Abstract
Steroid hormone receptor (SR) signaling leads to widespread changes in gene expression, and aberrant SR signaling can lead to malignancies including breast, prostate, and lung cancers. Chromatin remodeling is an essential component of SR signaling, and defining the process of chromatin and nucleosome remodeling during signaling is critical to the continued development of related therapies. The glucocorticoid receptor (GR) is a key SR that activates numerous promoters including the well defined MMTV promoter. The activation of MMTV by GR provides an excellent model for teasing apart the sequence of events between hormone treatment and changes in gene expression. Comparing hormone-induced transcription from stably integrated promoters with defined nucleosomal structure to that from transiently expressed, unstructured promoters permits key distinctions between interactions that require remodeling and those that do not. The importance of co-activators and histone modifications prior to remodeling and the formation of the preinitation complex that follows can also be clarified by defining key transition points in the propagation of hormonal signals. Combined with detailed mapping of proteins along the promoter, a temporal and spatial understanding of the signaling and remodeling processes begins to emerge. In this review, we examine SR signaling with a focus on GR activation of the MMTV promoter. We also discuss the ATP-dependent remodeling complex SWI/SNF, which provides the necessary remodeling activity during GR signaling and interacts with several SRs. BRG1, the central ATPase of SWI/SNF, also interacts with a set of BAF proteins that help determine the specialized function and fine-tuned regulation of BRG1 remodeling activity. BRG1 regulation comes from its own subdomains as well as its interactive partners. In particular, the HSA domain region of BRG1 and unique features of its ATPase homology appear to play key roles in regulating remodeling function. Details of the interworkings of this chromatin remodeling protein continue to be revealed and promise to improve our understanding of the role of chromatin remodeling during steroid hormone signaling.
Keywords: Chromatin, transcription, steroid receptor, BRG1, MMTV, Chromatin Remodeling, glucocorticoid receptor
1. Introduction
1.1 Chromatin
In eukaryotes, changes in gene expression are regulated not only by interactions between DNA and transcription factors but also by DNA accessibility. DNA wraps around histone octamers to form nucleosomes, the basic units of chromatin, and nucleosomes are further packed into condensed chromatin fibers [1, 2]. Each nucleosome contains approximately 146 base pairs of DNA and two copies each of histones H2A, H2B, H3, and H4 [1, 2]. Favorable electrostatic interactions between the acidic phosphodiester backbone of DNA and the basic surfaces of the histone octamer lead to the formation of the nucleosome core particle along with hydrogen binding between histones and DNA phosphates and non-polar contacts between histones and DNA deoxyribose groups [3]. Between nucleosomes, stretches of DNA bind the linker histone H1, which stabilizes chromatin and compacts 10nm strands of nucleosomes into higher order 30nm and 100nm fibers that can ultimately form chromosomes [4–7]. Not only does this system allow cells to pack almost two meters of DNA into a sphere with a radius of a few micrometers, it also regulates access to DNA. Densely packed chromatin can repress transcription and other processes that require protein-DNA interactions, such as replication, recombination, and repair [8–10]. In this manner, chromatin introduces a system for regulating gene expression and other DNA-dependent processes based on the interactions between DNA and histones that position nucleosomes and underlie chromatin structure.
1.2 Histone modifying and chromatin remodeling complexes
Several biological processes in eukaryotes are dedicated to modulating chromatin structure [11, 12]. The two major classes of chromatin remodeling/modifying complexes are the ATP-dependent remodelers and the histone modifying remodelers. While ATP-dependent remodelers mechanically separate DNA-histone contacts using the energy of ATP hydrolysis, histone modifying chromatin remodelers posttranslationally “mark” histone tails that protrude from nucleosomes. Histone marks include acetylation, phosphorylation, methylation, ubiquitination, and other covalent modifications that allow the tails to act as binding surfaces, often for chromatin-modifying factors and complexes [12, 13]. These modifications do not substantially impact the nucleosome core particle structure, but do affect higher order chromatin structure and gene expression [14–16].
The relationship between transcriptional activation and histone acetylation was established over 50 years ago, after it was observed that acetylation of histones results in increased transcription [17]. Since then, associations between specific histone marks and changes in transcription have been the focus of numerous investigations and led to the development of a histone code hypothesis [18, 19]. The extensive study of acetylation at specific lysine residues along histone tails has demonstrated how the histone acetylases (HATs) that catalyze this modification loosen chromatin by replacing a positively charged residue with a negatively charged functional group, reducing DNA-histone affinity and leading to increased transcription [20, 21]. This activating mark can be removed by histone deacetylases (HDACs), establishing a dynamic process that plays a key role in regulating gene expression. Many proteins involved in steroid hormone stimulated transcriptional activation have HAT activity, while repressors are associated with HDAC function [14].
Although acetylation is generally associated with active transcription, the effects of other histone tail modifications are more complex. Methylation, for example, can both enhance and repress transcription [18]. The context-specific consequences of individual marks as well as the importance of neighboring histone tail modifications in directing interactions between nucleosomes and additional histone modifying complexes leads to the complex “code” that continues to be characterized [13, 18, 22]. One mark of particular interest in hormone inducible transcriptional activation is histone H1 phosphorylation, which is required for active transcription from the GR-inducible MMTV promoter and is discussed further in section 3.2 of this review [23–28]. The impact of histone structure and biochemistry on gene expression can also be observed in the effects of variant histones, which can replace each of the core histones and H1 [11, 29]. Subtle structural differences in variant histones, like changes that occur through tail modifications, are associated with changes in both chromatin structure and gene expression [29]. The impact of histone modifications on gene expression demonstrates one system of regulation based on manipulating the structure of chromatin. Another system for gene regulation through chromatin remodeling comes from the direct mechanical action of ATPase-driven chromatin remodeling complexes, which often work in concert with histone modifying complexes to regulate transcription and other DNA-dependent processes in response to biological events and signals [30].
1.3 ATP-dependent Chromatin Remodeling complexes
ATP-dependent remodeling complexes work by directly breaking histone-DNA contacts to slide and reposition nucleosomes [31, 32]. To separate bonds between histones and DNA, the ATP-dependent class of remodelers uses energy derived from ATP hydrolysis, which is carried out by a central ATPase subunit. These proteins come from the SF2 helicase superfamily, which share a structural core consisting of two RecA helicase-like domains that bind and hydrolyze ATP [33, 34]. Within SF2 superfamily, the Snf2 family includes five major families of ATP-dependent remodelers including SWI/SNF, ISWI, Mi-2/NuRD, INO80, and SWR1 as well as the repair and recombination proteins RAD16, ERCC6 and RAD54 [35, 36]. The central ATPase domain region of Snf2 proteins consists of paired DEXHc and HELICc domains which include subregions regions of strong homology [37]. In addition to the ATPase domain, other common domains and motifs are found among some Snf2 proteins. Some of these domains are known to interact with histones (BROMO, CHROMO, SANT) and to bind DNA (AT hook, Zn finger), while others have functions less well defined (HSA, BRK) but which may be important for protein-protein interactions [38–40]. Yeast SWI2/SNF2 (mating type SWItching defective/Sucrose Non Fermenting) was the first Snf2 remodeler to be discovered through genetic studies that initially demonstrated roles in mating-type switching and sucrose fermentation [41]. These effects were later attributed to a role in opposing the inhibitory effect of chromatin [42, 43]. Biochemical assays revealed SWI2/SNF2 forms a large protein complex and remodels nucleosomes in an ATP-dependent manner [44, 45]. The human homologues of yeast SWI/SNF proteins have been identified through sequence homology and shown to similarly form complexes and remodel chromatin in an ATP-dependent manner [46, 47].
