient specimens over time also adds evidence supporting an inverse association between A3G and viremia level. The present study also extended analyses to provirus burden. A3G, and not A3F, expression was inversely associated with provirus burden. Hypermutation did not account for this latter association, consistent with the extant hypothesis that the deaminase-independent activities of A3G affect provirus formation. Our results are not consistent with the alternative hypothesis that A3 activity only adds genetic diversity that benefits replication of Vif-positive HIV-1 quasi-species without impairing replication (Simon et al., 2005; Mulder et al., 2008; Jern et al., 2009; Fourati et al., 2010; Kim et al., 2010).
It has been hypothesized that if greater functional A3 activities provide a selective advantage against Vif-positive HIV-1 quasi-species in vivo that is manifest over time, the distribution of A3 mRNA levels will be shifted toward higher levels in those untreated subjects who survive longer (Jin, et al., 2007). Indeed, if we consider the higher CD4 cell count median of the LTNP subjects (614 cells/μl) compared to that of the NC subjects (459 cells/μl) (Mann-Whitney U test, p=0.03) as a surrogate of duration of infection, this does suggest an expected longer survival of the LTNP subjects, compared to the NC subjects in this study. Thus, these results are consistent with Jin et al.’s hypothesis (Jin et al., 2007).
Although RNA levels of A3F were similar to, and highly correlated with, those of A3G, the predominance of A3G-targeted hypermutation suggests that either A3F protein levels or deaminase activity were lower than those of A3G. Only one subject, a LTNP, had hypermutation above that threshold level of HI>1 in the provirus dinucleotide targets of A3F as well as A3G. However, this one subject had the greatest magnitude of hypermutation of all the subjects studied, and also had markedly high A3F mRNA levels. One subject in an earlier report had a similar A3F predominance (Kiljak et al., 2008). The observed association of A3F RNA with A3G RNA might explain the finding that A3F expression was associated with viremia here. However, our data cannot exclude deaminase-independent effects of A3F on viremia that are separate from those of A3G.
A3G and A3F are found either in a low molecular mass form or in a high molecular mass complex in cells. Evidence suggests that virion A3G is packaged from the low molecular mass form (Soros et al., 2007). Further work will be needed to determine if increases in one specific form are responsible for the inverse association with HIV-1 replication, or if A3G differs from A3F in the proportion of each form at higher cellular levels of expression.
Earlier studies also suggested antiviral effects of A3s against Vif-positive HIV-1 in vivo. Jin et al. reported an inverse association between A3G RNA levels in PBMCs activated ex vivo by anti-CD3/CD28 antibodies and plasma HIV RNA levels in their analysis of 8 LTNP and 17 progressors who were antiretroviral naive (Jin et al., 2005). They also found a positive association between A3G expression and CD4 cell count (Jin et al., 2005). Ulenga et al. also found that A3G mRNA levels were higher among infected subjects with a low viral load set-point than those with a high viral load set-point (Ulenga et al., 2008). Associations of A3G-mediated PBMC DNA hypermutation with plasma HIV-1 RNA or CD4+ T cell level consistent with an antiviral effect were also identified (Pace et al. 2006; Land et al., 2008; Vazquez-Perez et al., 2009).
In our analyses, non-controllers had lower levels of A3 expression and fewer A3-hypermutated bases (after controlling for the number of bases sequenced and the estimated rate of RT-mediated mutation) than did subjects with spontaneous control of HIV viremia. Adding data from subjects with HI < 1 to our analysis of the association of plasma viral load with provirus hypermutation led to loss of the significant effect seen when the analysis was limited to subjects with HI > 1. This may help explain why some reports did not identify an association with A3G expression (Cho et al., 2006) or hypermutation (Piantadosi et al., 2009; Ulenga et al., 2008b). Each of those studies included unselected, antiretroviral naïve subjects among whom spontaneous controllers might be infrequently found. Studying predominantly non-controllers may have obscured an association.
