Abstract
Mutation of the p53 gene is the most common genetic alteration in human cancer and contributes to malignant process by enhancing transformed properties of cells and resistance to anticancer therapy. Mutant p53 is often highly expressed in tumor cells at least in part due to its increased half-life. However, whether mutant p53 expression is regulated by other mechanisms in tumors is unclear. Here, we found that histone deacetylase inhibitors suppress both wild-type and mutant p53 transcription in time- and dose-dependent manners. Consistent with this, the levels of wild-type and mutant p53 proteins are decreased upon treatment with HDAC inhibitors. Importantly, we found that upon knockdown of each class I HDAC, only HDAC8 knockdown leads to decreased expression of wild-type and mutant p53 proteins and transcripts. Conversely, we found that ectopic expression of wild-type but not mutant HDAC8 leads to increased transcription of p53. Furthermore, we found that knockdown of HDAC8 results in reduced expression of HoxA5 and consequently attenuated ability of HoxA5 to activate p53 transcription, which can be rescued by ectopic expression of HoxA5. Due to the fact that HDAC8 is required for expression of both wild-type and mutant p53, we found that targeted disruption of HDAC8 expression remarkably triggers proliferative defect in cells with a mutant, but not wild-type, p53. Together, our data uncover a regulatory mechanism of mutant p53 transcription via HDAC8 and suggest that HDAC inhibitors and especially HDAC8-targeting agents might be explored as an adjuvant for tumors carrying a mutant p53.
Keywords: p53, mutant p53, HDAC8, HDAC inhibitor, HoxA5, Transcription
Introduction
Histone deacetylases (HDACs), which remove the acetyl groups from histones and non-histone proteins, play a key role in the modulation of chromatin topology and gene expression. To date, 18 HDACs have been characterized in humans and classified into four groups based on homology to yeast HDACs. Class I HDACs consist of HDAC1-3 and 8. Class II HDACs consist of HDAC4-7 and 9–10. Class I–II HDACs are zinc-dependent enzymes. Class III HDACs, also called sirtuins 1–7, have an enzymatic activity that requires NAD+ and are insensitive to HDAC inhibitors of hydroxamic acid derivative, such as vorinostat (SAHA) which potently inhibits class I–II HDACs (1). Class IV HDAC has one member, HDAC11, which shares a conserved catalytic core with class I–II HDACs.
Aberrant expression of HDACs has been detected in various tumors. For example, HDAC1 is found to be overexpressed in prostate cancer (2) whereas HDAC2 overexpressed in gastric carcinomas, colorectal carcinomas, cervical dysplasias and endometrial stromal sarcomas (3–5). HDAC8 is found to be overexpressed in multiple tumors, especially neuroblastoma and glioma (6). It is well-characterized that overexpression of HDAC1-3 decreases, whereas knockdown of HDAC1-3 increases, p21 expression (3–5, 7). As a key cell cycle regulator, increased expression of p21 leads to tumor suppression. Thus, targeting HDACs is an ideal strategy for tumors addicted to HDACs. Indeed, HDAC inhibitors have been approved for treating cutaneous T cell lymphoma (8) and currently are being tested in phase I–II trials for advanced solid malignancies (9), hormone therapy-resistant breast cancer (10), and non-Hodgkin’s lymphoma and mantle cell lymphoma (11).
HDAC8 is a unique member of class I HDACs since it lacks the conserved C-terminal domain (12). The C-terminal domain in HDAC1-3 is necessary for interacting with other proteins and for cellular localization (13) and posttranslational modifications (14, 15). In addition, HDAC8 is found to be regulated by protein kinase A (PKA) whereas HDAC1-2 regulated by casein kinase-2 (16, 17). Furthermore, the expression profile of HDAC8 is distinct from that for other class I HDACs. HDAC8 is highly expressed in brain, kidney and prostate whereas HDAC1-3 are highly expressed in heart and placenta (6, 18). The difference might imply a distinct biological function for HDAC8. In neuroblastoma, HDAC8 is the only one that shows significant correlation with advanced disease stage (19). Knockdown of HDAC8 in cultured neuroblastoma cells results in inhibition of proliferation, reduced clonogenic growth, and cell cycle arrest (19). Interestingly, knockdown of HDAC8 has no effect on histone acetylation (19) although class I HDAC1-3 are potently regulators of histone acetylation. In addition, HDAC8-specific inhibitors induce a unique pattern of hyperacetylated proteins compared with the pan-HDAC inhibitor trichostatin A in human cells (20).
How p53 is regulated has been subject to intense scrutiny, including transcriptional regulation. The majority of transcriptional regulatory elements in the p53 promoter are found within the region upstream the transcription initiation site, including HoxA5 and p53 (21). Indeed, HoxA5 was found to increase p53 expression by binding to consensus Hox-binding sites in the p53 promoter (22). p53 activates its own expression through direct binding to a p53-responsive element in the p53 promoter under physiologic conditions or in response to cellular stress (23). However, whether mutant p53 is regulated is underexplored simply due to the perception that mutant p53 protein is hyper-stable. In fact, recent evidence suggests that mutant p53 protein is unstable and subject to polyubiquitination and proteasomal degradation (24). Thus, it is important to understand whether transcriptional regulation plays a role in mutant p53 expression. In this study, we analyzed mutant p53 transcription in tumor cells with HDAC inhibitors or specific knockdown of an individual HDAC. We found that HDAC8 is necessary for p53 transcription via HoxA5 transcription factor. Our study indicates that the use of HDAC inhibitors as a cancer therapeutic agent should be approached with caution since the status of the p53 gene may dictate the response of tumors to HDAC inhibitors alone or in combination with other chemotherapeutic agents.
Results
HDAC inhibitors decrease the level of mutant p53 protein in time- and dose-dependent manners
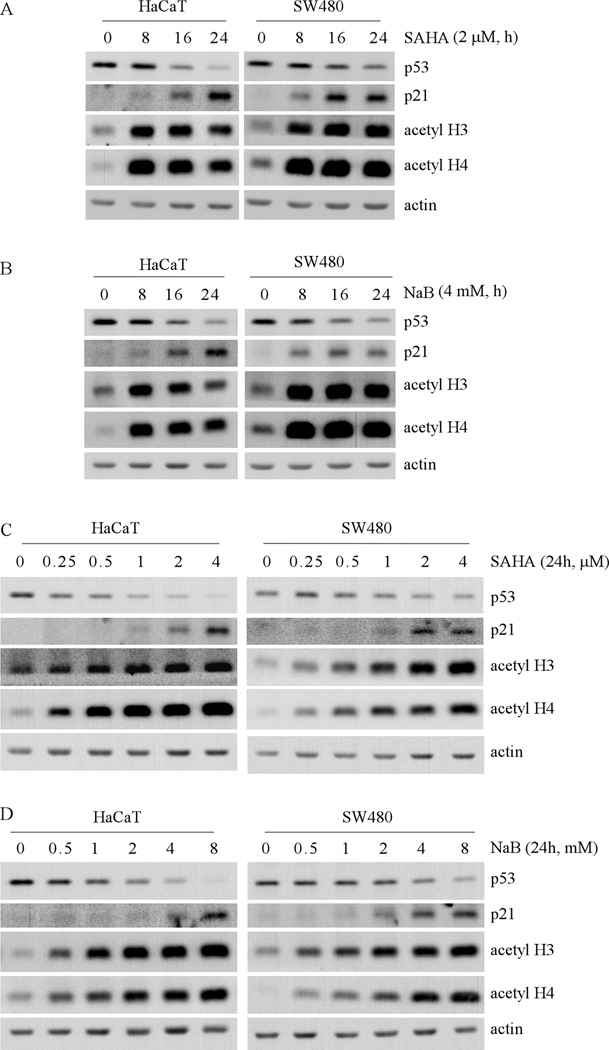
Non-histone targets of HDACs include transcription factors and other signaling proteins (25), some of which are involved in cancer development and progression. The tumor suppressor p53 is the first non-histone target for acetylation and deacetylation. HDACs can deacetylate p53 and affect its transcriptional activity (26–28). Knockdown of HDAC2 was found to alter p53 DNA-binding activity but not p53 expression or posttranslational modifications (29). A recent study showed that in tumor cells harboring a mutant p53, SAHA treatment can destabilize mutant p53 protein via inhibition of the HDAC6-HSP90 chaperone pathway (30). In this study, we explored transcriptional regulation of the p53 gene by HDACs. To confirm that mutant p53 expression can be decreased by pan-HDAC inhibitors, HaCaT and SW480 cells were treated with SAHA and sodium butyrate (NaB). We found that upon treatment of 2 µM SAHA, the level of mutant p53 protein was decreased in a time-dependent manner in HaCaT cells (Fig. 1A, left panel) and in SW480 cells (Fig. 1A, right panel). It is well-known that p21 is transcriptionally upregulated by HDAC inhibitors (31). Thus, the level of p21 protein was examined as a positive control. As expected, p21 expression in both cell lines was increased in a time-dependent manner (Fig. 1A). Consistent with SAHA treatment, the levels of acetylated histones H3 and H4 were significantly increased (Fig. 1A). Furthermore, we found that upon exposure to 4 mM NaB, the level of mutant p53 protein was decreased in HaCaT and SW480 cells whereas the level of p21 protein and acetylated histones H3 and H4 were increased (Fig. 1B).
