Abstract
Hepatitis C virus/human immunodeficiency virus (HCV/HIV) coinfected patients demonstrate accelerated progression to severe liver injury in comparison to HCV monoinfected patients, although the underlying mechanisms are unclear owing to infection of separate tissue compartments with two distinct viral pathogens. Microarray analysis of paired liver biopsy and peripheral blood mononuclear cell (PBMC) specimens from HCV/HIV coinfected and HCV monoinfected patients identified a gene expression signature associated with increased inflammation and immune activation that was present only in liver and PBMC samples from coinfected patients. We also identified in these samples liver- and PBMC-specific signatures enriched with fibrogenic/hepatic stellate activation and proinflammatory genes, respectively. Finally, Bayesian networks were constructed by assimilating these data with existing data from liver and PBMC samples from other cohorts, augmenting enrichment of biologically important pathways and further indicating that chronic immune activation in HCV/HIV coinfection may exacerbate liver disease progression in coinfected patients.
Keywords: hepatitis C virus, human immunodeficiency virus, HCV/HIV coinfection, liver, peripheral blood, hepatic fibrosis, systems biology, gene expression, pathogenesis
Introduction
Chronic HCV infection may lead to severe hepatic injury, including fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). Liver disease typically progresses slowly, with severe disease developing over 10–30 years following HCV infection. However, human immunodeficiency virus (HIV) coinfection can profoundly impact the natural history of HCV infection. Several studies have demonstrated that HCV/HIV coinfected patients exhibit increased hepatic inflammation (Di Martino et al., 2001; Martinez-Sierra et al., 2003; Pol et al., 1998), and progress more rapidly to severe liver disease (Graham et al., 2001; Soto et al., 1997) in comparison to HCV-monoinfected patients. Due to the effectiveness of highly active antiretroviral therapy (HAART), the incidence of HIV progression to acquired immune deficiency syndrome (AIDS) has been dramatically decreasing. However, HCV/HIV-coinfected patients are increasingly experiencing morbidity and mortality associated with HCV-induced liver disease.
The mechanisms underlying rapid liver disease progression in HCV/HIV co-infected patients are poorly understood and are likely multifactorial. Contributing factors which may accelerate liver disease progression include HIV-associated immune dysfunction (Gonzalez et al., 2009b; Sandberg, Falconer, and Gonzalez, 2010), oxidative stress (Baum et al., 2011; Lin et al., 2011; Wanchu et al., 2009), apoptosis (Jang et al., 2011), enhanced HCV replication (Lin et al., 2008), viral modulation of cytokine/chemokine responses (Allison et al., 2009), and HIV infection or activation of resident liver cells such as hepatic stellate cells or Kupffer cells (Tuyama et al., 2010). Unfortunately, no widely accepted experimental model of HCV/HIV coinfection is currently available. Therefore, detailed studies of liver disease pathogenesis have been limited, and are restricted to clinical samples.
Previously, we analyzed liver biopsies from HCV mono- and HCV/HIV coinfected patients and identified gene expression signatures indicative of dysregulation of apoptosis, increased immune cell activation, and decreased innate antiviral responses compared to normal liver reference samples (Walters et al., 2006). However, a specific intrahepatic signature distinguishing HCV/HIV coinfected from HCV monoinfected patients was not identified. In this study, we utilized core needle liver biopsies and a whole genome microarray with over twice the human transcriptome coverage of those used previously to identify intrahepatic gene signatures specifically associated with HCV/HIV. We also explored previous observations that HCV mono- and HCV/HIV coinfection induce unique immunologic gene expression profiles in peripheral mononuclear blood cells (PBMC) (Kottilil et al., 2009). In addition to liver biopsy specimens, PBMCs from the same subjects were evaluated by microarray to investigate how transcriptional programming of immune cells in the periphery might contribute to hepatic inflammation, cellular infiltration, and disease progression.
Expression profiling of clinical specimens can prove challenging due to numerous confounding variables, limitations on sample sizes, and heterogeneous subject characteristics. In an attempt to mitigate some of these challenges, and to expand the scope of our functional analysis, we used our transcriptomic data to construct Bayesian networks in conjunction with published liver (Lamb et al., 2011; Schadt et al., 2008; Zhong et al., 2010) and PBMC gene expression data (Emilsson et al., 2008) obtained from larger independent cohorts. These networks allowed us to identify additional genes in critical pathways functionally associated with HCV/HIV co-infection, enhancing our understanding of the complex biology of liver disease progression in HCV/HIV coinfected patients.
Results
Subject characteristics
Demographic and clinical characteristics of the subjects are shown in Table 1. In total, 10 HCV monoinfected and 13 HCV/HIV coinfected patients were recruited in this study. The two groups were well-matched for age, ethnic background, body mass index (BMI), duration of HCV infection, alcohol use, serum ALT levels, HCV genotype, HCV viral load, and Batts-Ludwig liver disease stage. The HCV/HIV coinfected group was predominantly male (92%) and demonstrated significantly lower CD4 T cell counts (mean 451 versus 1120, p=.0014) compared to the HCV monoinfected group. Also, the majority of coinfected patients (77%) received anti-retroviral treatment, which likely contributed to the lack of detectable HIV RNA in 63% of the subjects. Both groups were naïve to HCV therapy. Seventy percent of the subjects reported abstinence from alcohol use for at least one year prior to enrollment. We also performed a correlation analysis to assess which clinical parameters might confound our gene expression results, and identified HIV status and consequent CD4 cell counts as the only parameters significantly influencing gene expression (data not shown). It is likely that the correlation with CD4 T cell counts reflects the lower counts observed in the HIV-positive subjects. Due to the limited sample size, a multivariate analysis was not performed. We specifically did not find any association between gene expression and other HIV-specific covariates such as serum HIV quantitation (suppressed or not) and the use of antiretroviral therapy, though we note that in this study, these patient subgroups are too small to perform a statistically robust correlation analysis.
Table 1.