1.4 BRG1, the central ATPase of SWI/SNF
In humans and other mammals, SWI/SNF can have one of two possible ATPase core subunits, BRG1 or BRM [48–50]. These ATPases have significant sequence homology yet non-redundant roles. Loss of BRG1 is lethal prior to embryonic implantation in mice, and BRM cannot compensate for its loss [51–53]. BRG1 can compensate for BRM, however, suggesting a BRG1-specific role that is critical during development. Furthermore, murine embryos heterozygous for a BRG1 null mutation are predisposed to form tumors [51]. In humans, BRG1 mutations have been observed in a variety of tumor cells [54–59]. More recently, BRG1 has been identified as critical for inducing and maintaining cellular pluripotency [60, 61]. The domain structure of BRG1, the functions of individual BRG1 subdomains, and the importance of BRG1 protein-protein interactions in the context of hormone dependent, BRG1-mediated transcriptional regulation will be discussed in greater detail in Section 3.
1.5 SWI/SNF complex subunits
The BRG1 ATPase is found in complex with several subunits termed BRG1 associated factors (BAFs) that form a highly regulated, multi-functional structures. Some human BAFs have direct yeast orthologs; these include BAF170, BAF155, BAF60, BAF53, and BAF47. Other BAFs are unique to the mammalian complex, and these are BAF57, BAF45, BAF250, BAF200, and BAF180/Polybromo. B-actin, Brd7, and Brd9 are also subunits of SWI/SNF complexes [62]. BRG1 is found in remodeling complexes other than SWI/SNF, as well, including NUMAC, WINAC, NCoR, and mSin3A/HDAC complexes [50]. Many BAFs also appear in these complexes, which are associated with chromatin dependent processes such as transcription, elongation, and DNA replication. Other than B-actin, the only component of SWI/SNF found in a complex that does not contain BRG1 is BAF53, which is also found in the INO80 remodeling complex [63]. While some BAF proteins appear in all SWI/SNF complexes, others are changeable [64–68]. The list of “core” BAFs has changed over time as new SWI/SNF complexes that exclude certain subunits or include new subunits are discovered, although BAF155, BAF57, BAF47, and BAF53 are still considered essential [49, 68]. The first major distinction between SWI/SNF subtypes came with the discovery that BAF250 and BAF180 are not found in the same complexes, formulating the BAF (BAF250-containing) and PBAF (BAF180-containing) subclasses of SWI/SNF [65]. Brd7 and BAF200 are only associated with PBAF complexes, and BRM, the close and typically interchangeable homolog of BRG1, is not found PBAFs [66, 67]. Additionally, BAF170 is not found in SWI/SNF complexes purified from mouse embryonic stem cells (esBAFs), and, like pBAFs, esBAFs contain only BRG1 and not BRM [68].
Different SWI/SNF configurations have been associated with different functions during heat shock and immune responses. BRM-BAFs occupy IFNγ activated sequence (GAS) promoters before heat shock or interferon stimulation, but are exchanged for BRG1-BAFs following induction [69]. BAF isoforms bring additional modularity to SWI/SNF, as well. BAF250, BAF53, BAF45, and BAF60 have multiple isoforms shown to associate with the complex at different times, and these changes have been associated with different cell types, stages of development, or phases of cellular differentiation. For example, BAF60c is specifically associated with cardiac development, while the exchange of BAF53a for BAF53b and BAF45a for BAF45b/c is part of the changing SWI/SNF complex observed during neuronal development [70, 71]. This subunit modularity may allow the central activity of the ATPase to be modified by the specific set of BAF subunits present in the complex. Variability through subunit exchange helps fine-tune BRG1 function so that it can activate a variety of promoters and respond to a wide range of biological events including the cell cycle, cellular differentiation, and immune response as well as hormonal signaling.
1.6 BRG1 and steroid hormone signaling
One well-characterized activity of the BRG1 complex is to regulate hormone-stimulated transcription by interacting with Class 1 nuclear receptors [72–74]. Nuclear receptors (NRs) are ligand-dependent transcription factors, and Class 1 NRs respond to steroid hormone ligands by homodimerizing and binding inverted repeat DNA sequences known as hormone response elements (HREs) or at sites of ubiquitous transacting factors within the promoter regions of target genes, as shown in Figure 1 [75]. Glucocorticoid receptor (GR), progesterone receptor (PR), estrogen receptor (ER), and androgen receptor (AR) are all steroid hormone receptors (SRs) known to associate with the BRG1 complex and to require its chromatin remodeling activity for proper function [72, 76, 77]. Careful study of the process that begins with SR binding the HRE, leads to recruitment of the BRG1 complex, and results in global changes in gene expression has brought to light important aspects of nucleosome remodeling by the BRG complex during transcription process.
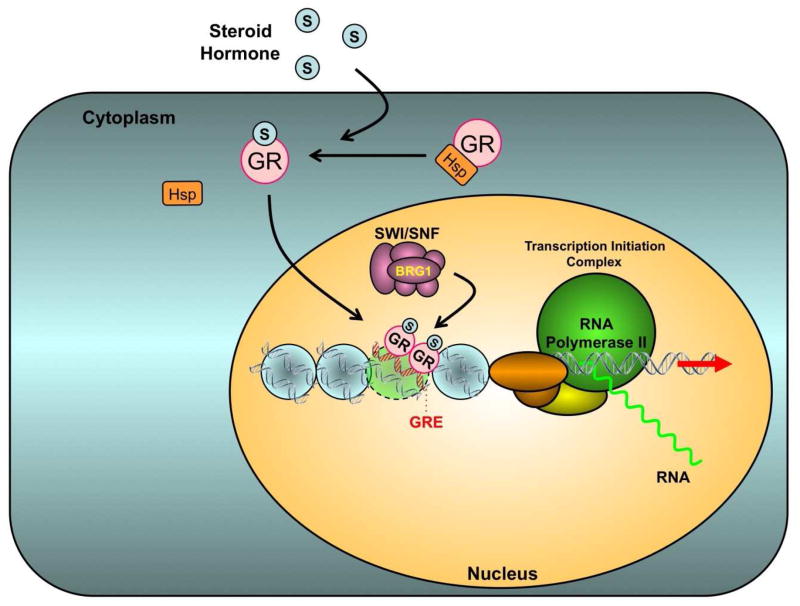
Figure 1. Hormone Signaling through the Glucorticoid Receptor (GR).
Glucocorticoid receptor (GR), like progesterone receptor (PR), estrogen receptor (ER), and androgen receptor (AR) responds to hormone by shedding heat shock protein, homodimerizing, and binding inverted repeat DNA sequences known as hormone response elements (HREs) or sites of ubiquitous transacting factors within the promoter regions of target genes. GR and other steriod hormone receptors recruit the BRG1 complex which provides an essential chromatin remodeling activity that facilitates formation of the transcription initiation complex and transcriptional activation.
2. Steroid hormone signaling and chromatin structure
2.1 SR Signaling and Nucleosome Remodeling
Observations of local chromatin alterations in response to hormone as well as increased sensitivity to nucleases at the promoters of hormone activated genes provided early clues that the mechanism of steroid hormone activated transcription depends upon tightly regulated chromatin remodeling [78]. The GR responsive promoter of the mouse mammary tumor virus (MMTV), in particular, has provided a useful model in dissecting key events of hormone response at this type of promoter, which operates similarly to the tyrosine aminotransferase (TAT) promoter and, when fused to a reporter gene, reflects the transcriptional responses of numerous GR regulated genes. By comparing the activation of a stably integrated MMTV promoter assembled in an ordered nucleosomal array to a transiently transfected promoter, which has a disordered chromatin state, it is possible to distinguish which events at the MMTV promoter require remodeling [79].