Fig. 1. HDAC inhibitors decrease the level of mutant p53 protein in time- and dose-dependent manners.
(A) Western blots were prepared with extracts from HaCat (left panel) and SW480 (right panel) cells untreated or treated with 2 µM SAHA for 8 to 24 h, and then probed with antibodies against p53, p21, acetyl-H3, acetyl-H4 and actin, respectively. (B) The experiments were performed as in (A) except that cells were treated with 4 mM NaB. (C) Western blots were prepared with extracts from HaCaT (left panel) and SW480 (right panel) cells untreated or treated with 0.25 to 4 µM SAHA for 24 h, and then probed with antibodies as in (A). (D) The experiments were performed as in (C) except that the cells were treated with 0.5 to 8 mM NaB for 24 h.
Next, we determined how much SAHA is required for inhibiting mutant p53 expression. To test this, HaCaT and SW480 cells were treated with various doses of SAHA for 24 h (Fig. 1C). We found that the level of mutant p53 protein in HaCaT cells was decreased by SAHA at a dose of as low as 0.25 µM (Fig. 1C, left panel). As the concentration was increased to 0.5–4 µM, SAHA further decreased the level of mutant p53 in HaCaT cells (Fig. 1C, left panel). Similarly, the levels of p21 protein and acetylated histones H3 and H4 were also increased in a dose-dependent manner (Fig. 1C, left panel). Consistent with the observations in HaCaT cells, the level of mutant p53 protein in SW480 cells was also decreased by SAHA in a dose-dependent manner (Fig. 1C, right panel). Thus, in cultured solid tumor cells, the dose of SAHA (0.25 to 4 µM), which efficiently suppresses mutant p53 expression, is within the range of the clinically relevant plasma peak level of 1.7 µM in advanced leukemia patients treated with 400 mg of SAHA daily (9). Similarly, we found that upon exposure to NaB for 24 h, the level of mutant p53 protein was decreased in HaCaT and SW480 cells in a dose-dependent manner (Fig. 1D).
HDAC inhibitors decrease the level of both mature and precursor mRNAs of mutant p53 in time- and dose-dependent manners
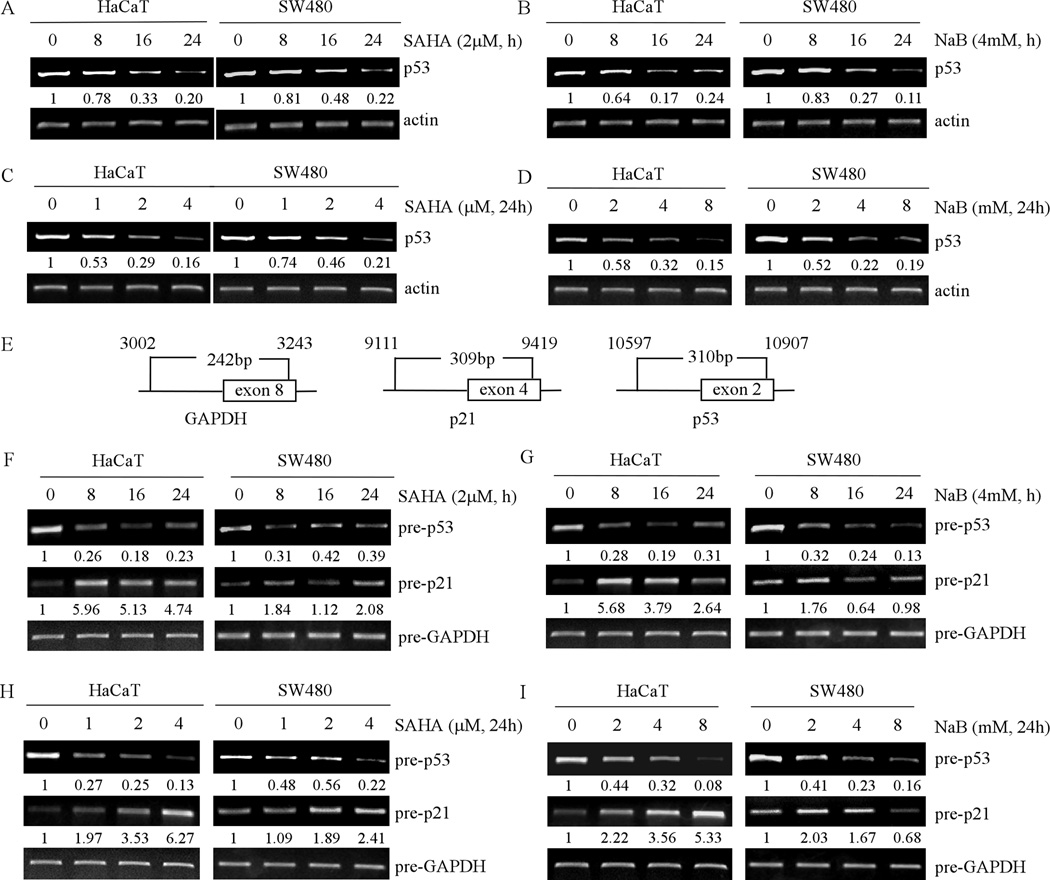
To examine whether p53 transcription is suppressed by HDAC inhibitors, RT-PCR was performed to measure the level of mutant p53 transcript in HaCaT and SW480 cells, which were untreated or treated with 2 µM SAHA or 4 mM NaB. Actin mRNA was measured as a control. We found that the level of mutant p53 mRNA in HaCaT and SW480 cells was reduced in a time-dependent manner by SAHA (Fig. 2A) and NaB (Fig. 2B). We also found that the level of mutant p53 mRNA in HaCaT and SW480 cells was reduced in a dose-dependent manner by SAHA (Fig. 2C) and NaB (Fig. 2D). Therefore, the decrease in the level of mutant p53 mRNA is likely responsible for the decrease in the level of mutant p53 protein.
Fig. 2. HDAC inhibitors decrease the level of mature and precursor mRNAs of mutant p53 in time- and dose-dependent manners.
(A) RT-PCR was performed with total RNAs isolated from HaCaT (left panel) and SW480 (right panel) cells untreated or treated with 2 µM SAHA for 8 to 24 h. Actin mRNA was amplified for a loading control. (B) The experiments were performed as in (A) except that the cells were treated with 4 mM NaB. (C) RT-PCR was performed with total RNAs isolated from HaCaT (left panel) and SW480 (right panel) cells untreated or treated with 0.25 to 4 µM SAHA for 24 h. (D) The experiments were performed as in (C) except that cells were treated with 0.5 to 8 mM NaB for 24 h. (E) Schematic presentation of primers used in (F–I) to amplify precursor mRNAs of p53, p21 and GAPDH. (F–I) RT-PCR was performed with total RNAs isolated from HaCaT (left panel) and SW480 (right panel) cells, which were untreated or treated as in (A–D). Prior to reverse transcription, total RNAs were treated with DNase I to remove genomic DNA.