Characteristics of HCV-Infected Subjects
| HIV- negative (n=10) | HIV-positive (n=13) | P value | |
|---|---|---|---|
| Age | 47.2 ± 7.5 | 42.6 ± 7.8 | .18 |
| Gender (% male) | 60 | 92 | .06 |
| Race (%) | .44 | ||
| Caucasian | 90 | 69 | |
| African American | 10 | 23 | |
| Native American | 0 | 7.7 | |
| HCV duration (years) | 24.5 ± 7.8 | 22.0 ± 11.0 | .57 |
| Alcohol use in lifetime (gm/d) (median, IQR) | 24.1, 40.7 | 17.0, 46.9 | .80 |
| Alcohol use in last 6 months (gm/d) (median, IQR) | 0, 0 | 0, .9 | .45 |
| ALT (U/L) | 92 ± 50 | 68± 36 | .23 |
| HCV genotype (% 1) | 80 | 77 | .86 |
| HCV RNA level (log10) | 6.2 ± 0.5 | 6.3 ± 1.0 | .60 |
| Antiretroviral therapy, % | NA | 77 | - |
| HIV RNA level (log10)* | NA | 4.5 ± 0.7 | - |
| HIV RNA level, % undetectable (<400 copies/mL) | NA | 54 | - |
| CD4 cell count (cells/μL) | 1120± 471 | 451 ± 292 | .0014 |
| HCV disease grade (0–4)† | 2.5 ± .7 | 1.8 ± .8 | .05 |
| HCV disease stage (0–4)† | 2.1 ± .3 | 2.1 ± 1.0 | .9 |
Values expressed as mean ± SD unless stated otherwise
NA, not applicable; IQR, interquartile range
In those with detectable virus
Batts-Ludwig scoring system
Common transcriptional signatures in liver and PBMC samples from HCV/HIV coinfected patients
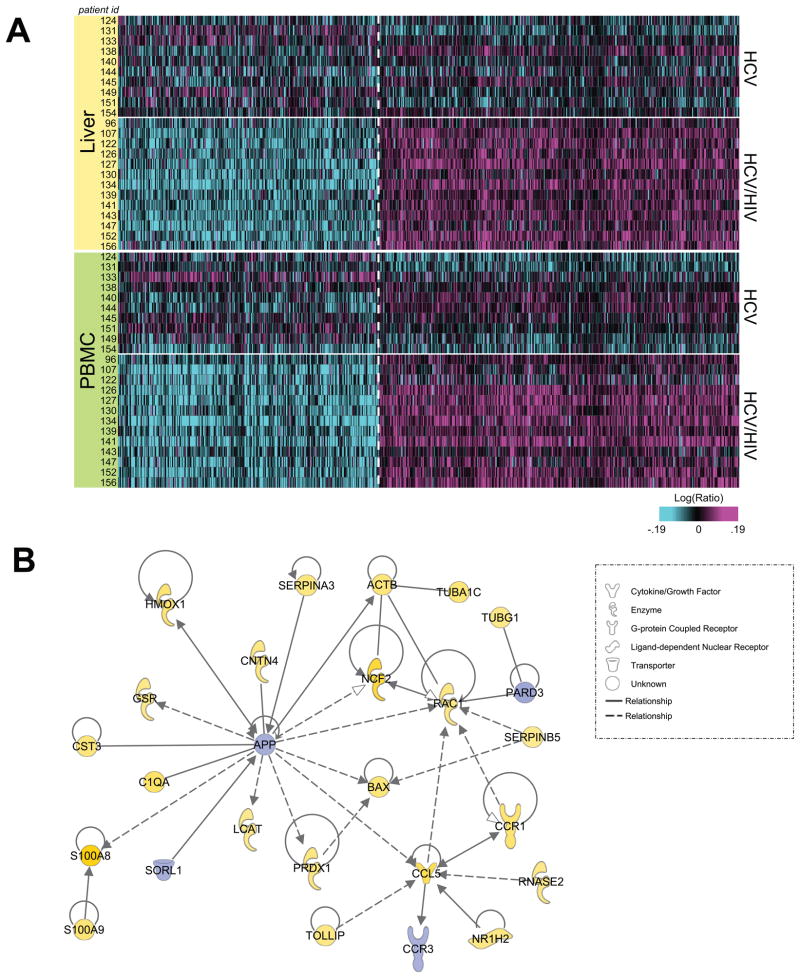
Using a two-way ANOVA approach, a molecular signature common to liver and PBMC samples from coinfected patients was identified. A total of 467 upregulated and 338 downregulated differentially expressed genes (DEG; p < 0.01) were identified in both samples (Figure 1A; Table S1). Common differential expression in both liver and PBMC may indicate that these pathways are comparably regulated separately in both tissues, and/or that many PBMC have migrated into and substantially contribute to the overall gene expression profile in liver. Functional analysis was performed on the 805 DEG common to both tissues using Ingenuity Pathway Analysis (IPA). Many of the top functional categories among the upregulated genes were associated with inflammation, indicating that distinct mechanisms may drive progression of hepatic inflammation in coinfected patients. Among 84 upregulated genes associated with inflammatory and immunological responses and disease (Table S1), we observed DEG associated with components of complement, chemokines, and antigen presentation and T cell activation (Figure 1B). The presence of gene expression changes associated with immune activation and migration may indicate enhanced trafficking of activated immune effector cells from the periphery to the liver in HCV/HIV coinfected patients.
Figure 1. Common gene signature distinguishing HCV/HIV coinfected patients.
A. Heatmap of 805 significant differentially expressed genes in liver and PBMC from HCV/HIV coinfected patients compared to HCV monoinfected as determined by two-way ANOVA (p<0.01) B. IPA network showing connected genes related to immune cell migration and inflammation from the common signature.