The initial binding of GR, which acts as a pioneering factor at the MMTV promoter, does not require remodeling, and nor do the p160/SRC coactivators that bind the receptor directly [14, 80, 81]. Without remodeling, the receptor and its coactivators can recruit HATs (p300/CBP) and histone methytransferases (CARM1, PRMT1) that both modify histones and recruit additional regulatory proteins [80, 82–84]. Some transcription factors, however, do require nucleosome disruption by ATPase driven remodeling - including nuclear factor 1 (NF1), the octamer transcription factors (OTFs), and Tata-binding protein (TBP) [85, 86]. The binding of NF-1, a transcription factor that interacts with an MMTV site that is nucleosome-occupied and inaccessible in the ordered promoter without remodeling, acts as a key indicator of remodeling activity. Mutating the NF-1 binding site of the MMTV promoter disrupts not only NF binding, but also the continued binding of hormone receptor, OTFs, and the chromatin remodeling complex, suggesting this transcription factor may also play a role in remodeling and stabilizing the promoter region. This work demonstrates the importance of both NF-1 and the remodeling of nucleosome B (required for its binding) in establishing promoter stability and accessibility during hormonal signaling [87].
Distinguishing between the remodeling requirement for GR/coactivator binding versus NF-1/OTF/TBP binding (and eventual recruitment of RNA PolII) demonstrates a bi-modal order of operations with GR/coactivator binding preceding recruitment of the SWI/SNF remodeling complex. SWI/SNF remodeling opens the NF-1 binding site and facilitates interactions with the OTFs and TBP - leading to enhanced transcription [88]. Time course studies of the MT2A promoter, which is GR responsive and sensitive to the GR inhibitor curcumin, demonstrate another transition point by correlating reduced GR activity with the loss of (PIC) members TFIIH and TFIIB from the promoter [89]. These studies show GR dependent transcriptional activation can also be inhibited following remodeling and at the level of PIC protein recruitment.
The placement of GR and transcription factor interactions along the MMTV promoter using an in vivo nuclease footprinting assay provides a spatial visualization to accompany the established order of operations extending from the receptor binding sites and TATA box [90, 91]. This elucidation of GR-signaling events demonstrates the requirement for nucleosome receptor-ligand binding, co-activator recruitment, the formation of the PIC, and the eventual transcriptional activity of RNA polymerase II at GR-responsive promoters. The order and spacing of key transtion points durng this process are depicted in Figure 2.
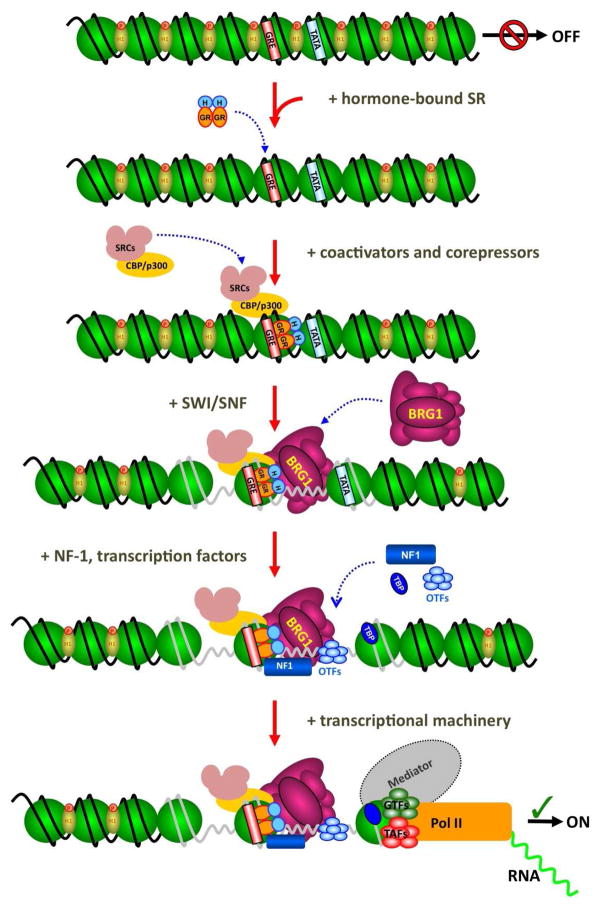
Figure 2. Key Transition Points During GR signaling.
After ligand binding dimerizes the Glucocorticoid Receptor (GR), it enters the nucleus and binds target sequences such as the Glucocorticoid Response Element (GRE) within chromatin. The receptor binds co-regulators such as NcoA1/SRC-1, NCoA2/TIF-2/GRP-1, and CBP/p300 which do not require remodeling to engage the receptor at the promoter. Next, the BRG1 complex SWI/SNF is recruited to the promoter through an interaction with GR and nucleosomes are repositioned. This allows transcription factors such as NF1 and the octamer transcription factors (OTFs) as well as the TATA-binding protein (TBP). Finally, mediator proteins are recruited and the preinitiation complex forms leading to transcripton by RNA Pol II.
2.2 The MMTV promoter
The mouse mammary tumor virus is a single-stranded RNA retrovirus that integrates into the host genome as a double-stranded DNA provirus [78]. Its long terminal repeat (LTR) has a typical eukaryotic promoter structure with hormone responsive elements (HREs) that allow it to respond to signals from steroid hormone receptors including the glucocorticoid receptor (GR), progestin receptor (PR), and androgen receptor (AR) [36, 92]. This ~1,300 base pair fragment is organized into a phased array of six nucleosomes designated A-F [93]. Mapping of the MMTV nucleosome boundaries shows alternate boundary frames in the proximal promoter region, a region containing the A and B nucleosomes as well as HREs and the binding sites for several transcription factors (NF1, OTFs, and TBP) followed by a TATA box [94]. Distinct positions identified for nucleosome B (−221 to −75 and −190 to −43) illustrate a shift in its position induced by hormone that results in nuclease sensitivity of the region surrounding the TATA box [85, 94, 95]. While sensitivity to specific restriction enzymes indicates nucleosome repositioning, sensitivity to general nucleases reveals the nucleosome footprint along MMTV, with the presence or absence of a distinct banding pattern indicative of both nucleosome order and linker length [90, 91, 96].
Interestingly, when MMTV is expressed in cells through transient transfection and not stably integrated into the genome, it does not assemble ordered nucleosomes [85, 95]. This important and unexpected feature of the transiently expressed promoter allows in vivo assays that compare protein binding and transcriptional activation of the integrated (ordered chromatin) form of MMTV versus the transient (disordered chromatin) form within the same cell [85, 88, 90]. This phenomena is not exclusive to the MMTV promoter and has also been used to study Myo-D mediated chromatin changes and transcriptional activation [97]. It is interesting to note the transiently transfected vector does not exclude nucleosomes, but rather lacks nucleosome positional stability [28]. Linker histones are also present, though at substochiometric levels, reflecting their role in producing ordered nucleosomal arrays that are amenable to remodeling-based regulation. Studies of the MMTV promoter and of differences between ordered and disordered MMTV promoters have been critical not only in the determining the order of events during SR signaling (and termination of signaling) but also in determining differences between GR and PR activity and, ultimately, the role of the BRG1 complex during hormone signaling [23, 86, 98].
