Abstract
The HMGN family of proteins binds to nucleosomes without any specificity for the underlying DNA sequence. They affect the global and local structure of chromatin, as well as the levels of histone modifications and thus play a role in epigenetic regulation of gene expression. This review focuses on the recent studies that provide new insights on the interactions between HMGN proteins, nucleosomes, and chromatin, and the effects of these interactions on epigenetic and transcriptional regulation.
Keywords: HMGN protein, epigenetics, transcription
1. Introduction
Chromatin-binding architectural proteins modulate nucleosome dynamics and changes in both global and local chromatin structure, but are still a poorly understood facet of chromosome biology. One such family of proteins, the HMGNs (High Mobility Group N), has been shown to play a role in epigenetic regulation of gene expression. ‘Epigenetics’ is the study of heritable non-sequence-based variations in DNA, and at the molecular level, epigenetic regulation of gene expression is mediated through changes in chromatin structure and nucleosome composition, as well as through chemical modifications in DNA and histones. HMGNs function by binding to the nucleosome core particle (CP), and the details of the interaction between the two have recently been elucidated. The binding of HMGNs to nucleosomes is a dynamic process that affects chromatin structure and post-translational histone modifications, and impacts the cellular transcription profile. HMGN proteins can therefore be considered to function as modulators of epigenetic processes that affect the fidelity of gene expression. This review focuses on the interactions between HMGN proteins, nucleosomes, and chromatin, and the effects of these interactions on epigenetic and transcriptional regulation. Other aspects of the HMGN protein family are reviewed in [1–10].
2. HMGN proteins bind specifically to nucleosomes
The HMGN protein family, which is found in all vertebrates, contains 5 members. All HMGN proteins contain a highly conserved nucleosomal binding domain (NBD), which is the hallmark of the HMGN family [8, 11, 12]. Within the NBD, the invariant sequence RRSARLSA is the core sequence that specifically anchors HMGN proteins to the 147 bp nucleosome core particle, the building block of the chromatin fiber [12]. In addition, all HMGNs have a bipartite nuclear localization signal (NLS) in their N-termini [13] and contain a negatively charged regulatory domain (RD) in their C-termini [14] (Figure 1a). This domain facilitates chromatin unfolding and plays a role in the effects of HMGNs on histone posttranslational modifications. HMGN1–4 have similar molecular weights, ~10kDa, whereas the newly discovered HMGN5 contains an unusually long C-terminus (more than 300 amino acids in the mouse protein, and about 200 in humans) [6, 15]. This long negatively charged C-terminal region of HMGN5 interacts with the positively charged linker histone H1, and in addition, targets HMGN5 to specific chromatin domains[16]. Among the 5 variants, HMGN2 is the most conserved member of the HMGN family according to sequence analysis. While HMGN1, 2, 3, and 5 are encoded by genes containing 6 exons and have been detected in all vertebrates that have been tested, the gene coding for HMGN4 is the product of a retroviral transposition and appears to be restricted to primates (J.K., unpublished).
Figure 1. Functional domains of of HMGN proteins and a model of the HMGN2-CP complex.
(a) Domain structure of HMGN proteins. All HMGN proteins contain three functional domains: a bipartite nuclear localization signal (NLS, shown in blue, a nucleosome binding domain, NBD, shown in red, and a C-terminal regulatory domain, RD, shown in black). The core binding domain within the NBD is indicated, and the four amino-acids essential for nucleosome binding are shown in red. (b) The HMGN2-core particle complex. One HMGN2 molecule binds to each side of the nucleosome core with its NBD domain. The highly conserved core of the NBD interacts with an acidic patch formed by histone H2A and H2B, while the C-terminal portion interacts with the DNA near or at the two major groves flanking the nucleosomal dyad axis. The binding of an HMGN to the nucleosome places the C-terminal tail of that HMGN near the site where the linker DNA exits and enter the CP, potentially affecting the binding of linker histone H1 to this region [48, 49]. This model supports earlier results that the N-terminal domain of HMGN targets a restricted region in histone H2B, whereas the C-terminal domain targets a restricted region in the N terminus of histone H3 (dotted oval). In addition, it also supports previous conclusions that HMGNs protect the ends of DNA flanking the nucleosomal dyad axis. Modified from [19].