Identification of hepatic signatures of coinfection
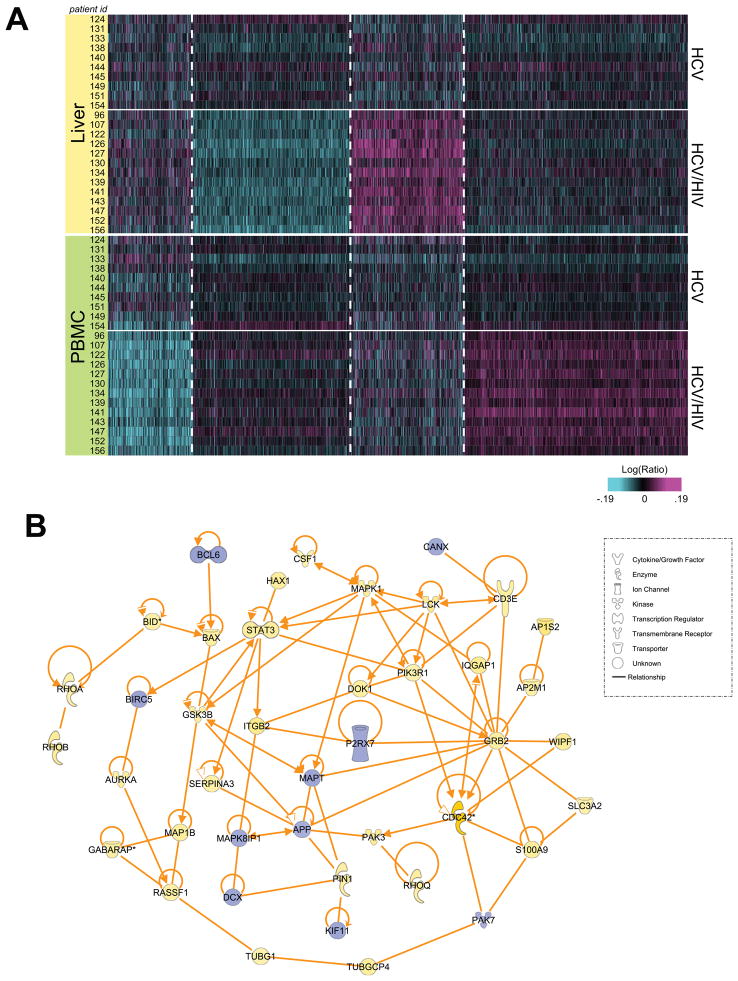
We also sought to identify significant hepatic DEG characteristic of HCV/HIV coinfected patients. Using one-way ANOVA (p<0.05, fold change >1.15 in at least 7 samples), we identified transcriptional signatures in the coinfected patient cohort using microarray data from liver samples only (Figure 2A; Table S2). Functional analysis by IPA demonstrated that in liver samples from coinfected patients, over 250 genes related to infectious disease and immune responses were upregulated, including 67 specifically related to HIV infection. Many of these genes are functionally similar to those identified in the common signature, and are associated with chemotaxis and cellular migration, including various chemokines, integrins (ITGAD, ITGA5, ITGA7, ITGB2), actin and tubulin cytoskeletal components (ACTB, ACTG, TUBA1A, TUBA1C, TUBA4A, TUBG1), and multiple Ras-like homolog members and related genes [(RHOA, RHOB, RHOD, RHOG, RHOQ, Ras-related C3 botulinum toxin 1, rho family small GTP binding protein 1 (RAC1)] involved in the recruitment of circulating immune cells to the liver. Many DEG associated with T cell activation processes, including human leukocyte antigens (HLA-DQA1, HLA-DQB1) that mediate antigen presentation, molecules associated with dendritic cell (DC) maturation (CD209), and T cell receptor signaling machinery (CD3, CD8A) were upregulated. This suggests that the T cells infiltrating the liver in coinfected patients may be transcriptionally distinct from T cells of HCV monoinfected patients, and may have a more activated phenotype due to chronic HIV infection. The substantial overlap (286 genes) between the hepatic signature and the signature common to both liver and PBMC (identified using two-way ANOVA) supports an important contributing role for infiltrating immune cells within the livers of coinfected patients.
Figure 2. Compartmental gene signatures distinguishing HCV/HIV coinfected patients.
A. Heatmap of 3039 significantly differentially expressed genes in liver or PBMC identified by one-way ANOVA (p<0.05, fold change >1.15 in at least 7 patients) and separated by K-means clustering. B. IPA network of intrahepatic signature showing directly connected molecules associated with inflammation and immunity.
We further observed the upregulation of genes functionally linked to hepatic stellate cell (HSC) activation including the Rho family GTPases, actins, myosin light chain 6 (MYL6), and cell division cycle 42 (CDC42), a major regulator of cytoskeletal remodeling previously associated with chronic liver injury and tumorigenesis (van Hengel et al., 2008) and HCV-induced hepatic fibrosis (Smith et al., 2003). This network of molecules (Figure 2B) links activation of HSCs, inflammatory signaling, and immune cell infiltration. We also identified several secreted factors that connect activated macrophages and Kupffer cells to HSC activation, including platelet-derived growth factor receptor, alpha polypeptide (PDGFRA) and vascular endothelial growth factor (VEGF), and the chemokines CCL2, CXCL3, CCL13, and CCL5. Additionally, genes known to be expressed by activated HSCs, such as Bcl-2 associated X (BAX), which are also related to chemoattraction and apoptotic induction were upregulated. This contrasts with the downregulation of genes associated with inhibition of apoptosis (such as baculovirus IAP repeat-containing 5 [BIRC5]) and cell cycle progression (such as p21-activated kinase 7 [PAK7] and kinesin family member 11 [KIF11] and is consistent with a report that HSC apoptosis is anticorrelated with HCV-induced fibrogenesis (Gonzalez et al., 2009a). Taken together, these findings suggest that in HCV/HIV coinfected patients, increased migration of activated immune effector cells likely contributes to increased hepatic inflammation. This in turn may lead to increased HSC activation, hastening collagen deposition and fibrogenesis and possibly contributing to more rapid liver disease progression in HCV/HIV coinfected patients.
Identification of peripheral signatures of coinfection
In the PBMC compartment, functional analysis using IPA revealed that many of the upregulated genes were associated with immune induction and immunological disease. Specifically, 271 genes associated with inflammatory responses, antigen presentation, and humoral immune responses, as well as hematological, immunological, and inflammatory disease were significantly upregulated (Table S3). Genes within this group include chemokine receptors and other molecules associated with chemotaxis and lymphocyte homing including the HIV receptor CCR5, complement components, and proinflammatory cytokines associated with T cell activation such as IL-11, IL-17RA, and IL-15. Additionally, we observed numerous gene expression changes associated with antigen presentation and CD4 T cell activation, including upregulation of the genes encoding costimulatory molecules CD86 and numerous HLA alleles. The molecular signature also featured concomitant downregulation of genes known to suppress T cell activation, such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4). These are consistent with the chronic activation of HIV-infected T cells.