To determine whether the level of mutant p53 mRNA is regulated at the transcriptional or post-transcriptional level by pan-HDAC inhibitors, we measured the level of the precursor mRNAs of p53, p21 and GAPDH with a specific set of primers (Fig. 2E). Interestingly, we found that the level of precursor mutant p53 mRNA in HaCaT and SW480 cells was markedly decreased in a time-dependent manner by SAHA (Fig. 2F) or NaB (Fig. 2G). We also found that upon exposure to SAHA (Fig. 2H) or NaB (Fig. 2I) for 24 h, the level of precursor mutant p53 mRNA was decreased in a dose-dependent manner. As expected, the level of precursor p21 mRNA was rapidly increased at 8 h upon treatment with SAHA or NaB (Fig. 2F–I), but then slightly decreased at 16 h, which is consistent with previous report (31). We would like to note that the long-term treatment with a high dose of NaB slightly decreased the level of precursor p21 mRNA in SW480 cells (Fig. 2I, left panel), which might be due to the cellular toxicity of NaB. Taken together, these findings suggested that mutant p53 expression is likely to be inhibited at the transcription level by HDAC inhibitors.
Knockdown of HDAC8 decreases the level of mutant p53 protein and transcript
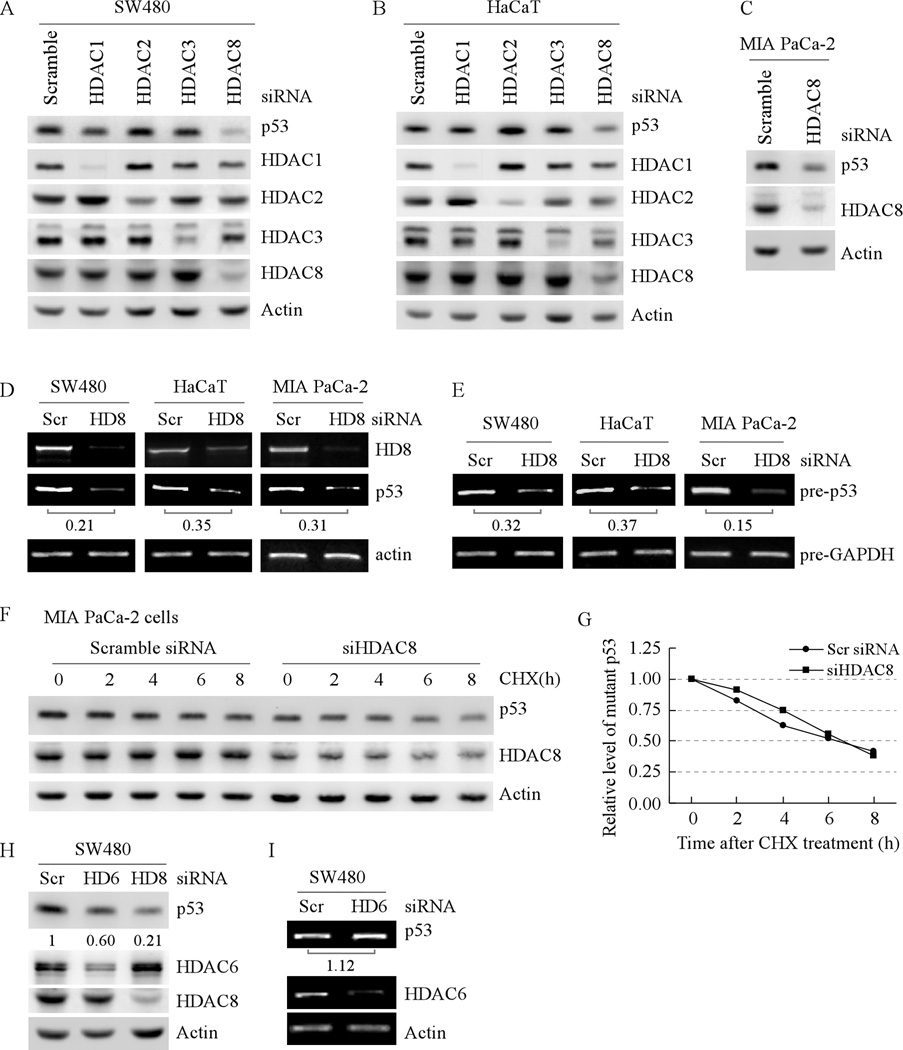
SAHA is a potent inhibitor of HDAC1-4 and 6–9 (32–34) whereas NaB inhibits most class I–II HDACs except HDAC6 and 10 (35, 36). Based on selective inhibition of HDACs by SAHA and NaB and cellular distribution of HDACs, it is likely that inhibition of HDAC1-3 and 8 is responsible for the decreased transcription of mutant p53. Since pharmacological inhibitors, including HDAC inhibitors, inherently possess non-specific or off-target effects, knockdown of an individual HDAC by siRNA was performed to identify a HDAC responsible for regulating mutant p53 transcription. We found that the level of HDAC1- 3 and 8 proteins in SW480 cells was significantly decreased by their respective siRNA but not scrambled siRNA (Fig. 3A, HDAC panels). Interestingly, we found that the level of mutant p53 protein was decreased only by HDAC8 knockdown (Fig. 3A, p53 panel). Similarly, we found that in HaCaT cells, the level of mutant p53 protein was decreased by knockdown of HDAC8 but not HDAC1-3 (Fig. 3B). Furthermore, we found that knockdown of HDAC8 led to decreased expression of mutant p53 in MIA PaCa-2 cells (Fig. 3C).
Fig. 3. Knockdown of HDAC8 decreases the level of mutant p53 protein and transcript.
(A) The level of mutant p53 protein in SW480 cells is decreased by knockdown of HDAC8 but not by HDAC1-3. Western blots were prepared with extracts from SW480 cells transfected with scrambled siRNA, siRNA against HDAC1-3, or siRNA against HDAC8 for 3 d. The blots were then probed with antibodies against p53, HDAC1-3, HDAC8 and actin, respectively. (B) The experiment was performed as in (A) except that HaCaT cells were used. (C) The experiment was performed as in (A) except that MIA PaCa-2 cells were used. (D) Knockdown of HDAC8 decreases the level of mature mutant p53 transcript in SW480 (left panel), HaCaT (middle panel) and MIA PaCa-2 (right panel) cells. RT-PCR was performed with total RNAs isolated from SW480, HaCaT, and MIA PaCa-2 cells, which were transfected with scrambled siRNA or siRNA against HDAC8 for 3 d. Actin mRNA was amplified for a loading control. (E) RT-PCR was performed as in (D) except that total RNAs were treated with DNaseI to remove genomic DNA prior to reverse transcription and that the primers for precursor mRNAs were used. The precursor mRNA of GAPDH was amplified as a loading control. (F) The half-life of mutant p53 protein is not changed by HDAC8 knockdown. Western blots were prepared with extracts from MIA PaCa-2 cells that were transfected with scrambled siRNA, or siRNA against HDAC8 for 3 d, and then treated with cycloheximide (50 µg/ml) for 0–8 h. The blots were then probed with antibodies against p53, HDAC8, and actin, respectively. (G) The relative levels of mutant p53 protein measured in (F) were normalized by levels of actin protein and then plotted versus time. (H) The level of mutant p53 protein in SW480 cells is decreased by knockdown of HDAC6 and HDAC8. Western blots were prepared with extracts from SW480 cells transfected with scrambled siRNA, or siRNA against HDAC6 or HDAC8 for 3 d. The blots were then probed with antibodies against p53, HDAC6, HDAC8 and actin, respectively. (I) Knockdown of HDAC6 has little if any effect on the level of mutant p53 transcript in SW480 cells. RT-PCR was performed as in (D).