HMGNs function by binding specifically to the 147 bp nucleosome core particle, independently of DNA sequence. Therefore, understanding the molecular mechanism whereby HMGNs affect various biological processes requires an understanding of the major features of the HMGN-CP complex. Previous studies have shown that two molecules of HMGN1/N2 specifically bind to the nucleosome core near its dyad axis and also protect the ends of DNA from hydroxyl radical cleavage, suggesting that HMGNs bridge two adjacent DNA strands on the surface of the particles [17]. By site-specific protein photocrosslinking, we also demonstrated that the N-terminal domain of HMGN1 targets a restricted region in histone H2B, whereas the C-terminal domain targets a restricted region in the N terminus of histone H3 [18]. Recent results obtained by Methyl-Transverse Relaxation Optimized Nuclear Magnetic Resonance Spectroscopy (methyl-TROSY) fully confirm these previous findings and provide additional details on the structure of the HMGN2-nucleosome complex [19]. One HMGN2 molecule binds to each side of the nucleosome core. The highly conserved core sequence of the NBD (see Fig 1a) interacts with the acidic patch formed by the H2A and H2B histone dimer. The C-terminal portion of the NBD interacts with the DNA near the nucleosomal entry/exit point while the unstructured C-terminal of HMGN2 protrudes beyond the periphery of the nucleosomal DNA and can interact with the adjacent N-terminal of H3 (Fig. 1B).
All HMGNs bind to chromatin with very similar affinities [12]. Both in vitro and in vivo, 2 molecules of an HMGN bind to a CP and form complexes containing a single type of HMGN variant [12, 20]. Under physiological conditions, two different types of HMGN variants never bind to the same nucleosome [20]. During metaphase, the two conserved serines in the core NBD of HMGNs are phosphorylated, preventing HMGN dimer-DNA complexes from forming, and the proteins associate with chromatin with low affinity as monomers [21, 22].
In addition to phosphorylation, the post-translational acetylation of HMGN proteins also affects their binding to nucleosomes. HMGN proteins were the first nonhistone protein shown to be acetylated by histone acetylases. The acetylase p300 acetylates both HMGN1 and HMGN2 at several different sites [23], whereas pCAF acetylates only HMGN2, predominantly at lysine 2 [24]. The acetylation of HMGN1 or HMGN2 reduces their nucleosome binding abilities, while phosphorylation of serine residues S6, S20 and S24 in HMGN1 or S24 and S26 in HMGN2 abolishes their binding to nucleosomes [25].
The binding of HMGN proteins to nucleosomes is highly dynamic [8, 26]. Fluorescence Recovery After Photobleaching (FRAP) experiments demonstrated that the mobility of GFP-labeled HMGNs is slower than that of GFP alone, but significantly faster than that of linker histone H1 [15, 16, 27, 28]. Significantly, point mutations that abolish the specific binding of HMGNs to the nucleosome in vitro increased their mobility in vivo, an indication that the FRAP results are a true reflection of their chromatin interactions [12, 15, 16, 25]. The emerging picture reveals that HMGNs bind transiently to nucleosomes and continuously exchange between chromatin binding sites. Nevertheless, because the time HMGNs spend in transit from one nucleosome to another is significantly shorter than the time spent bound to a nucleosome, most of the time, most of the HMGNs are bound to chromatin. The amount of HMGN proteins in the nucleus is sufficient to bind to only about 1% of the nucleosomes. The dynamic binding and rapid exchange of the proteins ensures that potentially all nucleosomes can associate with HMGNs.
3. HMGN proteins modulate global chromatin structure
Using simian virus 40 minichromosomes as an in vitro model system, it was found that HMGN1 partially disrupted histone H1-dependent chromatin condensation, and thus counteracted H1-mediated transcription inhibition [29]. In living cells, wild-type HMGN1 or HMGN2, but not their nucleosome-binding deficient mutants, increased the mobility of histone H1 [26], suggesting HMGNs compete with H1 by binding to the same nucleosomes. Thus, HMGNs compete with linker histone H1 for nucleosome binding and reverse H1 induced chromatin compaction (Figure 2).