2.3 Glucocorticoid and Progesterone Receptors (GR & PR)
The importance of chromatin structure varies between GR and PR stimulated transcriptional responses, which differ despite the high degree of sequence homology and identical DNA recognition sites of these receptors [98–100]. The observation that in T47D/A1-2 breast cancer cells expressing both GR and PR, GR activation leads to transcriptional upregulation from both stable and transiently transfected templates while PR activation leads only to increased transcription from transiently traransfected templates reveals a major difference between GR and PR activity [79, 101]. Nuclease hypersensitivity analyses of the promoter following hormone induction shows reduced remodeling correlates with impaired function in the case of PR signaling [99, 101]. Despite weak remodeling capability, PR is capable of inhibiting GR activation when both hormones and receptors are present [101]. Additionally, PR is able to remodel chromatin following stable introduction into a mouse GR+PR− cell line, highlighting the complexity and cell type specificity of PR activation and suggesting a system for selective receptor activation in cells such as T47D that endogenously express both receptors [98]. Consistent with this, stably integrated MMTV promoters in T47D cells that lack PR or both GR and PR assume an open and remodeling independent configuration, unlike the closed configuration found when both receptors are expressed [102]. Studies exploring the effects of GR and PR antagonists on GR activity in breast cancer versus osteosarcoma cells reveal how the hormones affect different events during receptor signaling as well as how the complement of co-activators and receptors in a given cell type determine their ultimate effect [98, 103]. Some of the dynamics between GR and PR in T47D cells, including the inhibitory effect of progestin during glucocorticoid signaling, can be explained by considering the interactions of both these receptors with the BRG1 chromatin remodeling complex (SWI/SNF), described in Section 3, which is essential for hormone driven transactivation [72].
3. GR stimulated chromatin remodeling requires BRG1
3.1 SRs interact with BRG1
SRs have been shown to associate with and require the remodeling function of the BRG1 complex for activity [72, 76, 77, 104]. Understanding the importance of the SR-BRG1 interaction during signaling and in the context of observed differences in GR versus PR remodeling activity helps distinguish between transcriptional activation and chromatin remodeling during hormone response. Association between both the glucocorticoid and progesterone receptors and the SWI/SNF complex reflects competition for the complex and explains the inhibition of remodeling dependent, but not remodeling independent, GR-mediated transcriptional activation in the presence of progestin and certain inhibitory PR ligands [72]. The notion that progestin competitively inhibits glucocorticoid signaling is further substantiated by evidence both receptors bind the through the same BAF60 interface to interact with the SWI/SNF complex [104]. The PR antagonist ORG31710, like progestin, triggers PR binding to HREs as well as BRG1. This sequesters the BRG1 complex, strongly diminishes the GR-BRG1 interaction, and inhibits GR-mediated transcriptional activation [105]. Binding of co-activators (NcoA1/SRC-1, NCoA2/TIF-2/GRP-1, and CBP/p300) to GR is not disrupted by this antagonist, however, while BRG1 binding is severely reduced. The co-activator interactions are important in transcriptional activation at templates that do not require remodeling as well as those that do, suggesting these interactions with GR occur independently from GR-BRG1 binding. From these studies and previous experiments that show transcription factors (including NF1 and the OTFs) bind subsequent to remodeling, a detailed view emerges of both the sequence of events during SR signaling and the promoter positioning of the hormone receptor, co-activators, BRG1 remodeling complex, and transcriptional PIC as shown in Figure 2.
3.2 Histone H1 and SR-mediated BRG1 dependent chromatin remodeling
The linker histone, H1, sits outside the nucleosome core particle on intervening stretches of DNA, which are found at variable lengths depending on cell state and cell type [5, 106]. The ordered, stably integrated MMTV promoter has inter-nucleosome linkers of roughly 50 base pairs, each occupied by a single histone H1 molecule [28]. During hormone-dependent transcriptional activation, H1 is displaced as part of the chromatin remodeling process, and H1 depletion is associated with general transcription activation [107]. Transiently transfected MMTV promoters are disordered, show no consistent linker length or nucelosome spacing, and have a reduced level of H1, which is decreased from its typical 1:1 stochiometry with the core nucleosomes independently of hormone signal [28]. Remodeling by BRG1 is not required at these disordered promoters, as demonstrated by their hormonal activation by PR signaling in T47D cells, an event which neither requires nor involves remodeling [79, 88, 93]. These H1-depleted promoters also show reduced NF-1 binding, which occurs during remodeling-dependent transcriptional activation following nucleosome repositioning [79, 85, 86, 90].
Changes in the phoshorylation of H1 that are associated with cell cycle progression and DNA repair also effect remodeling [7, 25, 27, 28, 79, 108, 109]. A refractory phase that follows long term (> 24 hour) induction by GR in stably integrated, ordered MMTV promoters is strongly correlated with H1 phosphorylation, but this refractory phase is not observed in disordered, transient promoters [79]. Likewise, kinase inhibitors that eliminate H1 phosphorylation at MMTV promoters lead to reduced transcription from ordered templates, but not from disordered templates [25]. The global decrease in H1 phosphorylation that occurs with long term GR signaling is seen at higher levels in H1 variants H1.3. H1.4, and H1.5, which are associated with greater affinity for and ability to compact chromatin [26, 96]. Loss of H1 phosphorylation, therefore, has greater effects at sites of dense chromatin, consistent with the observation disordered, transient MMTV promoters are resistant to loss of function with decreased H1 phosphorylation. Taken together, these results indicate the presence of H1 and the phosphorylation state of H1 are important to both chromatin order and chromatin remodeling by the BRG1 complex. Disordered chromatin that is not is remodeled is associated with reduced H1, while ordered chromatin that can be remodeled by BRG1 is associated with phosphorylated H1, and ordered chromatin that is prohibitive to productive BRG1 remodeling is associated with de-phosphorylated H1 [28, 79]. Interestingly, the overexpression of histone H1 leads to increased H1 incorporation in transient promoters but does not impose regular order, as shown by the micrococcal nuclease footprint of the promoter. The overall chromatin structure is more dense, but not more organized, and although general transcription is repressed, this occurs in neither in a SR-signaling nor a remodeling dependent fashion. The distinction here between general chromatin compaction and establishing ordered nucelosomes is important, as productive remodeling by BRG1 is exclusively associated with the latter.
3.3 Connecting BAFs bridge GR and BRG1
The association between GR and BRG1 is not direct and relies on connecting BAF proteins to bridge BRG1 and the hormone receptor. BAF proteins that participate in receptor bridging interactions include BAF250, BAF60a, BAF57, and BAF53a [65, 84, 104, 110]. BAF250, discovered initially through SWI/SNF purification by BRG1 immunoprecipitation, is a homolog of yeast SWI1 and the Drosophila Osa protein [48]. In addition to distinguishing a separate class of BRG1 complexes, BAF250 is able to bind GR through its C-terminal domain [65, 84]. This binding event is evident from both in vivo immunoprecipitation and in vitro pull down assays between GR and the BAF250 C-terminus [65]. GR-induced transcriptional activation is diminished in cells expressing the BAF250 C-terminal truncation mutant, suggesting the fragment is able to compete with full-length BRG1 for GR binding. A specific role for BAF250a during GR signaling is consistent with observed increases in GR-responsive transcription with BAF250a overexpression [111]. BAF60a and BAF57 have also been identified as GR-bridging BAF proteins [104]. Truncated GR (1–556 of 777), which lacks the C-terminal ligand-binding domain (LBD) but retains the N-terminal activating function (AF-1) and DNA binding (DBD) domains, can initiate transcription from the MMTV promoter in the absence of ligand and, like full length GR, can co-immunoprecipitate BRG1 and BAF155 [72]. Testing the GR-binding of in vitro translated SWI/SNF subunits reveals direct interactions occur with both BAF60a and BAF57. The BAF60a GR-binding region has been localized to its N-terminus (residues 4–128 of 515), which also binds the PR, ER, and FXR receptors. BAF57 has also been observed to bind ER, establishing a general role for this protein in binding steroid hormone receptors [112]. Notably, these receptors all contain a conserved “RRK” motif within the LBD, and the mutation of the primary arginine to glutamine in GR (GR R488Q) eliminates the BAF60a interaction, suggesting this basic patch is a critical feature of the interface between the receptor and BAF60a [104]. It is interesting to note the association between BRG1 and GR is not eliminated by the BAF60a R488Q mutation, consistent with binding between the receptor and SWI/SNF relying on several bridging subunits. Remodeling and transcription are diminished, however, indicating the mechanism of activation is compromised even though the association remains. The N-terminus of BAF60a is also responsible for its interaction with BRG1, while the central portion (residues 129–435) binds BAF170 and BAF155, showing multiple interfaces of BAF60a allow it to bridge not only BRG1 and GR but also BRG1 and other core BAF subunits [104].