Next, we examined whether knockdown of HDAC8 has an effect on mutant p53 transcription. We found that upon knockdown of HDAC8 in SW480, HaCaT, and MIA PaCa-2 cells, the level of mutant p53 mRNA was significantly decreased (Fig. 3D). To confirm the effect of HDAC8 knockdown on p53 transcription, the level of precursor mutant p53 transcript along with precursor GAPDH mRNA was measured. Again, we found that the level of precursor mRNA for mutant p53 was decreased by HDAC8 knockdown (Fig. 3E).
To rule out the possibility that decreased expression of mutant p53 protein is through a post-translational mechanism, MIA PaCa-2 cells were treated with cycloheximide (50 µg/ml) in the absence or presence of HDAC8 knockdown for 0–8 h. The relative levels of mutant p53 protein were quantified by Western blotting and normalized by levels of actin protein, which were then plotted versus time (h) to calculate the half-life of mutant p53. We found that the half-life for mutant p53 protein was about 6.5 h regardless of HDAC8 knockdown (Fig. 3F and G). This result suggests that HDAC8 knockdown does not target the stability of mutant p53 protein but rather inhibits mutant p53 transcription.
It is known that SAHA induces the degradation of mutant p53 protein via inhibiting HDAC6, a member of class IIb HDACs (30). However, it is still unclear whether HDAC6 regulates mutant p53 transcription. To rule out this possibility, SW480 cells were transfected with scrambled siRNA or siRNA against HDAC6. We found that HDAC6 knockdown decreased the level of mutant p53 protein (Fig. 3H), consistent with the previous report (30). However, the effect of HDAC6 knockdown on the expression of mutant p53 protein was weaker than that of HDAC8 knockdown (Fig. 3H). Furthermore, we found that knockdown of HDAC6 had little if any effect on the level of mutant p53 mRNA (Fig. 3I). Thus, our data suggest that HDAC8 but not HDAC6 is required for mutant p53 transcription.
HDAC inhibitors and knockdown of HDAC8 decrease the level of wild-type p53 protein and transcript
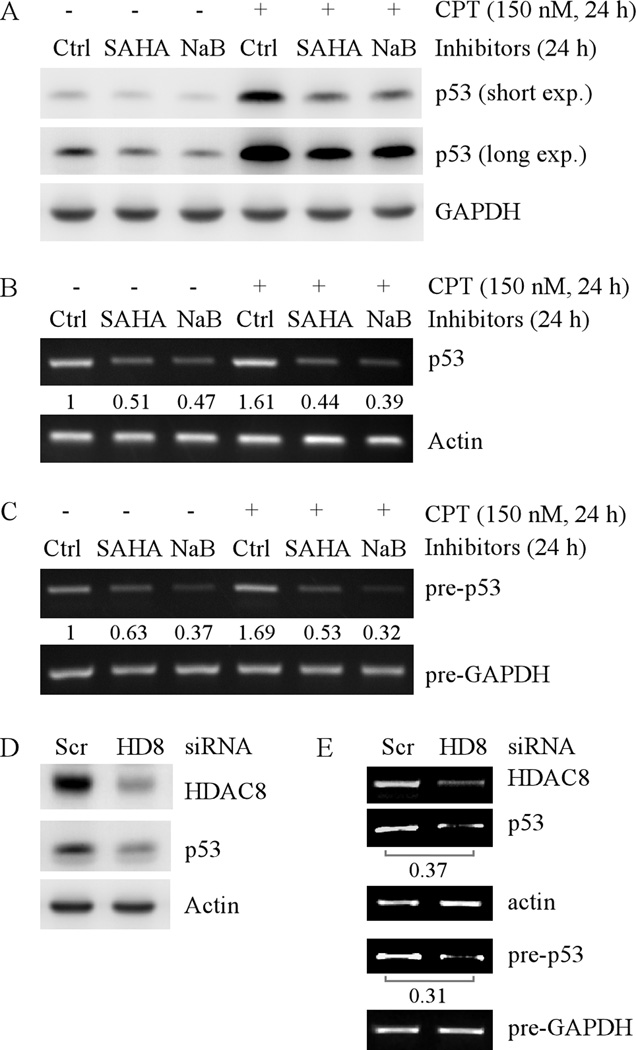
Since mutations of the p53 gene in SW480, HaCaT, and MIA PaCa-2 cell lines are located in the coding region but not the promoter region, it is possible that HDACs may regulate wild-type p53 expression via the same mechanism. To test this, HCT116 cells, which carry a wild-type p53, were treated with 2 µM SAHA or 4 mM NaB for 24h. We found that SAHA or NaB, alone or in combination with camptothecin (CPT) treatment, inhibited accumulation of wild-type p53 protein in HCT116 cells (Fig. 4A). This is consistent with an early observation in that HDAC inhibitors can suppress cisplatin-induced p53 expression in rat kidney proximal tubular cell line (37). Next, we examined the effect of SAHA or NaB on wild-type p53 transcription. We found that SAHA or NaB, alone or in combination with CPT treatment, decreased the level of both mature and precursor transcripts of wild-type p53 in HCT116 cells (Fig. 4B and C).
Fig. 4. HDAC inhibitors or knockdown of HDAC8 decreases the levels of wild-type p53 protein and transcript.
(A) Western blots were prepared with extracts from HCT116 cells untreated or treated with 2 µM SAHA or 4 mM NaB for 24 h in the absence or presence of CPT. The blots were then probed with antibodies against p53 and actin, respectively. (B) RT-PCR was performed with total RNAs isolated from HCT116 cells, which were treated as in (A). Actin mRNA was amplified for a loading control. (C) Experiments were performed as in (B), except that the total RNAs were treated with DNaseI to remove genomic DNA prior to reverse transcription and primers for precursor mRNA were used for PCR. (D) Western blots were prepared with extracts from HCT116 cells, which were transfected with scrambled siRNA or siRNA against HDAC8 for 3 d. The blots were then probed with antibodies against HDAC8, p53 and actin, respectively. (E) RT-PCR for mature and precursor mRNAs was performed with total RNAs isolated from HCT116 cells treated as in (D).
To determine whether HDAC8 is responsible for HDAC inhibitors to suppress wild-type p53 transcription, HDAC8 was knocked down in HCT116 cells. We found that the levels of wild-type p53 protein (Fig. 4D), mature and precursor transcripts (Fig.4E) were decreased by HDAC8 knockdown. Together, these data indicate that HDAC8 transcriptionally regulates p53 expression regardless of the mutation status of the p53 gene.
Ectopic expression of HDAC8 increases mutant p53 expression
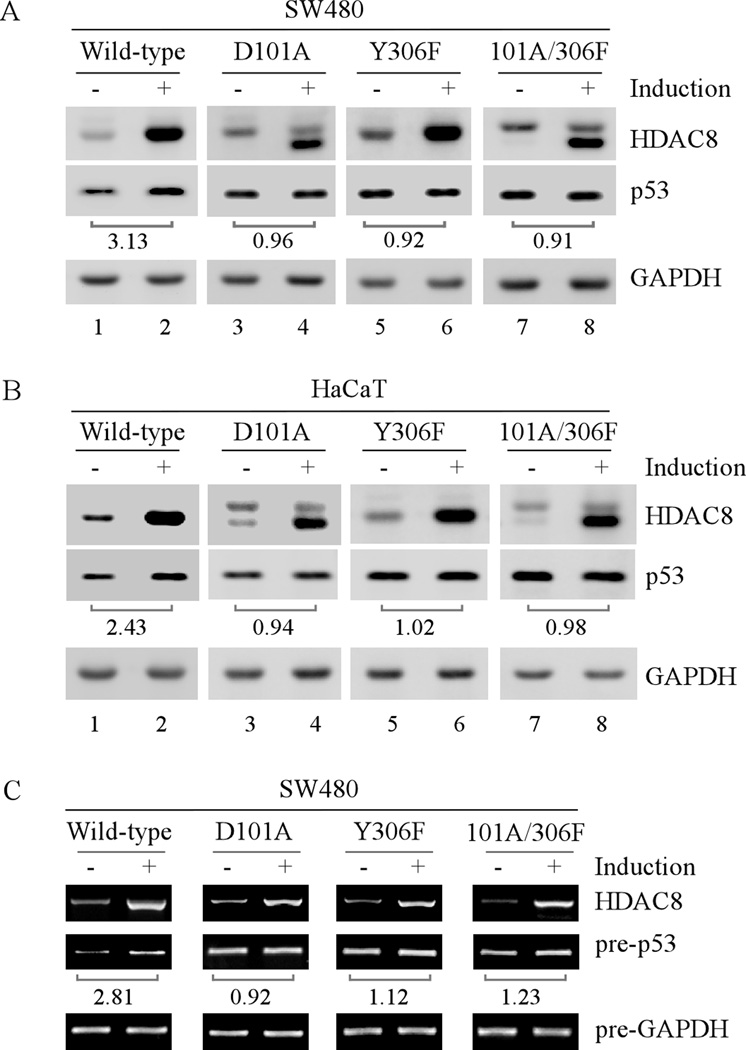
To demonstrate the effect of HDAC8 on mutant p53 expression, we examined whether ectopic expression of HDAC8 can alter mutant p53 expression in tumor cells. It has been reported that residues D101 and Y306 in HDAC8 are crucial for HDAC8 activity (38, 39). Thus, mutant HDAC8-D101A, -Y306F, and -D101A/Y306F, which carry one or both mutations in these two critical residues, were generated. We found that the level of mutant p53 protein was markedly increased by wild-type HDAC8 but not mutant D101A, Y306F, and D101A/Y306F in SW480 and HaCaT cells (Fig. 5A–B). In addition, the level of precursor mutant p53 transcript was increased upon ectopic expression of wild-type but not mutant HDAC8 (Fig. 5C). These data indicate that HDAC8 activity is required for p53 transcription.