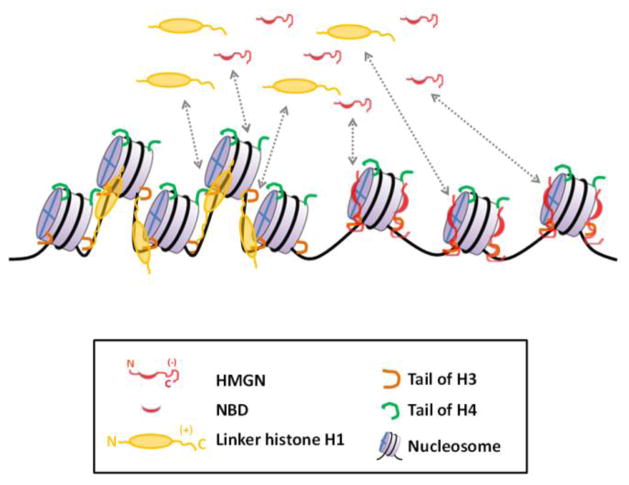
Figure 2. A model for the dynamic modulation of chromatin structure by HMGNs.
In living cells, HMGN proteins compete with linker histone H1 and bind dynamically to nucleosomes. HMGNs also interact with histone H3 and H4 tails. The C-terminal domain of HMGNs interacts with the N-terminal of histone H3, and the interaction of the NBD with the acidic patch in the histone octamer competes with the binding of the N-terminal of H4 from a neighboring nucleosome. The overall effect of HMGN binding is chromatin decompaction.
The structure of the HMGN2-nucleosome complex (Figure 1b) suggests a mechanism whereby HMGNs compete with histone H1. As discussed above, the C-terminal region of the NBD binds to nucleosomal DNA near the entry/exit point, placing the C-terminal tail of HMGN2 near the dyad axis and in proximity to the linker DNA, potentially interfering with the binding of linker histone H1 to this region. Indeed, unlike the full length protein, the HMGN1–43 deletion mutant, which lacks the C-terminal tail, had no effect on H1 binding [19]. The newly discovered HMGN5, which contains an unusually long and negatively charged C-terminus, interacts with the positively-charged H1 tail more efficiently than other HMGN proteins. This interaction interferes with the dynamic binding of H1 to chromatin; the ensuing lack of H1 binding leads to chromatin decompaction and modulation of the cellular transcription profile [15].
HMGNs reduce the compaction of chromatin not only by interfering with the binding of linker H1, but also by affecting the interaction of the N-termini of both H3 and H4 with neighboring nucleosomes. Thus, cross linking experiments indicated that the C-terminal domain of HMGNs interacts with the N-terminal of histone H3 [18], while the recent methyl-based NMR analysis revealed that the conserved core NBD binds to the H2A.H2B acidic patch, the known binding site of the N-terminal of H4 from a neighboring nucleosome. HMGNs therefore unfold chromatin by targeting the two main elements essential for maintaining chromatin compaction, linker histone H1 and the N-terminals of histones H3 and H4. Chromatin decompaction by HMGNs increases access of the nucleosomal DNA to regulatory factors and thus modulates DNA-dependent activities like transcription and DNA repair. HMGN-mediated chromatin decompaction can therefore either enhance or suppress transcription, by allowing access to either transcriptional activators or repressors [15, 30].
In addition, HMGN proteins seem to play a role in chromatin remodeling by affecting the action of ATP-dependent chromatin remodeling complexes. HMGN1 and N2, but not their mutants that do not bind to chromatin, reversibly and dynamically repress the chromatin remodeling mediated by some SWI/SNF complexes [31]. Further analysis demonstrated that HMGN1/N2 did not directly inhibit the ATPase activity of these factors but reduced their chromatin binding affinities [31]. However, another report suggested HMGN1 did not affect chromatin remodeling mediated by the SWI/SNF complex [32]. The specific function of HMGN proteins in chromatin remodeling remains to be elucidated.
4. HMGNs and Post-translational Histone Modifications
Post-translational modifications of histones are epigenetic marks shown to play an important role in modulating cellular processes such as gene expression and cell cycle progression [33]. These reversible modifications include methylation and acetylation of lysine residues, and phosphorylation of serine residues [33], all of which are added and removed from the histone tail in a dynamic manner by enzymes which modify specific histone residues in nucleosomes. Due to the frequent interactions between HMGNs and the nucleosome, and their specific interactions with the nucleosomal histones, HMGNs can be expected to affect the levels of some of the histone modifications.