Sequences of vif from high molecular weight PBMC DNA of LTNP subjects had a greater mean number of hypermutated bases in A3G target sequences than did sequences from NC subjects (determined using Hyperpack software) (Kijak et al. 2007; Kijak et al. 2008). These results extends findings of earlier comparisons of controllers versus non-controllers that associated hypermutation with spontaneous control of viremia (Vazquez-Perez et al., 2009; Pace et al., 2006; Land et al., 2008) with a more quantitative analysis comparing number of hypermutated positions. Another report found more HIV-infected children with substantial PBMC hypermutation among controllers than the non-controllers studied, although the difference in numbers of subjects was not statistically significant in that small sample with a predominance of A3F-mediated hypermutation; number of hypermutated positions was not compared (Amoedo et al., 2011). As in that report, the larger number of LTNP than NC subjects with HI > 1 did not reach statistical significance here. We speculate that the similarly small number of subjects in each of these studies limited power (6 LTNP and 11 NC in Amoedo et al, 2011; 12 LTNP and 7 NC in the present study).
In the current report, we compared analyses of population PCR product sequences to that of molecular clones of PCR products from 3 subjects. Almost all earlier studies of this topic used population PCR product sequences (Pace et al. 2006; Land et al., 2008; Vazquez-Perez et al., 2009; Ulenga et al., 2008b); only one of the earlier reports analyzed 10 clones of PCR products from each subject (Amoedo et al, 2011) and one sequenced multiple single copy amplicons from each subject (Piantadosi et al., 2009). As in the earlier report sequencing single copy amplicons that found a range of 5 to 43% hypermutated sequences (Piantadosi et al., 2009), we found 5 to 27% of cloned amplicons from high molecular weight PBMC DNA with HIs > 1. This frequency of hypermutated variants was detectable with population sequencing. Clonal sequence results thus closely agreed with, and validated the population PCR product sequencing. Because of this close agreement and detectable frequency of hypermutated variants, clonal sequencing was not performed on all subjects studied here. This may be a limitation of the current study.
An earlier analysis of A3-mediated hypermutation did not find a difference between untreated elite suppressor (ES) subjects versus subjects who were well suppressed on HAART (Gandhi et al. 2008). This earlier work did not compare hypermutation in HIV DNA in untreated subjects who spontaneously controlled viremia to untreated subjects without such control of viremia, as did our study and the others cited.
It has been suggested that hypermutation in proviruses may be more obscured by greater ongoing replication of HIV and that “the ease of detection of hypermutated HIV-1 is likely a consequence, rather than a cause, of LTNP status” (Simon et al., 2005). Indeed, linear and episomal forms of lower molecular weight, unintegrated HIV-1 DNA that are by-products of productive replication are 10-fold more abundant than integrated provirus in cells from untreated subjects (Koelsch et al., 2008). Physical separation of high molecular weight DNA prior to amplification for population sequence analyses was done here to minimize the possibility of artifactual underestimation of hypermutation in the untreated NC subjects with presumed greater replication, based on higher viremia levels, than the LTNP subjects. The GG HIs in vif population sequences were also strongly correlated with A3G expression levels (Fig. 3B), which would not be expected if the HI differences were an artifactual consequence of relatively higher levels of unintegrated DNA in NC subjects than LTNP subjects.
LTNP in the present study also had decreased provirus burden, relative to NC, in vivo. The latter finding is consistent with three recent reports (Buzon et al. 2011; Graf et al., 2011; Saez-Cirion et al., 2011) and was not studied in earlier work on A3s in vivo. Using an alu-PCR assay from physically-separated genomic PBMC DNA that is sensitive and specific for integrated genomes, a nearly 2-log difference in the median integrated provirus levels between the cells from LTNP and NC subjects was noted here. The amount of integrated HIV measured by alu-PCR here was stable over two time-points within 15 months apart. Changes in the amount of proviral DNA over time were detected in only 2 of the 13 subjects (1 LTNP and 1 NC). DNA from both of them harbored high levels of hypermutations, which suggests that interference with PCR primer annealing may have contributed to variability in repeated provirus estimates.