BAF60a also serves as a bridge between the SWI/SNF complex and other transcriptional activators including the tumor suppressor p53, which relies on SWI/SNF for transcription of several targets [113]. The BAF60a-p53 interaction, like the BAF60a-GR interaction, relies on the BAF60a N-terminus, though it also requires additional residues between 129 and 150. BAF60a has also been shown to bind fos/jun dimers and act as a bridge between this transactivation dimer and SWI/SNF [114]. In this circumstance, BAF60a binds the dimer using both N-terminal (1–129) and C-terminal (307–366) surfaces. Taken together, these data suggest that the ability of BAF60 to act as an adaptor and facilitate SWI/SNF recruitment not limited to SR super family of transcriptional activators.
Bridging between BAFs and the steroid hormone receptor has also been observed through BAF53, an actin-like protein and member of SWI/SNF that interacts with ER through the protein Flightless-1 (Fli-1) [110]. Fli-1 is an actin-binding protein that plays a role in cytoskeletal formations as well transcription and contains six gelsolin repeats, G1-G6, named for their structural homology to the actin-binding protein gelsolin [115]. The G1 repeat of flightless binds BAF53a, while G3 binds the LBD of ER, forming a bridge between ER and SWI/SNF. Both Fli-1 and SWI/SNF are necessary for ER-mediated transcription, and depletion of Fli-1 inhibits SWI/SNF recruitment to the promoter. In addition to ER and SWI/SNF, Fli-I binds members of the p160 coactivator complex, connecting three major components of hormone signaling through one multi-domain protein [116]. The role of actin and actin-related proteins in SWI/SNF (as well as other ATP-dependent remodeling complexes) is not well defined, and this work suggests one role is to connect these large complexes with transcriptional activators as well as other protein complexes involved in transcription.
3.4 BRG1 subdomains regulate ATPase function
The BRG1 molecule is over 1600 residues long and contains several conserved motifs and domains defined through sequence homology (Figure 3) [50]. Starting at the N-terminal end, a QLQ motif is thought to be involved in protein-protein interactions, consistent with the observation that N-terminal deletion mutants of BRG1 fail to bind several key BAF proteins [84]. The HSA domain that lies between the QLQ and ATPase is similar to domains found throughout BRG1 homologs as well as the related INO80 remodeler [117]. Adjacent to the HSA domain, the BRK domain is unique to multicellular organisms. BRK domains are also found in a subset of the chromodomain helicase remodeling proteins from the Snf2 family and often in pairs [118]. The functions of both the HSA and BRK domains of BRG1 are not well defined. The ATPase homology that follows them occupies approximately one third of the BRG1 sequence. This domain region performs the mechanical remodeling function of the SWI/SNF complex, and contains two separate Rec-A like domains (DEXHc and HelicC) that are found paired in all Snf2 family members joined by linkers of varying length [119]. A point mutation in the BRG1 ATP binding site, K798R, renders the SWI/SNF complex incapable of remodeling chromatin [51, 120]. Although there is no structural data for the BRG1 ATPase, ATPase homology regions for two related Snf2 family members (Rad54 and CHD1) have been reported, and these reflect common folds with variable loops that confer specificity [34]. Following the ATPase region, a small AT hook motif is thought to act as a DNA binding motif. Such motifs are found in many DNA binding and chromatin interacting proteins and often act in concert with additional DNA interacting domains [50]. Finally, the BROMO domain of BRG1 is able to interact with acetylated histones, though its affinity is weak when compared to BROMO domains from other proteins suggesting the BRG1 BROMO domain interacts with histones in the context of additional interactions [121]. With the exception of the ATPase, there is little domain-specific functional information for BRG1.
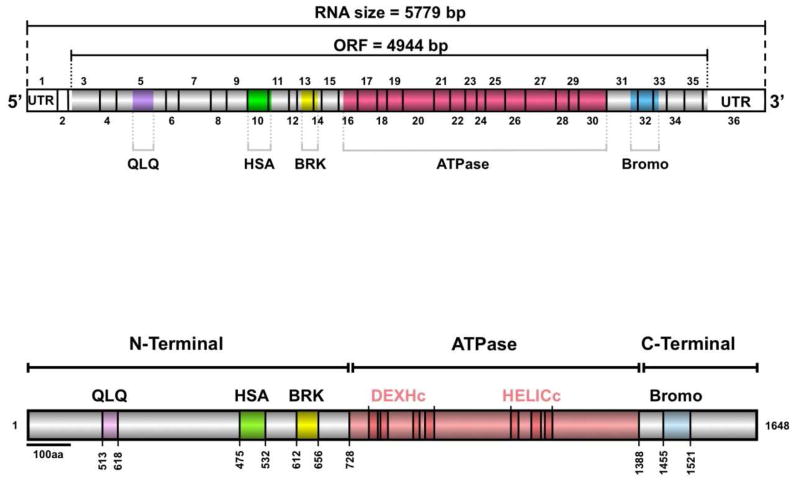
Figure 3. The BRG1 remodeling ATPase.
The BRG1 gene is located on Chromosome 19 and is transcribed and processed to produce a 5779 bp RNA with an open reading frame (ORF) of 4944 bp. The BRG1 protein consists of 1647 amino acids and contains several conserved domains such as the HSA, BRK, ATPase, and BROMO domains. Highly conserved regions within the ATPase homology are highlighted in bright pink.
A model system has been established for studying BRG1 mediated, glucocorticoid receptor-dependent transcription in vivo that allows the overall activity BRG1 to be dissected into specific domain functions including stable expression, BAF complex formation, recruitment to the promoter, remodeling capability, and transcriptional activation [122]. This system uses SW-13 cells, which do not express endogenous BRG1 but do express BAF proteins capable of reconstituting the BAF complex following exogenous BRG1 expression. An MMTV promoter is stably integrated that has been shown through previous work to take on an ordered chromatin structure. This promoter is fused to a luciferase reporter, so that luciferase signal as well as transcript levels of several endogenous hormone responsive BRG1-dependent genes indicate transcriptional upregulation. By comparing the behavior of wild type BRG1 to BRG1 chimeras and mutants in this system, more insight is gained to the mechanism of BRG1 ATPase activity and the role of its non-ATPase subdomains [74, 84, 123].