Interestingly, we also identified a subset of transcripts associated with fibrosis and extracellular matrix deposition upregulated in HCV/HIV coinfected PBMC, including several types of collagen (COL22A1, COL3A1, COL4A1) and galectins (LGALS1, LGALS3, LGALS8). Notably, both galectin-1 and galectin-3 have been implicated in HSC activation and hepatic fibrogenesis (Henderson et al., 2006; Jiang et al., 2011; Maeda et al., 2003), as have Rho GTPases and rearrangement of the actin cytoskeleton (Kato et al., 1999). Circulating PBMC in HCV/HIV coinfected patients thus display increased fibrogenic gene expression, and may subsequently enhance HSC activation and collagen deposition upon immune infiltration of the liver.
Overexpression of REG1A and REG3A secreted proteins
In addition to evaluating the global DEG signature associated with HCV/HIV coinfection, we sought to identify molecules highly expressed by the liver which may be candidates for evaluation as potential therapeutic or diagnostic targets. From DEG in the intrahepatic signature, we observed that regenerating islet- derived 1 alpha and 3 alpha (REG1A and REG3A) were among the most highly upregulated in a subset of 7 patients. Reg1α and Reg3α are both secreted growth factors associated with pancreatic islet cell regeneration, which were previously linked to a number of gastrointestinal cancers, including hepatocellular carcinoma (Cavard et al., 2006; Yuan et al., 2005). Reg1α and Reg3α accelerate liver regeneration (Lieu et al., 2005; Wang et al., 2009), and Reg3α has been implicated in the activation of pancreatic stellate cells, as well as deposition of collagen and associated extracellular matrix components (Li et al., 2009), suggesting that these molecules may be linked to liver disease progression. Therefore, we evaluated whether or not these molecules might have potential as prognostic indicators of HCV/HIV coinfection-associated liver disease.
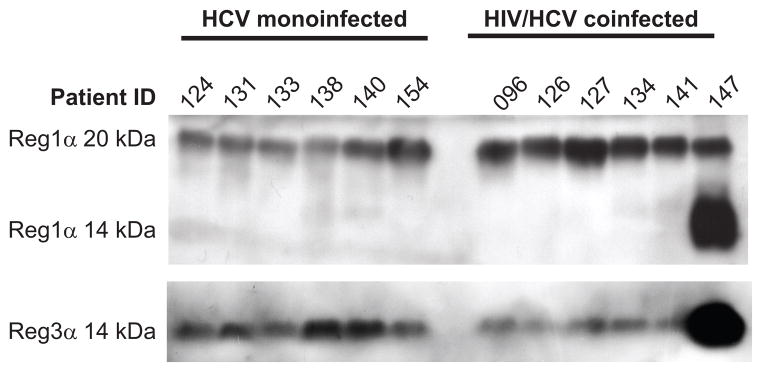
By assessing Reg1α and Reg3α levels in patient plasma specimens by immunoblot (Figure 3), we observed that HCV/HIV patients exhibited substantially higher levels of circulating Reg1α species compared to monoinfected individuals. In particular, a higher molecular weight form of Reg1α corresponding to either a precursor form or a heavily glycosylated protein, was consistently expressed at higher levels. Secreted Reg3α levels were not significantly different between the two patient conditions, although Reg3α was detectable in all tested samples. This suggests that the higher molecular weight form of circulating Reg1α may be a candidate for evaluating hepatic disease progression by a non-invasive, blood-based study, and warrants further evaluation.
Figure 3. Expression of Reg1α and Reg3α proteins.
Immunoblot analysis of Reg1α and Reg3α protein in patient plasma specimens. Molecular weight of the detected Reg1α and Reg3α proteins are indicated on the left side of the figure. Reg1α is detected in the 14–22 kDa range, consistent with differential proteolytic processing and/or glycosylation heterogeneity observed as a result of proteolytic processing and differential glycosylation than monoinfected patients. Additionally, a higher molecular weight band at approximately 20 kDa represents either the precursor form of Reg1α or one of the other glycosylated isoforms of this protein reported within the 17–22 kDa size range (De Reggi et al., 1995) present in all samples.
Expanding co-infected liver and PBMC signatures by a network approach
The cross-sectional design of this study restricted detection of liver and PBMC signature DEG to one time point. To identify additional genes which may contribute to liver disease progression in the coinfected tissues, we adopted a Bayesian network modeling approach to predict interactions between significant DEG in our study and those in networks built from previously published large data sets. Bayesian networks represent statistical dependencies between genes (“nodes”) as “edges,” which are calculated by multiple expression measurements. Leveraging networks generated from much larger datasets can increase the robustness of biological findings from a smaller dataset by further exploring the interrelated connectivity of DEG found in lower abundance.
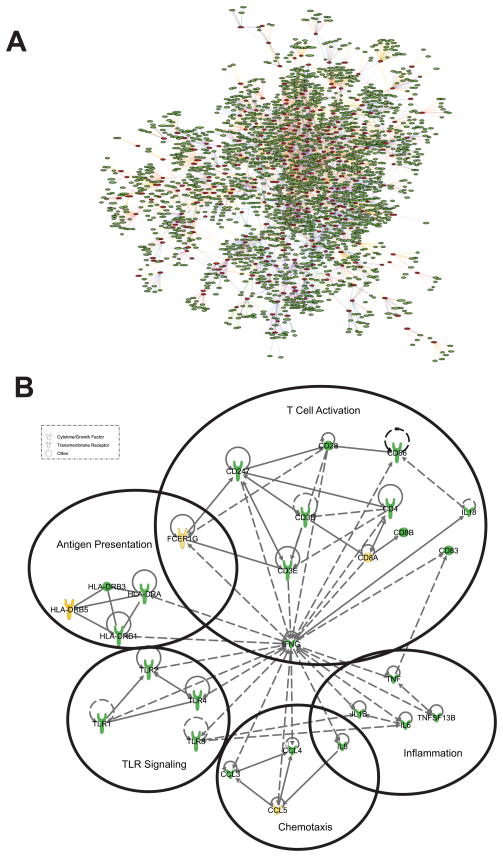
Combined replicates of liver signature sequences were used as the input to Bayesian networks built from three human liver cohorts including 400 normal samples (Schadt et al., 2008), 272 samples each of hepatocellular carcinoma (HCC) and adjacent normal livers (Lamb et al., 2011), and 950 livers from obese subjects (Zhong et al., 2010). Out of the 2,442 liver signature sequences, 924 genes were present in the three liver networks but only those subnetworks with at least 3 nodes (290 genes) were maintained. In subnetworks meeting these criteria, we expanded along one edge to identify additional connected nodes and obtain the network shown in Figure 4A with 2,544 genes.