Increased A3G, but not A3F expression, was significantly associated with decreased provirus burden. The degree of hypermutation was not significantly associated with provirus burden. This supports the hypothesis raised by in vitro studies (Luo et al., 2007; Mbias et al., 2010) that A3Gs’ deaminase-independent activity contributes to the confirmed cell-intrinsic block to HIV-1 provirus formation in PBMCs of LTNPs in vivo and ex vivo (Graf et al., 2011; Saez-Cirion et al. 2011; Buzon et al. 2011). Provirus burden was directly associated with plasma viral load, consistent with its role as a key measure of HIV replication. Since provirus GG HI > 1 was inversely associated with plasma viral load, both types of A3G’s functional activities (deaminase-independent and deaminase-dependent) can contribute to spontaneous control of viremia.
One of a number of factors may contribute in different individuals to spontaneous control of viremia. These factors include innate or acquired immune responses, defective viruses, defective host co-receptors and other host factors (Deeks and Walker, 2007; Lambotte et al., 2005). Our findings suggest that increased activity of A3G/F is one factor that can contribute to spontaneous control of HIV-1 in vivo.
Either decreased Vif levels/activity or increased A3 levels/activity could shift the balance in favor of deaminase-independent and -dependent A3 activities in the infected cell. Although decreased Vif activity due to mutation has been reported in clinical isolates and associated with spontaneous control of HIV-1 in case reports (Simon et al., 2005; Alexander et al. 2002; Farrow et al. 2005), mutations were not identified in residues associated with Vif function in any of the open Vif reading frames in replication competent HIV genomes in our subjects. This, the higher A3G and A3F expression levels in LTNP PBMCs relative to NC PBMCs, and the well established variation in expression levels of A3 family members in different tissues (Koning et al, 2009; Refsland et al., 2010), suggests that physiologically increased cellular A3 levels and activities can provide therapeutic benefit against HIV-1 harboring functionally competent Vif in vivo. A recent report that APOBEC3 induction during interferon-alpha treatment of subjects coinfected with hepatitis C virus and HIV-1 was correlated with degree of HIV DNA hypermutation adds support for this concept (Pillai et al., 2012). In conclusion, our study indicates that the higher expression levels, and both deaminase-dependent and -independent activities, of the host restriction factor A3G can contribute to long-term, spontaneous control of viremia in some individuals.
Materials and Methods
Study subjects and specimens
The study included 19 antiretroviral-naïve HIV-1 positive individuals with plasma viral loads ranging from below 50 copies/ml to 56,841 copies/ml. The Vanderbilt Meharry CFAR HIV Immunopathogenesis Core provided cryopreserved PBMC from all subjects. Twelve subjects were categorized as long-term non-progressors (LTNP) because plasma viral load was consistently < 5,000 copies/ml over the prior 5 years. Seven subjects were non-controllers (NC) with plasma VL > 10,000 copies/ml. Median (and IQR) VL for LTNP and NC were 229 copies/ml (50–911) and 27,400 copies/ml (17690–49770), respectively. Median (and IQR) CD4 counts for LTNP and NC were 614/μl (527–1023) and 459/μl (300–553), respectively. Median age was 44 yrs (31–49) for LTNP and 47 yrs (41–55) for NC. Specimens were available from 2 time-points within 15 months from 9 of the LTNP and 4 of the NC.
RNA isolation and Real-Time PCR (qRT-PCR)
Viral RNA was isolated from frozen plasma samples according to the instructions using QIAamp Viral RNA Kit (Qiagen, Valencia, CA). Samples with viral load < 1000 copies/ml were concentrated by centrifugation at 25,000g for 1hr before RNA isolation. Cellular RNA was isolated from PBMC using RNeasy kit (Qiagen, Valencia, CA) and quantified with RiboGreen RNA Quantitation Reagent and Kit (Molecular Probes, Eugene, OR). After normalization by RNA concentration, A3G/A3F transcripts were quantified by TaqMan qRT-PCR (Applied Biosystems Prism 7000 Sequence Detection System, Foster City, CA) using probes and primers developed in our laboratory (Vetter et al., 2009). Previous validation of the real-time PCR showed < 2.8 fold intra- and inter- assay variation. Values are expressed as copies of transcripts normalized by 18S and total RNA amount. It was previously noted that oligo-dT primed cDNA synthesis for quantitative RNA PCR can underestimate A3F RNA quantity (Refsland et al., 2010; Amoedo et al., 2011). In the present study, a specific primer was used to avoid this known problem in quantifying A3F RNA.