Comparing BRG1 and the Snf2 family protein SNF2h from the ISWI complex, one finds the ATPase proteins and their complexes have similar basic nucleosome remodeling capability, but are non-complementary [124, 125]. Additionally, they have different substrate specificity for histone modifications and BRG1 is better able to free DNA near the nucleosome dyad in addition to entry and exit points of the nucleosome, while SNF2h, unlike BRG1, is able to assemble and space nucleosomes [126, 127]. When the BRG1 ATPase domain region is exchanged for that of SNF2h, the SWI/SNF complex is retained, while activation of BRG1-dependent promoters is compromised, suggesting differences in the ATPase domain, itself, are important for BRG1 functional specificity [123]. Some BRG1-dependent promoters are more sensitive to the SNF2h chimera than others, including the MMTV promoter, which is not activated by the BRG1/SNF2h ATPase chimera [123]. Notably, promoter binding is not compromised - instead the main difference observed in the ATPase chimeras is weakened nucleosome repositioning capability. A chimera that exchanges the C-termini of BRG1 and SNF2h does not affect this assay, consistent with the deletion of the same exchanged C-terminus also showing little effect on transcriptional upregulation from the MMTV promoter [123]. While this does not rule out a role for the C-terminus of BRG1 in specific interactions at some promoters, such as interactions with acetylated histones via the BROMO domain, it does suggest the basic signal response and nucleosome remodeling functions do not require the C-terminus. Another BRG1/SNF2h chimera that exchanges the N-termini does have severe functional consequences. Replacing the BRG1 N-terminus with the SNF2h N-terminus results in loss of BRG1-dependent transcriptional activation, although the original ATPase domain region remains intact and functional in vitro [84].
The deletion of the BRG1 N-terminus also abolishes BRG1-dependent transcriptional activation from the MMTV promoter and other BRG1-dependent promoters, and both changes result in loss of core BAF subunits BAF155, BAF170, and BAF60a [84]. The loss of these subunits is not the sole cause of lost function, however, as a smaller deletion that excises only the HSA domain retains these BAFs yet remains incapable of stimulating transcription. HSA domain deletion does interfere with the binding the BAF250a subunit, however, suggesting the BAF250a interaction is more critical for transcriptional activation from the promoters observed than interactions with the core BAFs [84]. The HSA domain of BRG1 and the domains of related yeast remodelers (Sth1, Snf2, Swr1, Eaf1, and Ino80) have also been associated with actin-related proteins, such as Baf53 [117]. It is interesting to note that, like the ATPase chimera, the HSA domain deletion does not cause a diminished interaction with the promoter or a failure to form the basic SWI/SNF complex. Also, unlike the ATPase catalytic mutant (K798R), both these BRG1 variants retain in vitro nucleosome remodeling function [51, 84]. These studies reveal the importance of both the unique features of the BRG1 ATPase domain and the HSA domain for activity. More importantly, they allow events including BRG1 protein expression, SWI/SNF complex formation, promoter binding, in vitro remodeling capability, and transcriptional regulation by the SWI/SNF complex to be analyzed individually and according to BRG1 subdomain. They also demonstrate the ultimate requirement for nucleosome remodeling in the context of ordered chromatin structure, though the mechanistic details of productive remodeling that facilitates transcription are still unclear. Determining what roles the BRG1 HSA domain and unique features of its ATPase region play in this process will be an important next step in elucidating this mechanism.
4. Conclusions
The complex process of steroid hormone signaling can be viewed as a series of key events by analyzing the physical changes and transcriptional output of the hormone inducible MMTV promoter. SR signaling requires an extensive network of specific proteins and protein complexes including the steroid receptor, receptor coactivators with histone-modifying capability, transcription factors, and the transcriptional machinery. For GR and other SR-promoter interactions to progress toward productive transcriptional activation in the context of ordered chromatin, remodeling by the ATPase-driven SWI/SNF complex is required [11, 72, 73, 112]. With SWI/SNF remodeling of the ordered, integrated MMTV promoter as well as the disordered transiently transfected MMTV promoter, the transcriptional activator NF-1 can bind the promoter to stabilize the active state and facilitate binding of additional transcription factors [85, 88]. Recruitment of mediator complex and of the PIC follows, and each of these events is necessary for productive signaling [11, 83, 89].
During this process, the ability of GR versus PR to activate transcription is highly cell-type specific and may depend on both the relative order of a given promoter and the remodeling capability (both histone modification and ATPase driven) the receptor is able to recruit [93, 98, 99, 101]. All SRs require SWI/SNF when promoter remodeling is needed, and each has been shown to physically interact with the SWI/SNF complex [104, 112]. Disordered, transiently transfected MMTV promoters, however, do not require remodeling despite containing nucleosomes [28, 79, 91]. These transient vectors have reduced levels of histone H1, nucleosomes that lack regular order along the DNA sequence, and restriction sites that are exposed in absence of hormonal signaling and SWI/SNF remodeling. Promoters without ordered chromatin circumvent the requirement for remodeling by SWI/SNF for transcriptional activation and are active in situations when ordered promoters are not (eg: PR signaling in T47D cells, GR signaling in cells that do not express BRG1, and GR signaling after 24+ hours) [79, 99, 122, 128].
It is interesting to note that although a disordered promoter can incorporate overexpressed H1.3 and become more compact and resistant to transcription, this general compaction of chromatin is not correlated with precise ordering of nucleosomes [28]. Compaction and ordering are distinguished by reduced restriction enzyme sensitivity without increase in nucleosomal footprinting as well as a general decrease in transcription that is not specific to hormonal-inducible genes. These distinctions reveal the presence of nucleosomes and histone H1 is not sufficient to produce regulatory chromatin order (which depends on precise nucleosome positioning and repositioning) and suggest roles for both histone modifiers and ATPase remodelers in establishing that order. The interplay between histone modifications and the BRG1 ATPase in remodeling has been evaluated through studies showing the phosphorylation state of the H1 N-terminal tail is critical for GR dependent BRG1 mediated transcriptional activation [23, 25, 28, 129]. Loss of hormone-dependent, BRG1-mediated remodeling from ordered promoters (but not disordered promoters) occurs with loss of H1 phosphorylation and shows an important connection between the modification state of H1, as opposed to the level of H1, and productive, regulated remodeling.
To better understand the mechanics of GR regulated remodeling, the activity of BRG1 can be broken down into multiple functions, and the protein itself can be broken down into multiple subdomains. The N-terminal domain is distinguished for its BAF subunit binding ability, and has been shown to bind BAF155, BAF170, and BAF60a [84]. Deletion of the N-terminus prohibits BRG1 from activating GR-induced transcription; however, this loss of function can be mapped to the small HSA domain region (~100 residues) within the N-terminus that is not responsible for these BAF-binding interactions. Deletion of the HSA domain results in complete loss of BRG1 function similar to the ATPase catalytic mutant K798R and reveals a mutation that eliminates BRG1 activity without interfering with ATPase function, core BAF binding capability, or promoter recruitment. Instead, the loss of another BAF, BAF250a, is correlated with the HSA domain dependent loss of remodeling function at the GR responsive promoter. This protein, specific to a subset of BAFs, is also known to bridge BRG1 and GR, suggesting the loss of this interaction may play a role in loss of GR-dependent activation. Since HSA domains are also important in BAF53 binding, compromising this interaction may also contribute to the observed deletion effect.