The first evidence of HMGNs affecting histone modifications was the discovery that HMGN1 modulates the phosphorylation of histone H3 [34]. Hmgn1−/− cells have elevated levels of H3S10p, and in vitro studies verified that indeed HMGN1 reduces the rate of H3S10 phosphorylation in nucleosomes but not in purified histone H3 [34]. The binding of HMGN1 to the nucleosome might interfere with the ability of kinases to phosphorylate histone H3, either by steric hindrance or by inducing conformational changes in the histone tail [34]. Phosphorylation of S10 on histone H3 is known to cause changes in chromatin associated with transcriptional activation and is a marker for mitotic chromosomes [35, 36].
In addition, HMGN1 was shown to enhance the acetylation of lysine 14 [37], the acetylation and methylation of H3K9 and the phosphorylation of H2AS1. Hmgn1−/− cells have reduced levels of H3K14Ac, and exogenous expression of HMGN1, but not of its mutant that does not bind to chromatin, in these cells restored normal acetylation [37]. HMGN1 enhances the levels of H3K14Ac by stimulating HAT activity rather than by inhibiting HDAC activity [37]. Subsequent studies indicated that HMGN1-stimulated enhancement of H3K14Ac levels optimizes the cellular response to heat shock, as determined by the levels of Hsp70 transcripts [38]. HMGN1 also affects the levels of H3K9Me and H3K9Ac [34], modifications known to affect transcription [33]. HMGN1 has also been found to reduce the levels of H2AS1p [39], in much the same manner as it does in H3S10p [34], and both phosphorylated serines are markers for mitotic chromosomes [40].
Significantly, the effects of HMGN1 on the levels of H3K14Ac and H3S10p are distinct from those of HMGN2, an indication of HMGN variant-specific effects. The full spectrum of histone modifications affected by the various HMGNs is not known. Nevertheless, these studies establish the principle that HMGNs, although they have no enzymatic activity, can modulate specific histone modifications, thereby affecting epigenetic processes.
The ability of HMGN1 to affect these modifications suggests a more complicated function for architectural proteins than simply altering chromatin structure by competing with H1 for nucleosome binding. Indeed several studies indicate that the HMGN-mediated changes in histone modifications are physiologically significant. The cellular response to ionizing radiation (IR) includes a global increase in H3K14Ac, as well as the auto-phosphorylation of the protein kinase ataxia-telangiectasia mutated (ATM) [41]. These responses are affected by HMGN1; in HMGN1-deficient cells, marked decreases in H3K14Ac and in ATM auto-phosphorylation (and in activation of its downstream targets) are observed [41]. Treatment with HDAC inhibitors rescues the ATM activation in cells lacking HMGN1, indicating that HMGN1 affects ATM phosphorylation by enhancing the levels of histone acetylation [41]. Similarly, HMGN1 has been shown to affect the levels of H3K14Ac at the Hsp70 promoter [38]. Heat shock is an evolutionarily conserved process involved in protecting cells against various types of damage, and is well known to induce transcription of the Hsp70 gene [42]. Loss of HMGN1 reduces the level of H3K14Ac in the Hsp70 promoter and the number of Hsp70 transcripts produced upon heat shock [38]. These HMGN1 dependent effects can be reversed by treatment with histone deacetylase inhibitors, again underlining the general principle that HMGN1 can affect the cellular phenotype by modulating the levels of histone modifications [38].
5. Global Organization of HMGNs in Chromatin
HMGNs bind dynamically to chromatin and continuously turn over on the surface of the nucleosomes, without any specificity for the underlying DNA sequence. A major outstanding question is whether the proteins bind randomly to all nucleosomes or whether they preferentially turn over at selected sites. Global chromatin immunoprecipitation sequence analysis in human T cells revealed that HMGN1 co-localizes with DNase I hypersensitive sites, promoters, functional enhancers, and transcription factor binding sites, all of which are associated with chromatin regulatory sites and active transcription [43]. It is as yet unknown whether HMGN1 itself generates or maintains these sites, or whether it simply has a propensity for binding there. Experiments are currently underway to distinguish between these possibilities, to determine the global chromatin positioning of all HMGN variants, and to elucidate the role of these variants in generating and maintaining regulatory sites in chromatin.