Quantification of HIV-1 provirus
HIV-1 proviral DNA was isolated from 1 million PBMC after separating chromosomal and viral unintegrated DNA in the presence of 1M NaCl and 0.6% SDS (Hirt, 1967). Cellular genomic DNA was further purified with the Genomic DNA Purification Kit (Gentra Systems, Minneapolis, MN). A modified 2-step Alu PCR assay quantified integrated proviral copy numbers (O’Doherty et al., 2002). In the first step, integrated HIV-1 sequences were amplified with a mix of genomic Alu primers (5′-TCC CAG CTA CTC GGG AGG CTG AGG and 5′-GCC TCC CAA AGT GCT GGG ATT ACA G) and HIV-1 PBS primer (5′-TTT CAG GTC CCT GTT CGG GCG CCA) using 180 ng of human genomic DNA, 200 nM of each dNTP, and 2 U of rTth DNA polymerase (Applied Biosystems, Foster City, CA). The reaction was run initially for 3 min at 94°C and for 15 cycles of 30 s at 94°C, 30 s at 65°C and 3 min at 70°C, with a final extension for 10 min at 72°C. Negative and background one way control amplifications were performed either with no primers, or with only HIV-1 PBS primer. ACH-2 cells were used as HIV-1 DNA copy number standard in a constant DNA background. DNA prepared from CEM cells mixed with graded doses of ACH-2 cells (NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) was amplified with the primers used in the first step and used for the standards in the second quantitative PCR step. To increase the sensitivity and accuracy of the assay, 6 Alu-PCR reactions were performed with each DNA sample. The first-step amplicon mix was diluted 50-fold for the second step real-time PCR to quantify HIV LTR sequences. This used TaqMan Universal PCR Master Mix (Applied Biosystems Foster City, CA,) with 2 μl of the diluted first amplification product, 500 nM of each primer (forward primer: 5′-CCT GGG AGC TCT CTG GCT AA-3′, reverse primer: 5′-GCA CTC AAG GCA AGC TTT ATT GA-3′) and 100 nM probe (6FAM-TAGGGAACCCACTGCTTA-MGBNFQ). The template was denatured for 10 min at 95°C followed by 40 cycles consisting of 15 s at 95°C and 1 min at 58°C using ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). The coefficient of variation (CV) between replicates in the second step was < 50%.
Sequencing
Plasma HIV-1 RNA was reversed-transcribed with OmniScript RT (Qiagen, Valencia, CA) using specific primers for the region of interest. Nested PCRs were performed to separately amplify both the 999-bp fragment flanking Vif and the 1307-bp Pol region including protease and part of RT. Primers were designed to anneal to sites lacking A3G/F consensus targets and primer mixes including matches to both hypermutated and non-hypermutated positions were used when that was not possible. For population-based sequencing, PCR amplicons were directly sequenced using ABI Prism BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and specifically designed inner PCR primers. For clonal sequencing, PCR products were TA cloned into the TOPO-2.1 vector (Invitrogen, Carlsbad, CA) and 10–15 clones were sequenced for each sample. ABI Prism 3100 genetic analyzer was used for all sequencing and resulting electropherograms were assembled using Sequencher 4.8 software (Gene Codes Corp, Ann Arbor, MI). GeneBank accession numbers are JQ409018 to JQ409041.