Fig. 5. Ectopic expression of HDAC8 increases mutant p53 expression.
(A) Western blots were prepared with extracts from SW480 cells uninduced or induced to express wild-type or a mutant HDAC8 for 48 h. The blots were then probed with antibodies against HDAC8, p53 and GAPDH, respectively. The relative fold increase of mutant p53 protein by ectopic expression of HDAC8 over the control shown below the corresponding bands was calculated after normalized by levels of actin protein. (B) The experiment was performed as in (A) except that HaCaT cells were used. (C) RT-PCR was performed with total RNAs isolated from SW480 cells treated as in (A). Mature mRNA of HDAC8 and precursor mRNAs of p53 and GAPDH were amplified. The fold increase of precursor p53 transcript by ectopic expression of HDAC8 over the control shown below the corresponding bands was calculated similarly as that in (A).
HDAC8 knockdown inhibits the ability of HoxA5 to activate the p53 promoter via decreasing HoxA5 expression
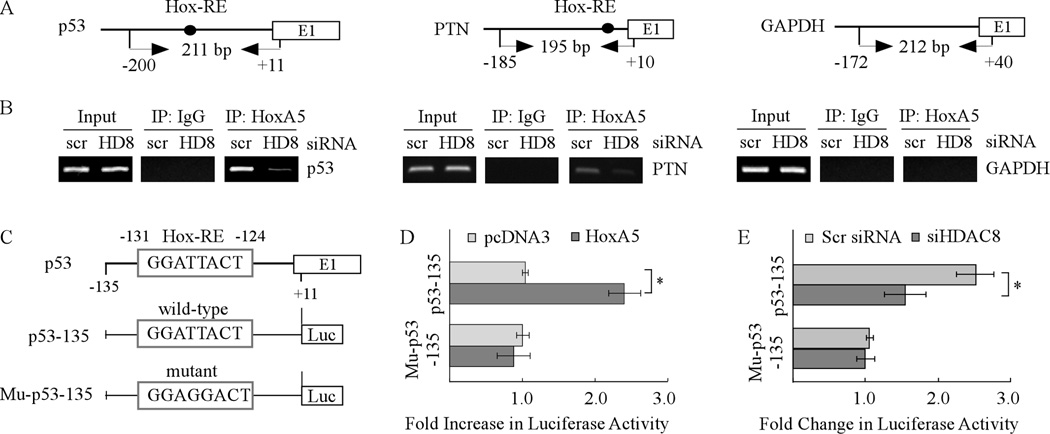
HoxA5 is a well-defined activator of p53 transcription (21). Thus, ChIP assay was performed to examine the binding of HoxA5 to the p53 promoter in cells with or without knockdown of HDAC8 (Fig. 6A). We found that while HoxA5 bound to the p53 promoter, knockdown of HDAC8 in SW480 cells markedly decreased the extent of HoxA5 associated with the p53 promoter (Fig. 6B, p53 panels). Similarly, we found that knockdown of HDAC8 obviously decreased the extent of HoxA5 binding to the promoter of PTN (Fig. 6B, PTN panels), a known transcriptional target of HoxA5 (40). As a negative control, HoxA5 was not found to associate with the GAPDH promoter (Fig. 6B, GAPDH panels). Next, we examined whether HDAC8 has an effect on HoxA5 to regulate the p53 promoter by luciferase assay. Consistent with a previous report (22), HoxA5 increased luciferase expression under the control of the p53 promoter with a consensus HoxA5-binding site but not a mutated one (Fig. 6C–D). In contrast, knockdown of HDAC8 attenuated the ability of endogenous HoxA5 to activate the p53 promoter (Fig. 6E).
Fig. 6. HDAC8 knockdown inhibits the ability of HoxA5 to activate the p53 promoter.
(A) Schematic presentation of the HoxA5 (left), PTN (middle), and GAPDH (right) promoters and the location of PCR primers used for ChIP assay. (B) HDAC8 knockdown inhibits the binding of HoxA5 to the p53 promoter. SW480 cells were transfected with scrambled siRNA or siRNA against HDAC8 for 3 d, and then HoxA5-DNA complexes were captured with anti-HoxA5 along with rabbit IgG as a control. The binding of HoxA5 protein to PTN or GAPDH promoter was measured as a positive or negative control. (C) Schematic presentation of luciferase reporter constructs including the location of a HoxA5 binding site in the p53 promoter. The reporter construct mu-p53-135 carries two nucleotide substitutions in which TT were substituted with GG. (D) Wild-type but not mutant HoxA5-binding site in the p53 promoter is responsive to HoxA5. The dual luciferase assay was performed as described in the Materials and Methods. (E) HDAC8 knockdown inhibits the luciferase activity under the control of the p53 promoter with wild-type but not mutant HoxA5-binding site. The experiment was performed as in (D) except that siRNA against HDAC8 or a scrambled siRNA was co-transfected with a luciferase reporter.
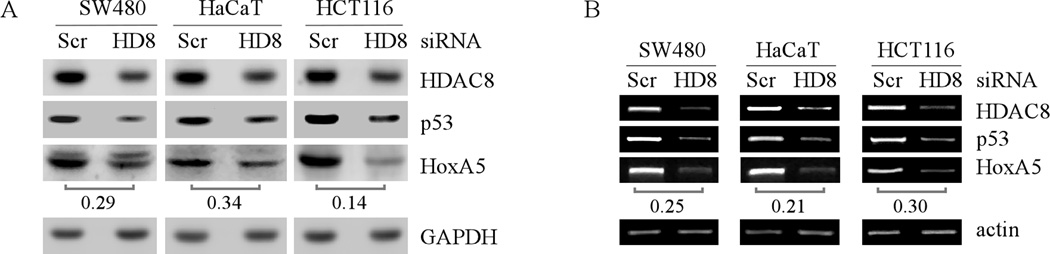
Next, to explore how HDAC8 knockdown inhibited the ability of HoxA5 to activate the p53 promoter, the level of HoxA5 protein was measured in cells with or without HDAC8 knockdown. We found that knockdown of HDAC8 obviously decreased the expression of HoxA5 protein and mRNA in SW480, HaCaT and HCT116 cells (Fig. 7A–B, HoxA5 panels). These data suggest that the inhibition of HDAC8 knockdown on p53 transcription is at least in part via decreasing the expression of HoxA5.
Fig. 7. Knockdown of HDAC8 decreases the levels of HoxA5 protein and transcript.