Figure 4. Construction of HCV/HIV co-infected liver Bayesian network.
(A) Liver network was constructed by overlaying 2,442 liver signature sequences onto the network formed from 3 existing liver cohorts as described in the Materials and Methods section. Only those sub-networks with at least 3 connected nodes (290 genes) were retained to grow one edge out to obtain the network shown with 2,544 genes. (B) Liver subnetwork of genes annotated as those involved in communication between innate and adaptive immune cells. Solid lines show directly interacting molecules, dashed lines show indirect interactions. Upregulated genes identified in the common signature in this study are colored yellow; genes identified in the network model are colored green.
We performed functional analysis of this liver network by using the Gene Annotator Module of the Merck Target Gene Information (TGI) Network Analysis and Visualization (NAV) tool, which finds related gene sets based on functional annotation, and ranks significance by calculating an expectation (E) value. Our analysis identified highly enriched pathways involved in chemokine signaling, dendritic cell maturation, interaction between innate and adaptive immunity, T cell receptor signaling, pattern recognition receptor-related pathway, NK cell signaling, and hepatic cholestasis/fibrosis (Table S4). This suggests that the genes added by the network analysis are related to cross-talk between the innate and adaptive immune system, and the interface between these different immune pathways may play an important role in HCV/HIV coinfection.
We identified similar functional categories and pathways using IPA. Some of these overlap with those already identified in Figure 1B whereas others are novel pathways found to be associated with this liver network. Using IPA, we generated a subnetwork using the genes annotated as involved in communication between innate and adaptive immune cells (Figure 4B). This subnetwork includes additional genes related to induction of innate antiviral responses (multiple Toll-like receptors; TLR) and inflammation (tumor necrosis factor alpha; TNFα), as well as induction of adaptive immunity via antigen presentation (HLA alleles) and T cell activation (CD3, CD8).
We built a PBMC network model using the similar approach of inputting 2,479 combined replicate signature sequences to the deCODE blood Bayesian network built from about 1,000 samples (Emilsson et al., 2008). We applied similar criteria as the liver network, maintaining only 131 genes (out of 561 present in the deCODE network) in sub-networks with at least 3 nodes and grew them one edge out to obtain a network with 1,117 genes. Annotation of genes in this PBMC network indicates an association with T and B cell signaling, interaction between dendritic and natural killer cells, and communication between innate and adaptive immune cells (Table S4). These results are consistent with findings shown in Figure 1B, and suggest that PBMC from coinfected patients exhibit the differential regulation of genes bridging innate and adaptive immunity.
Discussion
Understanding disease pathogenesis in HCV/HIV coinfected patients presents numerous challenges, one of which is discerning how distinct tissue compartments infected with two unique pathogens contribute separately and in concert to liver disease progression. Our data suggest that the chronic immune activation characteristic of HIV infection and progression to AIDS (Deeks et al., 2004; Hazenberg et al., 2003) is a distinguishing feature of the host response to HCV/HIV coinfection. We observed substantial overlap (104 DEG, 24 of which are associated with immune activation and differentiation) between both the individual liver and PBMC compartments and the signature common to both tissues. This further supports the notion that increased immune and inflammatory activity are present in both the liver and the periphery of coinfected patients and are likely important drivers of disease pathogenesis. The enhanced expression of inflammatory genes in both the common and individual compartment signatures suggests that HIV infection causes both circulating leukocytes, and those infiltrating the liver, to be more activated than those from HCV monoinfected patients. Consequently, the pro-inflammatory functions of these cells may enhance pathogenic processes and progression of fibrosis and HCV-induced liver injury via immune effector functions, cytokine and chemokine secretion, increased migration, and activation of HSC and other cellular mediators of fibrogenesis.
Another effect of HIV-infected infiltrating immune cells on HCV-infected hepatic cells may be to stimulate expression of genes with as yet undescribed roles in liver disease progression. The growth factors Reg1α and Reg3α have not been associated with HCV or HIV infection or pathogenesis. However, their apparent role in HCC tumorigenesis and metastasis (Cavard et al., 2006; Yuan et al., 2005), as well as our data indicating that these proteins are secreted, merits further evaluation of these proteins in a larger patient cohort as serum biomarkers with prognostic or diagnostic utility.
Recent systems biology approaches incorporating high-throughput data types, such as genome-wide DNA genotyping and RNA expression profiling, has allowed the construction of various types of genetic networks. Characterizing networks that underlie complex phenotypes such as clinical disease can provide more comprehensive understanding of the disease pathophysiology and in turn identify or validate disease susceptibility genes (Chen et al., 2008; Derry et al., 2010; Emilsson et al., 2008; Schadt et al., 2008; Yang et al., 2009). In the current study, the gene signatures we obtained were from a single time point (at which disease progression could be heterogeneous) and from a relatively small number of patients. In an effort to address these constraints and expand potential biological coverage, we used coinfected liver and PBMC gene signatures to build Bayesian statistical causal networks based on published large liver and blood data sets (Emilsson et al., 2008; Lamb et al., 2011; Schadt et al., 2008; Zhong et al., 2010). From these networks, we obtained greater enrichment based on functional annotation for the resulting liver network (Figure 4A and Table 2) and the PBMC network (Figure 4B and Table 2) compared to our functional analysis using the significant DEG identified in this study alone. These expanded networks solidified the pathway connections between genes in both the HCV/HIV coinfected liver and PBMC molecular signatures, and provided greater insight into functionally related genes that might also be significant for liver disease pathogenesis in a larger population of HCV/HIV coinfected patients.