Hypermutation analysis
The extent of hypermutation was quantified by 3 different parameters: hypermutation index (HI) using Hyperpack, a statistically significant difference from the level in the reference sequence determined with Hypermut 2.0, and premature stop codons in HIV-1 open reading frames due to G to A mutation using Hyperpack (Rose, et al., 2000; Kijak et al., 2007; Kijak et al., 2008). Hyperpack calculates a hypermutation index (HI) as G-to-A substitutions per base pair minus the A-to-G substitutions per base pair divided by the sequence length in base pairs. This uses the level of A-to-G substitution to correct for background mutation rate due to factors other than A3 activity. Dinucleotide contexts of G-to-A hypermutations were also analyzed with the Hyperpack software (Kijak et al., 2007). GG HI and GA HI were determined by the levels of GG to AG and GA to AA substitutions respectively per 100 bp sequences. G-to-A hypermutation was also analyzed using Hypermut 2.0 (Rose et al., 2000 and www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) which compares the hypermutated sequence to a reference sequence and evaluates statistical significance of a difference from the reference by Fisher’s exact test. It does not specifically compare G-to-A changes to A-to-G changes to adjust for background mutation, nor does it normalize number of mutated G’s over a specific number of bases.
G to A mutations were not found in sequences derived from plasma HIV-1 RNA, as others have noted (Kieffer et al., 2005; Land et al., 2008). Therefore, we used a non-hypermutated sequence from plasma HIV-1 RNA from the same subject as a comparison, except for one subject, a LTNP with undetectable viremia, for whom a non-hypermutated sequence derived from PBMC proviral DNA was used. The non-hypermutated reference sequence for each subject was confirmed to be phylogenetically related to the same parental virus as the hypermutated variants. Hypermutation was greater by both HI and the Hypermut analysis criteria when the single reference HXB2 sequence, rather than an indivdual subject’s reference, was used (data not shown).
Statistical analysis
Correlations were assessed by Spearman’s rank correlation coefficients. Two-tailed, unpaired Student’s t test compared distributions of continuous variables between two groups with statistical significance at p < 0.05. Medians of hypermutated positions in vif sequences were compared between LTNP and NC subjects by one-tailed Mann-Whitney U test with statistical significance at p< 0.05 because the hypothesis suggested by the majority of earlier studies was that there would be more hypermutated positions among LTNP subjects and this was supported by our results showing higher A3 expression among LTNP subjects. Fisher’s exact test compared the number of subjects with HI>1 among LTNP versus NC.
Supplementary Material
(A) Hypermutation index (HI) in vif population sequences from PBMC high molecular weight DNA of 10 LTNP (circles) and 7 NC (filled triangles) subjects. Subjects that were cloned are labeled. (B) Number of stop codons introduced in PBMC vif population sequences from LTNP (circles) and NC (filled triangles) subjects by G-to-A hypermutation.. (C) HI in PBMC vif clonal sequences of subjects LTNP 31 (filled circles), LTNP 40 (open circles), and NC 44 (filled triangles). 12–16 clones were sequenced for each subject. Gray symbols represent clones with HI>1 (D) Number of stop codons introduced in PBMC vif clonal sequences of subjects LTNP 31 (filled circles), LTNP 40 (open circles) and NC 44 (filled triangles) by G-to-A hypermutation.
There was no significant variation of HIV integrated copies between 2 time points in 13 subjects, 9 LTNP (filled circles) and 4 NC (open circles), who had longitudinal specimens collected within a 15-months period (Paired t test: p=0.8).
Highlights.
A3G mRNA levels strongly correlates to A3G hypermutation index in proviral Vif.
Mutational activity of A3G and A3F is higher in cells of LTNP in comparison to NC.
A3G mRNA levels in PBMCs correlates with fewer integration events.
A3G and A3F RNA levels in PBMC are inversely associated with HIV-1 viral load.
A3G hypermutation index correlates inversely with plasma viral load.
Acknowledgments
We thank Lorraine Sutton for help with sequencing, Gustavo Kijak for providing the Hyperpack software and Ambra Pozzi for critically reading the manuscript. This work was supported by Vanderbilt-Meharry Center for AIDS Research (CFAR) Developmental Core Awards (to YK and MPD) and has been facilitated by the infrastructure and resources provided by the Vanderbilt-Meharry CFAR, an NIH funded program # P30 AI 54999. Funding from an investigator-initiated grant from Merck to RD is also appreciated. We are thankful to the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH for providing ACH-2 cells.