6. Transcriptional Effects of HMGNs
While the structure of the various members of the HMGN protein family is quite uniform, except for HMGN5, which has a large c-terminal domain [15], their biological effects are not identical. Part of the biological specificity of some of the variants, such as HMGN3, can be attributed to tissue-specific expression [2, 44, 46]. Since HMGN proteins affect transcription [29, 30], their biological effects are most likely due to their effects on the cellular transcription profile.
The extent to which HMGN variants have specific effects on transcription has been recently tested by systematically analyzing mouse embryonic fibroblasts (MEFs) isolated from HMGN knock-out mice and their wild type littermates. In addition, the transcription profile of MEFs in which the amount of various HMGN variants was doubled has also been analyzed. When over-expressed or knocked-down, HMGN1, HMGN2, HMGN3 and HMGN5 each affect the transcription of a unique set of genes [30]. The target genes identified generally do not share any common functions and thus cannot be categorized into any specific pathway, and while the changes were statistically significant the magnitude of change was not very high. Most likely HMGN proteins are not major regulators of transcription, but instead serve to optimize transcription from chromatin templates. Nevertheless, the effects are sufficient to affect the cellular phenotype, since loss of HMGNs impairs the cellular ability to withstand various stresses such as DNA damage or heat shock.
One cell type in which HMGN function is clear is pancreatic islet cells; in these cells, HMGN3 is highly expressed, and loss of this expression leads to a mildly diabetic phenotype [44, 45]. Further studies led to the identification of two specific gene targets in pancreatic β-cells, the products of which were known to affect insulin secretion: the Glut2 glucose transporter, and Kir6.2, a component of the potassium channel which affects insulin secretion [44]. Subsequent analysis revealed that HMGN3 and the transcription factor PDX1 mutually reinforce each other’s binding to the Glut2 promoter, increasing its histone acetylation and enhancing its transcriptional activity [44]. Loss of HMGN3 has also been observed to down-regulate glucagon levels in pancreatic β-cells, although the mechanism by which it does so remains unknown [45].
Although the HMGN proteins have no enzymatic activity, their presence in the nucleus is important for proper transcriptional regulation. HMGN proteins modulate local and global chromatin structure, and affect transcriptional fidelity. This transcriptional fine-tuning affects important processes, such as DNA repair, heat shock response and metabolic regulation. Each HMGN variant affects the expression of a distinct set of genes, suggesting that they are not fully redundant. The presence of all HMGNs seems therefore to be necessary to ensure proper cellular function, especially when cells must respond rapidly to stress.
7. Future Directions
Although the biochemical properties of HMGN proteins and their interactions with chromatin have been extensively studied, the physiological functions of HMGN proteins are still largely unknown, and are currently being studied in knockout mice. As elaborated above, analysis of Hmgn1−/− mice revealed that loss of this variant leads to an increase in tumorigenicity, hypersensitivity to stress such as heat shock or DNA damage induced by either UV or ionizing radiation, and mild developmental abnormalities [46]. The gene coding for HMGN1 is located in the critical Down syndrome region of chromosome 21, and recent studies revealed that either up-regulation or loss of HMGN1 leads to altered behavior [47]. The HMGN3 variant is highly expressed in beta cells, and Hmgn3−/− mice are mildly diabetic [45]. The phenotype of mice lacking additional HMGN variants is currently under investigation. A full discussion of the phenotypes found thus far is outside the scope of this review. Future study may be focused not only on discovering the further physiological functions of HMGN proteins, but also on the molecular mechanisms underlying these functions. The emerging picture suggests that by modulating epigenetic regulatory mechanisms, such as global chromatin structure and histone modifications, HMGN variants fine-tune the fidelity of transcription and affect the cellular phenotype.
Highlights.