NK cells and DCs mediate both effector and regulatory functions in innate immunity and the induction of adaptive responses (Altfeld et al., 2011; Gonzalez, Landay, and Sandberg, 2010; Kim and Chung, 2009); however, the HCV/HIV coinfected liver signature did not capture these functions as significantly associated with coinfection (Figure 1B). The expanded liver network, however, did identify significant associations with NK and DC functions (Table 2). Innate immunity-associated genes present in the network, but not in the co-infected liver signature, include TLR1, TLR2, TLR4, and TLR8. TLR signaling provides a critical link between innate and adaptive immunity in numerous viral infections. In chronic HCV infection, reduced TLR responsiveness is associated with liver dysfunction (Chung et al., 2011) and chronic inflammation consequent to reduced induction of adaptive immune responses (Dental et al., 2012; Yonkers et al., 2011b). Furthermore, aberrant TLR signaling has been implicated in T cell activation defects in HIV infection (Yonkers et al., 2011a), indicating that these molecules may be important mediators of chronic immune activation and inflammation in the HCV/HIV coinfection setting. In addition, the liver network (Table 2) includes some key fibrogenic genes such as transforming growth factor-β1,2, and 3 (TGFB1, TGFB2 and TGFB3), which have been implicated both in Rho GTPase-mediated HSC activation (Shimada, Staten, and Rajagopalan, 2011) and enhancement of HCV replication and associated liver injury (Lin et al., 2008). This network allowed us to bridge gaps present in the molecular signatures from our dataset, and establish a more comprehensive expression profile characteristic of liver disease pathogenesis in HCV/HIV coinfection.
The observed host transcriptional signatures and networks characteristic of liver and/or PBMC in HCV/HIV coinfected patients indicate that these compartments are distinguished by increased immune activation consistent with HIV infection. This environment may in turn contribute to increased hepatic upregulation of pro-fibrogenic genes, and reveals a possible mechanism for the accelerated liver disease progression observed in coinfected patients. Future studies employing longitudinal samples from a larger patient cohort should be directed at understanding how these genes are temporally regulated throughout the course of liver disease progression. Additionally, follow up studies should determine the clinical utility of potential biomarkers such as Reg1α to identify patients at risk of accelerated severe liver injury.
Materials and methods
Study subjects and specimen collection
Approval for this study was obtained from the University of Washington Human Subjects Division, and experiments were performed in compliance with legal, institutional, and ethical standards for human subjects research. All samples were obtained under informed consent.
Ten subjects infected with HCV and 13 subjects coinfected with HCV and HIV were included in this study. Exclusion criteria included chronic hepatitis B infection, alcohol use >1 drink per day in women and >2 drinks per day in men, and acute infection of any type. An experienced hepatopathologist (L.V.T.) blinded to all clinical data including HIV status reviewed all liver biopsies. Liver fibrosis stage was assessed using the Batts-Ludwig system: F0, no fibrosis; F1, portal fibrosis; F2, periportal fibrosis with rare bridges; F3, bridging/septal fibrosis; or F4, cirrhosis (Batts and Ludwig, 1995). All samples included were deemed adequate for staging. At the time of liver biopsy, 2–3 mm of liver tissue was stored in RNALater (Life Technologies, Mountain View, CA) at 4°C overnight, and then stored at −80°C until RNA processing. Whole blood was obtained by venipuncture within 30 days of the liver biopsy (typically within one week of the biopsy). Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density centrifugation (Ficoll-Paque Plus; GE Healthcare Biosciences, Piscataway, NJ) within 24 hours of blood draw, and 2 × 106 freshly isolated PBMC were pelleted and resuspended in Trizol (Life Technologies, Carlsbad, CA) prior to flash freezing in liquid nitrogen. Samples were stored at −80°C until RNA was prepared according to the manufacturer’s instructions. Samples were extracted according to manufacturer’s protocol and resuspended in RNAse-free water prior to use in microarray experiments.
Microarray platform
Samples were amplified and labeled using a custom automated version of the RT/IVT protocol and reagents provided by Affymetrix. Hybridization, labeling and scanning were completed following the manufacturer’s recommendations using Merck Affymetrix Human Custom Arrays 1.0 containing 43,737 unique probe sequences (Affymetrix, Santa Clara, CA). Sample amplification, labeling, and microarray processing were performed by the Rosetta Inpharmatics Gene Expression Laboratory in Seattle, WA (acquired by Merck and Co., Whitehouse Station, NJ). Expression data were loaded into the Resolver (Rosetta proprietary software database) for transformation, normalization, and error modeling.
Microarray data analysis
Microarray data from each individual liver or PBMC sample was compared back to a reference pool composed of all corresponding samples of liver or PBMC, respectively, from HCV monoinfected subjects. HCV/HIV-coinfected gene sets were established based on their statistically significant up- or down-regulation (absolute fold change ≥ 1.15 and ANOVA or combined p-value ≤ 0.01 in at least 7 samples) when compared to the reference pool. Signature genes from each tissue were identified and compared. Unsupervised clustering methods, including hierarchical and k-means clustering, were performed to identify genes with similar expression patterns. Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) software was used for functional analysis of significant genes identified by ANOVA.
Immunoblotting
Patient plasma samples were diluted in M-PER (Thermo Scientific, Rockford, IL) and protein concentration was quantified by BCA assay (Thermo Scientific) per the manufacturer’s instructions. A Criterion TGX Any kD Tris-glycine polyacrylamide gel (Bio-Rad, Hercules, CA) was loaded with 20 μg protein per sample, run using standard SDS-PAGE methods, and transferred to a PVDF membrane. Membranes were probed with anti-Reg1α (Abcam, Cambridge, MA) and anti-Reg3α (Abcam) primary antibodies, followed by peroxidase-conjugated donkey anti-rabbit secondary (Jackson ImmunoResearch Labs, West Grove, PA), and detected using ECL Plus reagent (GE Healthcare Biosciences). There is no protein reported to be constitutively secreted at similar levels in plasma or serum samples which could act as a loading control, therefore protein loading per well was normalized by loading equal quantities of protein per lane.
Liver and PBMC network construction
Networks were constructed as described previously (Zhu et al., 2004; Zhu, Zhang, and Schadt, 2008) and were visualized using the Target Gene Information (TGI) Network Analysis and Visualization (NAV) desktop application developed at Rosetta Inpharmatics. We utilized previously published data sets for liver (Lamb et al., 2011; Schadt et al., 2008; Zhong et al., 2010) and PBMC (Emilsson et al., 2008) network constructions. To construct liver and PBMC networks based on HCV/HIV coinfected gene signatures, the two gene signatures were loaded into the database as described in the results section and the TGI NAV tool was used to extract all edges from this network involving the included signature genes of interest.
The TGI NAV tool allows rapid, real-time, graphical analysis of pathway network models built from a comprehensive and fully integrated set of public and proprietary interaction databases available through a back-end central database. In addition, it supports experimentally generated systems biology data such as the statistical associations and causal relationships described in the manuscripts cited above and applied in the current study.