Footnotes
Disclosure statement
Each of the authors acknowledges that they do not have any conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander L, Aquino-DeJesus MJ, Chan M, Andiman WA. Inhibition of human immunodeficiency virus type 1 (HIV-1) replication by a two-amino-acid insertion in HIV-1 Vif from a nonprogressing mother and child. J Virol. 2002;76:10533–10539. doi: 10.1128/JVI.76.20.10533-10539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoedo ND, Afonso AO, Cunha SM, Oliveira RH, Machado ES, Soares MA. Expression of APOBEC3G/3F and G-to-A hypermutation levels in HIV-1-infected children with different profiles of disease progression. PLoS One. 2011;6:e24118. doi: 10.1371/journal.pone.0024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S, Goedert JJ, Phair J, Buchbinder S, O’Brien SJ, Telenti A, Winkler CA. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S, Mercenne G, Tournaire C, Marquet R, Paillart JC. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2011;39:2404–2415. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, Seiss K, Weiss R, Brass AL, Rosenberg ES, Pereyra F, Yu XG, Lichterfeld M. Inhibition of HIV-1 integration in ex vivo-infected CD4 T cells from elite controllers. J Virol. 2011;85:9646–50. doi: 10.1128/JVI.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, Davidson NO. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Donahue JP, Vetter ML, Mukhtar NA, D’Aquila RT. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Farrow MA, Somasundaran M, Zhang C, Gabuzda D, Sullivan JL, Greenough TC. Nuclear localization of HIV type 1 Vif isolated from a long-term asymptomatic individual and potential role in virus attenuation. AIDS Res Hum Retroviruses. 2005;21:565–574. doi: 10.1089/aid.2005.21.565. [DOI] [PubMed] [Google Scholar]
- Fourati S, Malet I, Binka M, Boukobza S, Wirden M, Sayon S, Simon A, Katlama C, Simon V, Calvez V, Marcelin AG. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. Aids. 2010;24:2313–2321. doi: 10.1097/QAD.0b013e32833e515a. [DOI] [PubMed] [Google Scholar]
- Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf EH, Mexas AM, Yu JJ, Shaheen F, Liszewski MK, Di Mascio M, Migueles SA, Connors M, O’Doherty U. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathogens. 2011;7(2):e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. Journal of molecular biology. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. Journal of molecular biology. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Iwabu Y, Kinomoto M, Tatsumi M, Fujita H, Shimura M, Tanaka Y, Ishizaka Y, Nolan D, Mallal S, Sata T, Tokunaga K. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J Biol Chem. 2010;285:35350–35358. doi: 10.1074/jbc.M110.173286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Wu H, Smith H. APOBEC3G levels predict rates of progression to AIDS. Retrovirology. 2007;4:20. doi: 10.1186/1742-4690-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijak GH, Janini LM, Tovanabutra S, Sanders-Buell E, Arroyo MA, Robb ML, Michael NL, Birx DL, McCutchan FE. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology. 2008;376:101–111. doi: 10.1016/j.virol.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Kijak GH, Janini M, Tovanabutra S, 512 E, Sanders-Buell E, Birx DL, Robb ML, Michael NL, McCutchan FE. HyperPack: a software package for the study of levels, contexts, and patterns of APOBEC-mediated hypermutation in HIV. AIDS research and human retroviruses. 2007;23:554–557. doi: 10.1089/aid.2006.0279. [DOI] [PubMed] [Google Scholar]
- Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfel SA, Di Giallonardo F, Daumer M, Thielen A, Metzner KJ. In-depth analysis of G-to-A hypermutation rate in HIV-1 env DNA induced by endogenous APOBEC3 proteins using massively parallel sequencing. Journal of Virological Methods. 2010;171:329–38. doi: 10.1016/j.jviromet.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-So to P, Little SJ, Shafer R, Robbins GK, D’Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infectious Diseases. 2008;197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–85. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O, Boufassa F, Madec Y, Nguyen A, Goujard C, Meyer L, Rouzioux C, Venet A, Delfraissy JF. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- Land AM, Ball TB, Luo M, Pilon R, Sandstrom P, Embree JE, Wachihi C, Kimani J, Plummer FA. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature medicine. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin proteasome pathway. The Journal of biological chemistry. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Kao S, Goila-Gaur R, Khan MA, Miyagi E, Takeuchi H, Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi A, Humes D, Chohan B, McClelland RS, Overbaugh J. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J Virol. 2009;83:7805–7814. doi: 10.1128/JVI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SK, Abdel-Mohsen M, Guatelli J, Skasko M, Monto A, Fujimoto K, Yukl S, Greene WC, Kovari H, Rauch A, Fellay J, Battegay M, Hirschel B, Witteck A, Bernasconi E, Ledergerber B, Gunthard HF, Wong JK. Role of retroviral restriction factors in the interferon-alpha-mediated suppression of HIV-1 in vivo. Proc Natl Acad Sci U S A. 2012;109:3035–3040. doi: 10.1073/pnas.1111573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Research 2010. 2010;38:4274–84. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G -> A hypermutation. Bioinformatics. 2000;16(4):400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Melard A, Boufassa F, Barre-Sinoussi F, Lambotte O, Rouzioux C, Pancino G ANRS CO18 Cohort. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood. 2011;118:955–64. doi: 10.1182/blood-2010-12-327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005 Sep;1(1):e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HC. APOBEC3G: a double agent in defense. Trends in Biochemical Sciences. 2011;36:239–244. doi: 10.1016/j.tibs.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathogens. 2007;3(2):e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Research. 2006;34:4677–84. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhang YXW, Wang 558T, Xu R, Yu XF. Differential Requirement for conserved Tryptophans in HIV-1 vif for the selective suppression of Apobec3G and Apobec3F. J Virology. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulenga NK, Sarr AD, Hamel D, Sankale JL, Mboup S, Kanki PJ. The level of APOBEC3G (hA3G)-related G-to-A mutations does not correlate with viral load in HIV type 1-infected individuals. AIDS Res Hum Retroviruses. 2008a;24:1285–1290. doi: 10.1089/aid.2008.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulenga NK, Sarr AD, Thakore-Meloni S, Sankale JL, Eisen G, Kanki PJ. Relationship between human immunodeficiency type 1 infection and expression of human APOBEC3G and APOBEC3F. J Infect Dis. 2008b;198:486–492. doi: 10.1086/590212. [DOI] [PubMed] [Google Scholar]
- Vazquez-Perez JA, Ormsby CE, Hernandez-Juan R, Torres KJ, Reyes-Teran G. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology. 2009;6:23. doi: 10.1186/1742-4690-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter ML, Johnson ME, Antons AK, Unutmaz D, D’Aquila RT. Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog. 2009;5:e1000292. doi: 10.1371/journal.ppat.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter MLRT. D’Aquila. Cytoplasmic APOBEC3G Restricts Incoming Vif-positive HIV-1 and Increases 2-LTR Circle Formation In Activated T Helper Subtype Cells. J Virol. 2009;83:8646–54. doi: 10.1128/JVI.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature Structural and Molecular Biology. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Hypermutation index (HI) in vif population sequences from PBMC high molecular weight DNA of 10 LTNP (circles) and 7 NC (filled triangles) subjects. Subjects that were cloned are labeled. (B) Number of stop codons introduced in PBMC vif population sequences from LTNP (circles) and NC (filled triangles) subjects by G-to-A hypermutation.. (C) HI in PBMC vif clonal sequences of subjects LTNP 31 (filled circles), LTNP 40 (open circles), and NC 44 (filled triangles). 12–16 clones were sequenced for each subject. Gray symbols represent clones with HI>1 (D) Number of stop codons introduced in PBMC vif clonal sequences of subjects LTNP 31 (filled circles), LTNP 40 (open circles) and NC 44 (filled triangles) by G-to-A hypermutation.
There was no significant variation of HIV integrated copies between 2 time points in 13 subjects, 9 LTNP (filled circles) and 4 NC (open circles), who had longitudinal specimens collected within a 15-months period (Paired t test: p=0.8).