HMGN proteins bind to nucleosomes and affect epigenetic processes
The mechanism of HMGN binding to the Core Particle has recently been elucidated
HMGN proteins affect global and local chromatin organization
Each HMGN variant affects transcription of a separate set of genes
Acknowledgments
The research described was supported by the Center for Cancer Research, intramural program of the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Catez F, Hock R. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim Biophys Acta. 1799:15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Furusawa T, Cherukuri S. Developmental function of HMGN proteins. Biochim Biophys Acta. 1799:69–73. doi: 10.1016/j.bbagrm.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlitz G. HMGNs, DNA repair and cancer. Biochim Biophys Acta. 1799:80–85. doi: 10.1016/j.bbagrm.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pogna EA, Clayton AL, Mahadevan LC. Signalling to chromatin through post-translational modifications of HMGN. Biochim Biophys Acta. 1799:93–100. doi: 10.1016/j.bbagrm.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Postnikov Y, Bustin M. Regulation of chromatin structure and function by HMGN proteins. Biochim Biophys Acta. 1799:62–68. doi: 10.1016/j.bbagrm.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta. 1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N, Hansen U. Transcriptional regulation by HMGN proteins. Biochim Biophys Acta. 1799:74–79. doi: 10.1016/j.bbagrm.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 9.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Crippa MP, Alfonso PJ, Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J Mol Biol. 1992;228:442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- 12.Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol Cell Biol. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hock R, Scheer U, Bustin M. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J Cell Biol. 1998;143:1427–1436. doi: 10.1083/jcb.143.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trieschmann L, Alfonso PJ, Crippa MP, Wolffe AP, Bustin M. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malicet C, Rochman M, Postnikov Y, Bustin M. Distinct properties of human HMGN5 reveal a rapidly evolving but functionally conserved nucleosome binding protein. Mol Cell Biol. 31:2742–2755. doi: 10.1128/MCB.05216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 18.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci U S A. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci U S A. 108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postnikov YV, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 21.Cherukuri S, Hock R, Ueda T, Catez F, Rochman M, Bustin M. Cell cycle-dependent binding of HMGN proteins to chromatin. Mol Biol Cell. 2008;19:1816–1824. doi: 10.1091/mbc.E07-10-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlitz G, Hock R, Ueda T, Bustin M. The dynamics of HMG protein-chromatin interactions in living cells. Biochem Cell Biol. 2009;87:127–137. doi: 10.1139/O08-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, Martin B, Bustin M. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J Biol Chem. 2000;275:11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- 24.Herrera JE, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, Bustin M. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol Cell Biol. 2001;21:5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 28.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 29.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rochman M, Taher L, Kurahashi T, Cherukuri S, Uversky VN, Landsman D, Ovcharenko I, Bustin M. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res. 39:4076–4087. doi: 10.1093/nar/gkq1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill DA, Peterson CL, Imbalzano AN. Effects of HMGN1 on chromatin structure and SWI/SNF-mediated chromatin remodeling. J Biol Chem. 2005;280:41777–41783. doi: 10.1074/jbc.M509637200. [DOI] [PubMed] [Google Scholar]
- 33.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 34.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Davie JR, Spencer VA. Signal transduction pathways and the modification of chromatin structure. Prog Nucleic Acid Res Mol Biol. 2001;65:299–340. doi: 10.1016/s0079-6603(00)65008-0. [DOI] [PubMed] [Google Scholar]
- 36.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 37.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belova GI, Postnikov YV, Furusawa T, Birger Y, Bustin M. Chromosomal protein HMGN1 enhances the heat shock-induced remodeling of Hsp70 chromatin. J Biol Chem. 2008;283:8080–8088. doi: 10.1074/jbc.M709782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber CM, Turner FB, Wang Y, Hagstrom K, Taverna SD, Mollah S, Ueberheide B, Meyer BJ, Hunt DF, Cheung P, Allis CD. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma. 2004;112:360–371. doi: 10.1007/s00412-004-0281-9. [DOI] [PubMed] [Google Scholar]
- 41.Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, Rochman M, Bustin M, Zhao K. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol Cell Biol. 31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurahashi T, Furusawa T, Ueda T, Bustin M. The nucleosome binding protein HMGN3 is expressed in pancreatic alpha-cells and affects plasma glucagon levels in mice. J Cell Biochem. 109:49–57. doi: 10.1002/jcb.22377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic beta cells and affects insulin secretion. Mol Cell Biol. 2009;29:5264–5276. doi: 10.1128/MCB.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak M, West KL, Postnikov YV, Haines DC, Bustin M. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005;65:6711–6718. doi: 10.1158/0008-5472.CAN-05-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abuhatzira L, Shamir A, Schones DE, Schaffer AA, Bustin M. The Chromatin Binding Protein HMGN1 Regulates the Expression of Methyl CpG Binding Protein 2 (MECP2) and Affects the Behavior of Mice. J Biol Chem. doi: 10.1074/jbc.M111.300541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]