Although protein-protein interactions are correlated with the HSA domain, its adjacency to the ATPase homology introduces the possibility of a direct interaction or allosteric effect on ATPase activity that is important in vivo, since remodeling of the ordered MMTV promoter is lost with this deletion. Another key structural feature of BRG1 during GR signaling comes from within the ATPase itself, and these are domain features unique from the conserved DEXHc and HELICc folds that BRG1 shares with Snf2h and revealed by differences in activity seen in BRG1/Snf2h chimeras [84, 123]. Although both ATPase proteins remodel chromatin and have in vitro ATPase activity, the Snf2h ATPase domain cannot be substituted for the BRG1 domain in GR-dependent remodeling and activation. These results are interesting in light of structural data for several SNF2 family proteins that shows how loops and inserts within the ATPase as well adjacent regulatory domains are essential to the specific mode of DNA manipulation and processivity carried out by a given ATPase [34]. The continued pursuit of mechanistic details in BRG1-mediated GR dependent signaling will be important to the revealing the nature of how this protein and its complex manipulate regulatory chromatin structure during signal response.
In closing, it is important to appreciate that the detailed mechanistic analysis described above has been conducted on model promoters and verified with exemplar steroid responsive genes but not on a genomic scale [74]. Recent genome wide receptor binding studies reveal a multiplicity of binding regions relative to the preceding gene expression studies [130–132]. Studies focused on GR binding show receptor interactions across of range of nuclease-accessible sites that become receptive to GR binding though numerous pathaways and have differing requirements for BRG1-dependent remodeling [133, 134]. Such studies also highlight the cell type specificity of receptor chromatin status and remodeling requirements [135]. These binding sites have not been systematically linked to functional transcriptional outcomes. They have, however, revealed exciting correlations with histone modifications and the underlying chromatin architectures of the binding regions that confirm a prominent role for chromatin in regulating GR transcriptional activity [136]. Major challenges ahead include: a) understanding the action of binding at sites many tens of kilobases from transcription initiation sites, b) the functional linking of binding with a specific affected gene, c) the functional role of histone modifications, and d) the modifying enzymes and the contribution of chromatin remodeling complexes to the underlying chromatin architecture and its structure upon hormone activation. What is abundantly clear is that the preceding detailed molecular characterizations of promoters such as the MMTV provide an excellent guide and standard for understanding these important biological questions [137].
Highlights.
Chromatin remodeling is an essential part of steroid hormone signaling
The glucocorticoid receptor is a particularly well explored member of the steroid receptor super family
The hormone inducible MMTV promoter provides a model for signal responsive remodeling
BRG1 mediated transactivation depends upon the makeup of the BAF complex
BRG1 activity is also regulated by its own subdomains and their interacting partners
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Kasinsky HE, Lewis JD, Dacks JB, Ausio J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 5.Wolffe AP. Histone H1. Int J Biochem Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 6.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 9.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 10.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 11.Aoyagi S, Trotter KW, Archer TK. ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm. 2005;70:281–307. doi: 10.1016/S0083-6729(05)70009-1. [DOI] [PubMed] [Google Scholar]
- 12.Edmondson DG, Roth SY. Chromatin and transcription. FASEB J. 1996;10:1173–1182. doi: 10.1096/fasebj.10.10.8751719. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Aoyagi S, Archer TK. Dynamic histone acetylation/deacetylation with progesterone receptor-mediated transcription. Mol Endocrinol. 2007;21:843–856. doi: 10.1210/me.2006-0244. [DOI] [PubMed] [Google Scholar]
- 15.Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JJ, Clark DJ, Wolffe AP. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allfrey VG, Mirsky AE. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science. 1964;144:559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 18.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 19.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 20.Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding-- wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 21.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HL, Archer TK. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banks GC, Deterding LJ, Tomer KB, Archer TK. Hormone-mediated dephosphorylation of specific histone H1 isoforms. J Biol Chem. 2001;276:36467–36473. doi: 10.1074/jbc.M104641200. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharjee RN, Banks GC, Trotter KW, Lee HL, Archer TK. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol Cell Biol. 2001;21:5417–5425. doi: 10.1128/MCB.21.16.5417-5425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deterding LJ, Banks GC, Tomer KB, Archer TK. Understanding global changes in histone H1 phosphorylation using mass spectrometry. Methods. 2004;33:53–58. doi: 10.1016/j.ymeth.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharjee RN, Archer TK. Transcriptional silencing of the mouse mammary tumor virus promoter through chromatin remodeling is concomitant with histone H1 phosphorylation and histone H3 hyperphosphorylation at M phase. Virology. 2006;346:1–6. doi: 10.1016/j.virol.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 28.Hebbar PB, Archer TK. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J Biol Chem. 2008;283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan G, Zhu B. Histone variants and epigenetic inheritance. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 31.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 32.Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene. 2001;20:3076–3085. doi: 10.1038/sj.onc.1204332. [DOI] [PubMed] [Google Scholar]
- 33.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauk G, Bowman GD. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol. 2011 doi: 10.1016/j.sbi.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Kinyamu HK, Archer TK. Changes in attitude, changes in latitude: nuclear receptors remodeling chromatin to regulate transcription. Mol Endocrinol. 2006;20:1–13. doi: 10.1210/me.2005-0192. [DOI] [PubMed] [Google Scholar]
- 37.Tang L, Nogales E, Ciferri C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol. 2010;102:122–128. doi: 10.1016/j.pbiomolbio.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodwin GH, Nicolas RH. The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene. 2001;268:1–7. doi: 10.1016/s0378-1119(01)00428-0. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tailbinding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 40.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24:1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 41.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 43.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 44.Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 47.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 49.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 50.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 52.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunduz E, Gunduz M, Ouchida M, Nagatsuka H, Beder L, Tsujigiwa H, Fukushima K, Nishizaki K, Shimizu K, Nagai N. Genetic and epigenetic alterations of BRG1 promote oral cancer development. Int J Oncol. 2005;26:201–210. [PubMed] [Google Scholar]
- 55.Hasselblatt M, Gesk S, Oyen F, Rossi S, Viscardi E, Giangaspero F, Giannini C, Judkins AR, Fruhwald MC, Obser T, Schneppenheim R, Siebert R, Paulus W. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol. 2011;35:933–935. doi: 10.1097/PAS.0b013e3182196a39. [DOI] [PubMed] [Google Scholar]
- 56.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 57.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 58.Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland AM, Smith R, Salada G, Carillo A, Laity K, Gupte J, Swedlund B, Tavtigian SV, Teng DH, Lees E. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 59.Schneppenheim R, Fruhwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, Oyen F, Vater I, Siebert R. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 61.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 62.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 64.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Cheng MB, Zhang YJ, Zhong X, Dai H, Yan L, Wu NH, Shen YF. A switch from hBrm to Brg1 at IFNgamma-activated sequences mediates the activation of human genes. Cell Res. 2010;20:1345–1360. doi: 10.1038/cr.2010.155. [DOI] [PubMed] [Google Scholar]