(A) Western blots were prepared with extracts from SW480, HaCaT, and HCT116 cells, which were transfected with scrambled siRNA or siRNA against HDAC8 for 3 d. The blots were then probed with antibodies against HDAC8, p53, HoxA5, and actin, respectively. (B) RT-PCR was performed with total RNAs isolated from SW480, HaCaT, and HCT116 cells, which were treated as in (A). Actin mRNA was amplified for a loading control.
Ectopic expression of HoxA5 restores p53 expression in HDAC8-knockdown cells
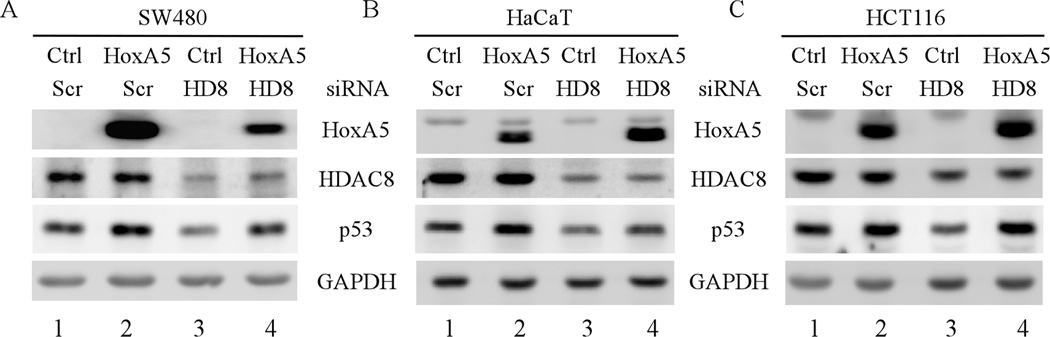
Since decreased expression of p53 by HDAC8 knockdown is possibly mediated by HoxA5, we examined whether ectopic expression of HoxA5 is sufficient to restore mutant p53 expression in HDAC8-knockdown cells. First, we found that upon ectopic expression of HoxA5, the level of mutant p53 in SW480 and HaCaT cells and wild-type p53 in HCT116 cells was increased (Fig. 8A–C, compare lanes 1–2), consistent with the data above that HoxA5 is a potent transcription activator of the p53 promoter. Next, we showed that knockdown of HDAC8 led to decreased expression of both mutant and wild-type p53 (Fig 8A–C, compare lane 1 with 3), consistent with the observations above (Figs. 3–4). Interestingly, we found that ectopic expression of HoxA5 markedly decreased the effect of HDAC8 knockdown on mutant and wild-type p53 expression (Fig. 8A–C, compare lanes 3–4). It should be noted that the level of HDAC8 protein was not significantly altered by ectopic expression of HoxA5 (Fig. 8A–B, compare lanes 1 and 3 with 2 and 4, respectively).
Fig. 8. Ectopic expression of HoxA5 restores p53 expression in HDAC8-knockdown cells.
(A) Western blots were prepared with extracts from SW480 cells which were transfected with scrambled siRNA or siRNA against HDAC8 along with or without overexpression of HoxA5 for 3 d. The blots were probed with antibodies against HoxA5, HDAC8, p53 and GAPDH, respectively. (B–C) The experiments were performed as in (A), except that HaCaT (B) and HCT116 (C) cells were used.
Targeted disruption of HDAC8 expression triggers proliferative defect in cells carrying a mutant p53
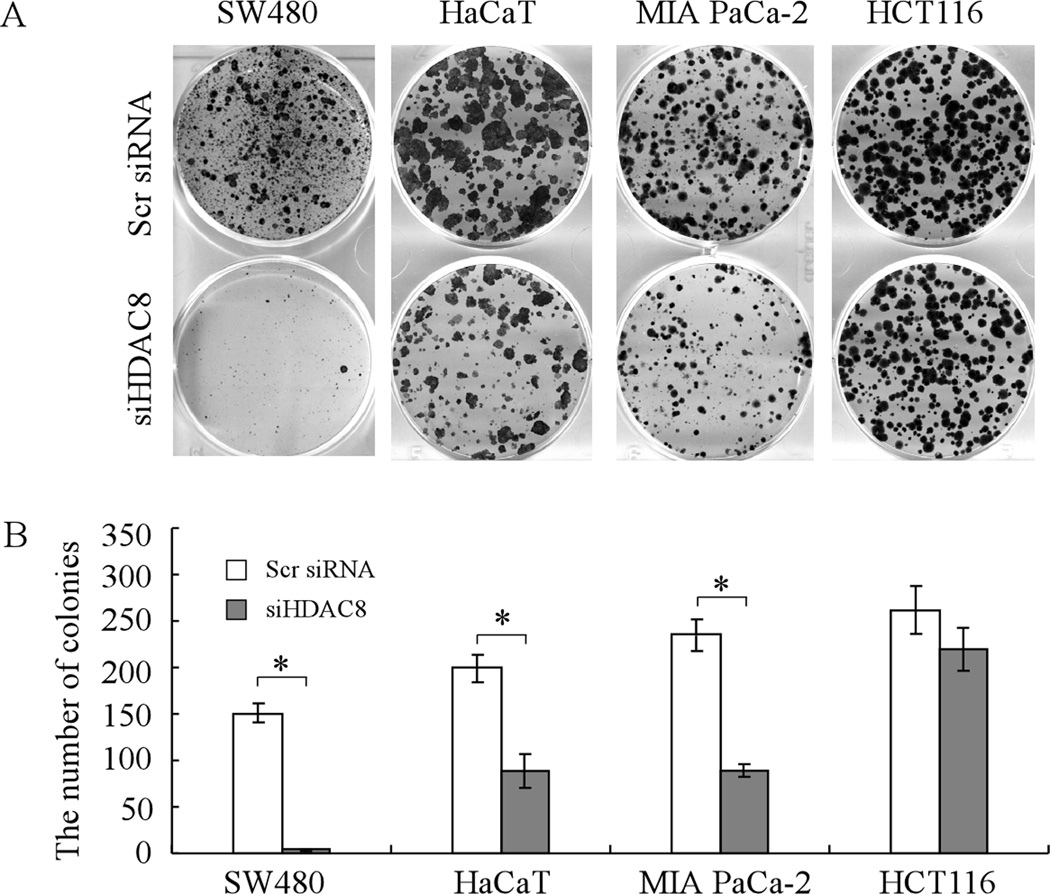
Since mutant p53 has been implicated in promoting cell survival (41, 42) and knockdown of HDAC8 leads to decreased expression of mutant p53, we tested whether knockdown of HDAC8 is capable of inhibiting the proliferation of cells containing a mutant p53. For this purpose, colony formation assays were performed with SW480, HaCaT, and MIA PaCa-2 cell lines, in which scrambled siRNA or siRNA against HDAC8 was transfected. We found that knockdown of HDAC8 significantly inhibited the proliferation of cells with a mutant p53 (Fig. 9A–B). Interestingly, we found that compared to mutant p53 cell lines, wild-type p53 cell line HCT116 was less sensitive to knockdown of HDAC8 (Fig. 9A–B). Together, these results suggest that the effect of HDAC8 knockdown on cell proliferation might be dependent on the status of the p53 gene in cells.
Fig. 9. Knockdown of HDAC8 inhibits cell proliferation of tumor cells harboring a mutant p53.
(A) Mutant p53 cell lines SW480, HaCaT, and MIA PaCa-2 and wild-type p53 cell line HCT116 were transfected with scrambled siRNA or siRNA against HDAC8 for 1 d, and then split and cultured in fresh medium for the next 15–20 days. The colonies were fixed with methanol/glacial acetic acid (7:1) and stained with 0.1% of crystal violet. (B) Quantification of the number of colonies with a diameter of >0.5 mm from three separate experiments. * represents p<0.05.