- 70.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 71.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 72.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 73.Hebbar PB, Archer TK. Chromatin remodeling by nuclear receptors. Chromosoma. 2003;111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]
- 74.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265–266:162–167. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Markov GV, Laudet V. Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol. 2011;334:21–30. doi: 10.1016/j.mce.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 76.Ichinose H, Garnier JM, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 77.Li J, Fu J, Toumazou C, Yoon HG, Wong J. A role of the amino-terminal (N) and carboxyl-terminal (C) interaction in binding of androgen receptor to chromatin. Mol Endocrinol. 2006;20:776–785. doi: 10.1210/me.2005-0298. [DOI] [PubMed] [Google Scholar]
- 78.Hager GL, Archer TK, Fragoso G, Bresnick EH, Tsukagoshi Y, John S, Smith CL. Influence of chromatin structure on the binding of transcription factors to DNA. Cold Spring Harb Symp Quant Biol. 1993;58:63–71. doi: 10.1101/sqb.1993.058.01.010. [DOI] [PubMed] [Google Scholar]
- 79.Lee HL, Archer TK. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kinyamu HK, Archer TK. Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim Biophys Acta. 2004;1677:30–45. doi: 10.1016/j.bbaexp.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 81.Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford R, Warren BS, Hager GL. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–3263. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Archer TK, Deroo BJ, Fryer CJ. Chromatin modulation of glucocorticoid and progesterone receptor activity. Trends Endocrinol Metab. 1997;8:384–390. doi: 10.1016/s1043-2760(97)00159-8. [DOI] [PubMed] [Google Scholar]
- 83.Aoyagi S, Archer TK. Dynamics of coactivator recruitment and chromatin modifications during nuclear receptor mediated transcription. Mol Cell Endocrinol. 2008;280:1–5. doi: 10.1016/j.mce.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol Cell Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Archer TK. Nucleosomes modulate access of transcription factor to the MMTV promoter in vivo and in vitro. Ann N Y Acad Sci. 1993;684:196–198. doi: 10.1111/j.1749-6632.1993.tb32282.x. [DOI] [PubMed] [Google Scholar]
- 87.Hebbar PB, Archer TK. Nuclear factor 1 is required for both hormone-dependent chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter. Mol Cell Biol. 2003;23:887–898. doi: 10.1128/MCB.23.3.887-898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 89.Aoyagi S, Archer TK. Differential glucocorticoid receptor-mediated transcription mechanisms. J Biol Chem. 2011;286:4610–4619. doi: 10.1074/jbc.M110.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Archer TK, Lee HL. Visualization of multicomponent transcription factor complexes on chromatin and nonnucleosomal templates in vivo. Methods. 1997;11:235–245. doi: 10.1006/meth.1996.0410. [DOI] [PubMed] [Google Scholar]
- 91.Mymryk JS, Fryer CJ, Jung LA, Archer TK. Analysis of chromatin structure in vivo. Methods. 1997;12:105–114. doi: 10.1006/meth.1997.0452. [DOI] [PubMed] [Google Scholar]
- 92.Adom J, Carr KD, Gouilleux F, Marsaud V, Richard-Foy H. Chromatin structure of hormono-dependent promoters. J Steroid Biochem Mol Biol. 1991;40:325–332. doi: 10.1016/0960-0760(91)90198-e. [DOI] [PubMed] [Google Scholar]
- 93.Archer TK, Fryer CJ, Lee HL, Zaniewski E, Liang T, Mymryk JS. Steroid hormone receptor status defines the MMTV promoter chromatin structure in vivo. J Steroid Biochem Mol Biol. 1995;53:421–429. doi: 10.1016/0960-0760(95)00088-h. [DOI] [PubMed] [Google Scholar]
- 94.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pina B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 96.Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATPdependent remodeling by SWI/SNF or NURF. PLoS One. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 98.Smith CL, Archer TK, Hamlin-Green G, Hager GL. Newly expressed progesterone receptor cannot activate stable, replicated mouse mammary tumor virus templates but acquires transactivation potential upon continuous expression. Proc Natl Acad Sci U S A. 1993;90:11202–11206. doi: 10.1073/pnas.90.23.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Archer TK, Lee HL, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol. 1994;8:568–576. doi: 10.1210/mend.8.5.8058066. [DOI] [PubMed] [Google Scholar]
- 100.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Archer TK, Zaniewski E, Moyer ML, Nordeen SK. The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol Endocrinol. 1994;8:1154–1162. doi: 10.1210/mend.8.9.7838148. [DOI] [PubMed] [Google Scholar]
- 102.Kinyamu HK, Fryer CJ, Horwitz KB, Archer TK. The mouse mammary tumor virus promoter adopts distinct chromatin structures in human breast cancer cells with and without glucocorticoid receptor. J Biol Chem. 2000;275:20061–20068. doi: 10.1074/jbc.M001142200. [DOI] [PubMed] [Google Scholar]
- 103.Fryer CJ, Kinyamu HK, Rogatsky I, Garabedian MJ, Archer TK. Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J Biol Chem. 2000;275:17771–17777. doi: 10.1074/jbc.M908729199. [DOI] [PubMed] [Google Scholar]
- 104.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fryer CJ, Nordeen SK, Archer TK. Antiprogestins mediate differential effects on glucocorticoid receptor remodeling of chromatin structure. J Biol Chem. 1998;273:1175–1183. doi: 10.1074/jbc.273.2.1175. [DOI] [PubMed] [Google Scholar]
- 106.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 107.Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosidi B, Wang M, Wu W, Sharma A, Wang H, Iliakis G. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res. 2008;36:1610–1623. doi: 10.1093/nar/gkn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst) 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 110.Jeong KW, Lee YH, Stallcup MR. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J Biol Chem. 2009;284:29298–29309. doi: 10.1074/jbc.M109.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277:41674–41685. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 112.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008;283:11924–11934. doi: 10.1074/jbc.M705401200. [DOI] [PubMed] [Google Scholar]
- 114.Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- 115.Davy DA, Campbell HD, Fountain S, de Jong D, Crouch MF. The flightless I protein colocalizes with actin- and microtubule-based structures in motile Swiss 3T3 fibroblasts: evidence for the involvement of PI 3-kinase and Rasrelated small GTPases. J Cell Sci. 2001;114:549–562. doi: 10.1242/jcs.114.3.549. [DOI] [PubMed] [Google Scholar]
- 116.Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Allen MD, Religa TL, Freund SM, Bycroft M. Solution structure of the BRK domains from CHD7. J Mol Biol. 2007;371:1135–1140. doi: 10.1016/j.jmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 119.Hauk G, Bowman GD. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol. 2011;21:719–727. doi: 10.1016/j.sbi.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell. 2008;19:3308–3322. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh M, Popowicz GM, Krajewski M, Holak TA. Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex. Chembiochem. 2007;8:1308–1316. doi: 10.1002/cbic.200600562. [DOI] [PubMed] [Google Scholar]
- 122.Trotter KW, Archer TK. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol. 2004;24:3347–3358. doi: 10.1128/MCB.24.8.3347-3358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 124.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 125.Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 127.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mymryk JS, Archer TK. Influence of hormone antagonists on chromatin remodeling and transcription factor binding to the mouse mammary tumor virus promoter in vivo. Mol Endocrinol. 1995;9:1825–1834. doi: 10.1210/mend.9.12.8614418. [DOI] [PubMed] [Google Scholar]
- 129.Deroo BJ, Archer TK. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene. 2001;20:3039–3046. doi: 10.1038/sj.onc.1204328. [DOI] [PubMed] [Google Scholar]
- 130.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 131.Biddie SC, John S, Hager GL. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol Metab. 2010;21:3–9. doi: 10.1016/j.tem.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baek S, Sung MH, Hager GL. Quantitative analysis of genome-wide chromatin remodeling. Methods Mol Biol. 2012;833:433–441. doi: 10.1007/978-1-61779-477-3_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 134.Grontved L, Hager GL. Impact of chromatin structure on PR signaling: Transition from local to global analysis. Mol Cell Endocrinol. 2011 doi: 10.1016/j.mce.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cheung E, Kraus WL. Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol. 2010;72:191–218. doi: 10.1146/annurev-physiol-021909-135840. [DOI] [PubMed] [Google Scholar]
- 137.Trotter KW, Archer TK. Assaying chromatin structure and remodeling by restriction enzyme accessibility. Methods Mol Biol. 2012;833:89–102. doi: 10.1007/978-1-61779-477-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]