Discussion
HDAC inhibitors have been shown to induce growth suppression via increased apoptosis and decreased cell proliferation in vitro and in vivo (32, 43, 44). In addition, many studies reported that HDAC inhibitors preferentially regulate expression of genes involved in cell survival and cell death (45–51). In this study, we examined the effect of SAHA and NaB on mutant p53 expression in tumor cells. We found that in SW480 and HaCaT cell lines, HDAC inhibitors decrease the level of mutant p53 protein and mRNA in time- and dose-dependent manners. The inhibition apparently occurs at the level of transcription because precursor p53 transcript is consistently decreased by HDAC inhibitors. In addition, we found that ectopic expression of HDAC8 increases, whereas knockdown of HDAC8 decreases, the level of mutant p53 protein and transcript. Similarly, we observed that the levels of wild-type p53 protein and transcripts are decreased by HDAC inhibitors or HDAC8 knockdown. Furthermore, we demonstrated that knockdown of HDAC8 decreases the expression of HoxA5 and consequently attenuates the ability of HoxA5 to activate the p53 promoter, whereas ectopic expression of HoxA5 is able to restore p53 expression in HDAC8-knockdown cells. Together, our data provide evidence that HDAC inhibitors decreases p53 transcription via HDAC8 and HoxA5.
In this study, although the expression of mutant p53 was found to be transcriptionally regulated by HDAC8 via HoxA5, we are still challenged with unresolved questions, such as whether HoxA5 is a direct target of HDAC8 and then how HDAC8 regulates the expression of HoxA5. In contrast to class I HDAC-selective inhibitors, HDAC8-selective inhibitor do not alter histone acetylation (19, 52). In line with this, knockdown of HDAC8 has no effect on histone acetylation (19). Therefore, HDAC8 may not act via global changes of histone acetylation, but rather act specifically at certain promoter sites, one of which may be the HoxA5 gene. Thus, the future studies are warranted to explore whether HDAC8 regulates the acetylation level of histone at the site of the HoxA5 promoter or the gene itself.
The promising use of pan-HDAC inhibitors to treat cancer in the clinic is exciting (8–11). However, considering the function of HDACs and pleiotropic effects of pan-HDAC inhibitors, the application of HDAC inhibitors in the clinic should be approached with caution. In this study, we found that SAHA and NaB clearly decrease the basal level and DNA damage-induced stabilization of wild-type p53 protein in tumor cells. Furthermore, knockdown of HDAC8 leads to the decreased level of wild-type p53 protein, and mature and precursor transcripts. It's worth noting that targeted disruption of HDAC8 expression triggers proliferative defect in cells with a mutant, but not wild-type, p53. Therefore, an effective personalized cancer therapy by HDAC inhibitors requires not only a thorough understanding of the mechanism of action for HDACs and their inhibitors but also a thoughtful consideration of genetic and epigenetic alterations for each individual cancer, such as the status of the p53 gene. Otherwise, the use of HDAC inhibitors with an agent that can activate wild-type p53 for cancer therapy may actually cause antagonistic or adverse effects rather than increase the efficacy against tumors with wild-type p53. Nevertheless, we would like to emphasize that due to the high frequency of p53 mutation and the requirement of HDAC8 for mutant p53 transcription, HDAC inhibitors and especially HDAC8-targeting agents might be explored as an adjuvant for tumors carrying a mutant p53.
Materials and Methods
Cell Culture
Human keratinocyte HaCaT cell line (containing mutant H179Y/R282W), human colorectal adenocarcinoma SW480 cell line (containing mutant R273H/P309S), human pancreatic cancer MIA PaCa-2 cell line (containing mutant R248W), colorectal carcinoma HCT116 cell line, and human lung carcinoma H1299 cell line were cultured in DMEM (Invitrogen) medium supplemented with ~10% fetal bovine serum (Hyclone). To generate cell lines that inducibly express wild-type HDAC8, mutant HDAC8(D101A), HDAC8(Y306F) or HDAC8(D010A/Y306F), pcDNA4 carrying cDNA encoding wild-type or a mutant HDAC8 was transfected into SW480 or HaCaT cells, which express a tetracycline repressor by pcDNA6. The resulting wild-type or mutant HDAC8 producing cell lines were selected with zeocin and then confirmed by Western blot analysis with anti-HDAC8.
Plasmids
To generate a construct expressing wild-type HDAC8, an 1,149-bp DNA fragment encoding aa 1–377 was amplified with forward primer P1, 5'-AAGCTTACCATGGAGGAGCCGGAGGAACCG-3', and reverse primer P2, 5'-CTCGAGCTAGACCACATGCTTCAGATTC-3'. The fragment was confirmed by sequencing and then cloned into pcDNA4. To generate a construct expressing mutant HDAC8(D101A), HDAC8(Y306F) or HDAC8(D010A/Y306F), a two-step PCR-based method was used. For HDAC8(D101A), fragment 1 was amplified with forward primer P1 and reverse primer 5`-GCTGGGCAGGCATAACCTAGCCCATATTCTA-3`; fragment 2 was amplified with forward primer 5'-GCTAGGTTATGCCTGCCCAGCCACTGAAG-3` and reverse primer P2. For HDAC8(Y306F), fragment 1 was amplified with forward primer P1 and reverse primer 5`-TGGCAAGGTTAAAGCCTCCTCCTCCCAAAATGA-3`; fragment 2 was amplified with forward primer 5'-GAGGAGGCTTTAACCTTGCCAACACGGCTC-3` and reverse primer P2. Then, fragments 1–2 were mixed together as a template and amplified with primers P1 and P2. To generate cDNA encoding HDAC8(D010A/Y306F), the same procedure and primers used to generate cDNA for HDAC8(Y306F) were used except that HDAC8(D101A) was used as template for generation of fragments 1–2. The resulting fragments encoding HDAC8(D101A), HDAC8(Y306F), and HDAC8(D010A/Y306F) were confirmed by sequencing and cloned into pcDNA4, respectively.
To generate a construct expressing HoxA5, an 828-bp DNA fragment encoding aa 1–270 was amplified with forward primer, 5'- AAGCTTACCATGAGCTCTTATTTTGTAAACTCA-3', and reverse primer, 5'- CTCGAGTCAGGGACGGAAGGCCCCTC -3'. The fragment was confirmed by sequencing and then cloned into pcDNA3.
To generate a luciferase reporter under the control of the p53 promoter with a HoxA5 binding site (nt −131 to −124), an 146-bp DNA fragment containing the p53 promoter (nt −135 to +11) was amplified using genomic DNA with forward primer 5`-GGTACCAGGCGGATTACTTGCCCTTACTTGTCATG -3` and reverse primer 5`-CTCGAGTGGCTCTAGACTTTTGAGAAGCTCA-3`. The PCR product, p53-135, was confirmed by DNA sequencing and cloned into pGL2-Basic vector. The resulting luciferase reporter was designated as pGL2-p53-135. The luciferase reporter under the control of the p53 promoter with point mutations in the HoxA5 binding site was generated with forward primer, 5`-GGTACCAGGCGGAGGACTTGCCCTTACTTGTCATG-3`, and reverse primer, 5`-CTCGAGTGGCTCTAGACTTTTGAGAAGCTCA -3`. The TT in the HoxA5-binding site (nt −128 to −127) were replaced with GG.
Small interference RNA oligos
21-bp small interference RNAs (siRNAs) specific against HDAC1, 5’-CAGCGACUGUUUGAGAACCdTdT-3’, HDAC2, 5’-GCAUCAGGAUUCUGUUACGdTdT-3, HDAC3, 5’-GGCUUCACCAAGAGUCUUAdTdT-3’, HDAC6, 5’-GGCCAAGGAUAUACCAUCdTdT-3’, HDAC8, 5’-GUCCCGAGUAUGUCAGUAUdTdT-3’, and a scrambled siRNA, 5’-GCAGUGUCUCCACGUACUAdTdT-3’, were synthesized by Dharmacon RNA Technologies.
Antibodies
Rabbit anti-p53 (FL-393), anti-HDAC8 (H-145), anti-HDAC1 (H-51), anti-HDAC2 (H-54), anti-GAPDH (FL-335), and anti-p21 (C-19) were purchased from Santa Cruz Biotechnology Inc. Rabbit anti-HoxA5 (Mid) was purchased from Invitrogen. Rabbit anti-acetyl-histone H3, anti-acetyl-histone H4, and anti-HDAC3 was purchased from Upstate Biotechnology. Rabbit anti-actin was purchased from Sigma.
Reverse transcription PCR (RT-PCR) assay
Total RNA was isolated from cells using TRIzol reagent (Invitrogen). cDNA was synthesized using an Iscript™ cDNA Synthesis kit (Bio-Rad). To measure mature p53 mRNA, RT-PCR was performed with forward primer 5`-GACCGGCGCACAGAGGAAG-3`and reverse primer 5`-GAGTTTTTTATGGCGGGAGG-3`. Mature HDAC8 mRNA was amplified with forward primer 5`- GATCAGAGGAGCAGGAACTG-3` and reverse primer 5`- CTGCTTAT GCAGTGCATATGC -3`. Mature p21 mRNA was amplified with forward primer 5`-CATCTTCTGCCTTAGTCTCAG-3` and reverse primer 5`-TCAAATGAAAAAGAATTCAGGTC -3`. Mature actin mRNA was amplified with forward primer 5`-TCCATCATGAAGTGTGACGT-3` and reverse primer 5`-TGATCCACATCTGCTGGAAG-3`.
To measure precursor p53 mRNA, RT-PCR was performed with forward primer 5`-GGAAGTCCCTCTCTGATTGTC-3` and reverse primer 5`-CTCGACGCTAGGATCTGACT-3`. precursor p21 mRNA was amplified with forward primer 5`-GACACTCCATAATACCCCTC-3`and reverse primer 5`-CATCTTCTGCCTTAGTCTCAG-3`. Precursor GAPDH mRNA was amplified with forward primer 5`-GGACTGGCTTTCCCATAATTTC-3`and reverse primer 5`-AAGGTCATCCCTGAGCTGAAC-3`.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed as previously described (53, 54). SW480 cells, transfected with scrambled siRNA or HDAC8 specific siRNA for 72 h, were cross-linked by 1% formaldehyde for 10 min at room temperature. Nuclear extracts were prepared and chromatin was sonicated to generate 200-to 1000-bp DNA fragments. Protein-DNA complexes were immunoprecipitated with HoxA5 antibody and control IgG. The DNA-protein cross-links were reversed by heating at 65°C for 4 h. After phenol and chloroform extraction, DNA was purified by ethanol precipitation. To amplify the region from nt −200 to +11 in the p53 promoter, PCR was performed with forward primer 5`-CAGGTCGGCGAGAATCCTG-3` and reverse primer 5’-TGGCTCTAGACTTTTGAGAAGC-3’. A region in the PTN promoter was amplified to serve as a positive control with forward primer 5`-TTTGCACTCATCTGAAGAATAG-3` and reverse primer 5`-CTTTGCACTCGAGAGCTGAT-3`. A region in the GAPDH promoter was amplified to serve as a negative control with forward primer 5`-AAAAGCGGGGAGAAAGTAGG-3` and reverse primer 5`-AAGAAGATGCGGCTGACTGT-3`.
Luciferase Assay
A dual luciferase assay was performed in triplicate according to the manufacturer’s instructions (Promega, Madison, WI, USA). Briefly, 0.25 µg of a luciferase reporter, 0.25 µg of pcDNA3 or pcDNA3-HoxA5 that expresses HoxA5 protein, and 3 ng of Renilla luciferase assay vector pRL-CMV were cotransfected into H1299 cells. The fold increase in relative luciferase activity is a product of the luciferase activity induced by HoxA5 divided by that induced by an empty pcDNA3 vector. For luciferase assay with HDAC8 knockdown, 0.25 µg of a luciferase reporter, 0.32 µg of scrambled siRNA or siRNA against HDAC8, and 3 ng of Renilla luciferase assay vector pRL-CMV were cotransfected into SW480 cells. The fold change in relative luciferase activity is a product of the luciferase activity induced by siRNA against HDAC8 divided by that induced by scrambled siRNA.
Statistics
All experiments were performed in triplicates. Two-group comparisons were analyzed by two-sided Student's t test. p values were calculated, and p< 0.05 was considered significant.
Acknowledgement
This work is supported in part by National Institutes of Health Grant R01 CA121137. We thank S. Townson (Merck Pharmaceuticals) for SAHA.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 2.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 3.Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 4.Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 5.Hrzenjak A, Moinfar F, Kremser ML, Strohmeier B, Staber PB, Zatloukal K, et al. Valproate inhibition of histone deacetylase 2 affects differentiation and decreases proliferation of endometrial stromal sarcoma cells. Mol Cancer Ther. 2006;5:2203–2210. doi: 10.1158/1535-7163.MCT-05-0480. [DOI] [PubMed] [Google Scholar]
- 6.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 7.Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 8.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 9.Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13:3605–3610. doi: 10.1158/1078-0432.CCR-07-0162. [DOI] [PubMed] [Google Scholar]
- 10.Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–1835. doi: 10.1038/bjc.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschbaum M, Frankel P, Popplewell L, Zain J, Delioukina M, Pullarkat V, et al. Phase II study of vorinostat for treatment of relapsed or refractory indolent non-Hodgkin's lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29:1198–1203. doi: 10.1200/JCO.2010.32.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Ayer DE. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 14.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 15.Tsai SC, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 19.Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 20.Krennhrubec K, Marshall BL, Hedglin M, Verdin E, Ulrich SM. Design and evaluation of 'Linkerless'hydroxamic acids as selective HDAC8 inhibitors. Bioorg Med Chem Lett. 2007;17:2874–2878. doi: 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 21.Reisman D, Loging WT. Transcriptional regulation of the p53 tumor suppressor gene. Semin Cancer Biol. 1998;8:317–324. doi: 10.1006/scbi.1998.0094. [DOI] [PubMed] [Google Scholar]
- 22.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66:6982–6989. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- 24.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, et al. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 28.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 29.Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007;67:3145–3152. doi: 10.1158/0008-5472.CAN-06-4397. [DOI] [PubMed] [Google Scholar]
- 30.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 34.Noureen N, Rashid H, Kalsoom S. Identification of type-specific anticancer histone deacetylase inhibitors: road to success. Cancer Chemother Pharmacol. 2010;66:625–633. doi: 10.1007/s00280-010-1324-y. [DOI] [PubMed] [Google Scholar]
- 35.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 36.Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 2011;3:166–179. [PMC free article] [PubMed] [Google Scholar]
- 37.Dong G, Luo J, Kumar V, Dong Z. Inhibitors of histone deacetylases suppress cisplatin-induced p53 activation and apoptosis in renal tubular cells. Am J Physiol Renal Physiol. 2010;298:F293–F300. doi: 10.1152/ajprenal.00410.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertrand P. Inside HDAC with HDAC inhibitors. Eur J Med Chem. 2010;45:2095–2116. doi: 10.1016/j.ejmech.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, Rubin E, Zhang H, Chung S, Jie CC, Garrett E, et al. Identification of transcriptional targets of HOXA5. J Biol Chem. 2005;280:19373–19380. doi: 10.1074/jbc.M413528200. [DOI] [PubMed] [Google Scholar]
- 41.Yan W, Chen X. Identification of GRO1 as a critical determinant for mutant p53 gain of function. J Biol Chem. 2009;284:12178–12187. doi: 10.1074/jbc.M900994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan W, Chen X. Characterization of functional domains necessary for mutant p53 gain of function. J Biol Chem. 2010;285:14229–14238. doi: 10.1074/jbc.M109.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15:3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers AE, Banerjee S, Chaplin T, Dunne J, Debernardi S, Joel SP, et al. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur J Cancer. 2003;39:1165–1175. doi: 10.1016/s0959-8049(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 46.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 47.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasakawa Y, Naoe Y, Sogo N, Inoue T, Sasakawa T, Matsuo M, et al. Marker genes to predict sensitivity to FK228, a histone deacetylase inhibitor. Biochem Pharmacol. 2005;69:603–616. doi: 10.1016/j.bcp.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peart MJ, Tainton KM, Ruefli AA, Dear AE, Sedelies KA, O'Reilly LA, et al. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2003;63:4460–4471. [PubMed] [Google Scholar]
- 52.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 53.Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Biol. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]