Supplementary Material
Using the same approach as that used for the liver network, the PBMC network containing 1,117 genes was derived from inputting 2,479 signature sequences to deCODE blood Bayesian network built from about 1,000 samples, and retaining 131 genes with at least 3 connected nodes to grow out.
Acknowledgments
The authors gratefully acknowledge David Gretch and Minjun Chung Apodaca for accessing patient plasma samples for immunoblot analysis. This work was funded in part by National Institute on Drug Abuse grant 1P30DA01562501.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison RD, Katsounas A, Koziol DE, Kleiner DE, Alter HJ, Lempicki RA, Wood B, Yang J, Fullmer B, Cortez KJ, Polis MA, Kottilil S. Association of interleukin-15-induced peripheral immune activation with hepatic stellate cell activation in persons coinfected with hepatitis C virus and HIV. J Infect Dis. 2009;200(4):619–23. doi: 10.1086/600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Fadda L, Frleta D, Bhardwaj N. DCs and NK cells: critical effectors in the immune response to HIV-1. Nat Rev Immunol. 2011;11(3):176–86. doi: 10.1038/nri2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts KP, Ludwig J. Chronic hepatitis. An update on terminology and reporting. Am J Surg Pathol. 1995;19(12):1409–17. doi: 10.1097/00000478-199512000-00007. [DOI] [PubMed] [Google Scholar]
- Baum MK, Sales S, Jayaweera DT, Lai S, Bradwin G, Rafie C, Page JB, Campa A. Coinfection with hepatitis C virus, oxidative stress and antioxidant status in HIV-positive drug users in Miami. HIV Med. 2011;12(2):78–86. doi: 10.1111/j.1468-1293.2010.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavard C, Terris B, Grimber G, Christa L, Audard V, Radenen-Bussiere B, Simon MT, Renard CA, Buendia MA, Perret C. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with beta-catenin mutations. Oncogene. 2006;25(4):599–608. doi: 10.1038/sj.onc.1208860. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Watanabe T, Kudo M, Chiba T. Correlation between hyporesponsiveness to Toll-like receptor ligands and liver dysfunction in patients with chronic hepatitis C virus infection. J Viral Hepat. 2011;18(10):e561–7. doi: 10.1111/j.1365-2893.2011.01478.x. [DOI] [PubMed] [Google Scholar]
- De Reggi M, Capon C, Gharib B, Wieruszeski JM, Michel R, Fournet B. The glycan moiety of human pancreatic lithostathine. Structure characterization and possible pathophysiological implications. Eur J Biochem. 1995;230(2):503–10. doi: 10.1111/j.1432-1033.1995.tb20589.x. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narvaez AB, Hunt P, Martin JN, Kahn JO, Levy J, McGrath MS, Hecht FM. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104(4):942–7. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- Dental C, Florentin J, Aouar B, Gondois-Rey F, Durantel D, Baumert TF, Nunes JA, Olive D, Hirsch I, Stranska R. Hepatitis C virus fails to activate NF-kappaB signaling in plasmacytoid dendritic cells. J Virol. 2012;86(2):1090–6. doi: 10.1128/JVI.05444-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry JM, Zhong H, Molony C, MacNeil D, Guhathakurta D, Zhang B, Mudgett J, Small K, El Fertak L, Guimond A, Selloum M, Zhao W, Champy MF, Monassier L, Vogt T, Cully D, Kasarskis A, Schadt EE. Identification of genes and networks driving cardiovascular and metabolic phenotypes in a mouse F2 intercross. PLoS One. 2010;5(12):e14319. doi: 10.1371/journal.pone.0014319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino V, Rufat P, Boyer N, Renard P, Degos F, Martinot-Peignoux M, Matheron S, Le Moing V, Vachon F, Degott C, Valla D, Marcellin P. The influence of human immunodeficiency virus coinfection on chronic hepatitis C in injection drug users: a long-term retrospective cohort study. Hepatology. 2001;34(6):1193–9. doi: 10.1053/jhep.2001.29201. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Gonzalez SA, Fiel MI, Sauk J, Canchis PW, Liu RC, Chiriboga L, Yee HT, Jacobson IM, Talal AH. Inverse association between hepatic stellate cell apoptosis and fibrosis in chronic hepatitis C virus infection. J Viral Hepat. 2009a;16(2):141–8. doi: 10.1111/j.1365-2893.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez VD, Falconer K, Blom KG, Reichard O, Morn B, Laursen AL, Weis N, Alaeus A, Sandberg JK. High levels of chronic immune activation in the T-cell compartments of patients coinfected with hepatitis C virus and human immunodeficiency virus type 1 and on highly active antiretroviral therapy are reverted by alpha interferon and ribavirin treatment. J Virol. 2009b;83(21):11407–11. doi: 10.1128/JVI.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez VD, Landay AL, Sandberg JK. Innate immunity and chronic immune activation in HCV/HIV-1 co-infection. Clin Immunol. 2010;135(1):12–25. doi: 10.1016/j.clim.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Graham CS, Baden LR, Yu E, Mrus JM, Carnie J, Heeren T, Koziel MJ. Influence of human immunodeficiency virus infection on the course of hepatitis C virus infection: a meta-analysis. Clin Infect Dis. 2001;33(4):562–9. doi: 10.1086/321909. [DOI] [PubMed] [Google Scholar]
- Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, Hamann D, Prins M, Miedema F. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17(13):1881–8. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- Henderson NC, Mackinnon AC, Farnworth SL, Poirier F, Russo FP, Iredale JP, Haslett C, Simpson KJ, Sethi T. Galectin-3 regulates myofibroblast activation and hepatic fibrosis. Proc Natl Acad Sci U S A. 2006;103(13):5060–5. doi: 10.1073/pnas.0511167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Shao RX, Lin W, Weinberg E, Chung WJ, Tsai WL, Zhao H, Goto K, Zhang L, Mendez-Navarro J, Jilg N, Peng LF, Brockman MA, Chung RT. HIV infection increases HCV-induced hepatocyte apoptosis. J Hepatol. 2011;54(4):612–20. doi: 10.1016/j.jhep.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JX, Chen X, Hsu DK, Baghy K, Serizawa N, Scott F, Takada Y, Fukada H, Chen J, Devaraj S, Adamson R, Liu FT, Torok NJ. Galectin-3 Modulates Phagocytosis-induced Stellate Cell Activation and Liver Fibrosis in vivo. Am J Physiol Gastrointest Liver Physiol. 2011 doi: 10.1152/ajpgi.00257.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Iwamoto H, Higashi N, Sugimoto R, Uchimura K, Tada S, Sakai H, Nakamuta M, Nawata H. Role of Rho small GTP binding protein in the regulation of actin cytoskeleton in hepatic stellate cells. J Hepatol. 1999;31(1):91–9. doi: 10.1016/s0168-8278(99)80168-8. [DOI] [PubMed] [Google Scholar]
- Kim AY, Chung RT. Coinfection with HIV-1 and HCV--a one-two punch. Gastroenterology. 2009;137(3):795–814. doi: 10.1053/j.gastro.2009.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottilil S, Yan MY, Reitano KN, Zhang X, Lempicki R, Roby G, Daucher M, Yang J, Cortez KJ, Ghany M, Polis MA, Fauci AS. Human immunodeficiency virus and hepatitis C infections induce distinct immunologic imprints in peripheral mononuclear cells. Hepatology. 2009;50(1):34–45. doi: 10.1002/hep.23055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Zhang C, Xie T, Wang K, Zhang B, Hao K, Chudin E, Fraser HB, Millstein J, Ferguson M, Suver C, Ivanovska I, Scott M, Philippar U, Bansal D, Zhang Z, Burchard J, Smith R, Greenawalt D, Cleary M, Derry J, Loboda A, Watters J, Poon RT, Fan ST, Yeung C, Lee NP, Guinney J, Molony C, Emilsson V, Buser-Doepner C, Zhu J, Friend S, Mao M, Shaw PM, Dai H, Luk JM, Schadt EE. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PLoS One. 2011;6(7):e20090. doi: 10.1371/journal.pone.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Bachem MG, Zhou S, Sun Z, Chen J, Siech M, Bimmler D, Graf R. Pancreatitis-associated protein inhibits human pancreatic stellate cell MMP-1 and -2, TIMP-1 and -2 secretion and RECK expression. Pancreatology. 2009;9(1–2):99–110. doi: 10.1159/000178880. [DOI] [PubMed] [Google Scholar]
- Lieu HT, Batteux F, Simon MT, Cortes A, Nicco C, Zavala F, Pauloin A, Tralhao JG, Soubrane O, Weill B, Brechot C, Christa L. HIP/PAP accelerates liver regeneration and protects against acetaminophen injury in mice. Hepatology. 2005;42(3):618–26. doi: 10.1002/hep.20845. [DOI] [PubMed] [Google Scholar]
- Lin W, Weinberg EM, Tai AW, Peng LF, Brockman MA, Kim KA, Kim SS, Borges CB, Shao RX, Chung RT. HIV increases HCV replication in a TGF-beta1-dependent manner. Gastroenterology. 2008;134(3):803–11. doi: 10.1053/j.gastro.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Lin W, Wu G, Li S, Weinberg EM, Kumthip K, Peng LF, Mendez-Navarro J, Chen WC, Jilg N, Zhao H, Goto K, Zhang L, Brockman MA, Schuppan D, Chung RT. HIV and HCV cooperatively promote hepatic fibrogenesis via induction of reactive oxygen species and NFkappaB. J Biol Chem. 2011;286(4):2665–74. doi: 10.1074/jbc.M110.168286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kawada N, Seki S, Arakawa T, Ikeda K, Iwao H, Okuyama H, Hirabayashi J, Kasai K, Yoshizato K. Stimulation of proliferation of rat hepatic stellate cells by galectin-1 and galectin-3 through different intracellular signaling pathways. J Biol Chem. 2003;278(21):18938–44. doi: 10.1074/jbc.M209673200. [DOI] [PubMed] [Google Scholar]
- Martinez-Sierra C, Arizcorreta A, Diaz F, Roldan R, Martin-Herrera L, Perez-Guzman E, Giron-Gonzalez JA. Progression of chronic hepatitis C to liver fibrosis and cirrhosis in patients coinfected with hepatitis C virus and human immunodeficiency virus. Clin Infect Dis. 2003;36(4):491–8. doi: 10.1086/367643. [DOI] [PubMed] [Google Scholar]
- Pol S, Lamorthe B, Thi NT, Thiers V, Carnot F, Zylberberg H, Berthelot P, Brechot C, Nalpas B. Retrospective analysis of the impact of HIV infection and alcohol use on chronic hepatitis C in a large cohort of drug users. J Hepatol. 1998;28(6):945–50. doi: 10.1016/s0168-8278(98)80341-3. [DOI] [PubMed] [Google Scholar]
- Sandberg JK, Falconer K, Gonzalez VD. Chronic immune activation in the T cell compartment of HCV/HIV-1 co-infected patients. Virulence. 2010;1(3):177–9. doi: 10.4161/viru.1.3.11206. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Staten NR, Rajagopalan LE. TGF-beta1 mediated activation of Rho kinase induces TGF-beta2 and endothelin-1 expression in human hepatic stellate cells. J Hepatol. 2011;54(3):521–8. doi: 10.1016/j.jhep.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Smith MW, Yue ZN, Korth MJ, Do HA, Boix L, Fausto N, Bruix J, Carithers RL, Jr, Katze MG. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology. 2003;38(6):1458–67. doi: 10.1016/j.hep.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Soto B, Sanchez-Quijano A, Rodrigo L, del Olmo JA, Garcia-Bengoechea M, Hernandez-Quero J, Rey C, Abad MA, Rodriguez M, Sales Gilabert M, Gonzalez F, Miron P, Caruz A, Relimpio F, Torronteras R, Leal M, Lissen E. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26(1):1–5. doi: 10.1016/s0168-8278(97)80001-3. [DOI] [PubMed] [Google Scholar]
- Tuyama AC, Hong F, Saiman Y, Wang C, Ozkok D, Mosoian A, Chen P, Chen BK, Klotman ME, Bansal MB. Human immunodeficiency virus (HIV)-1 infects human hepatic stellate cells and promotes collagen I and monocyte chemoattractant protein-1 expression: implications for the pathogenesis of HIV/hepatitis C virus-induced liver fibrosis. Hepatology. 2010;52(2):612–22. doi: 10.1002/hep.23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel J, D’Hooge P, Hooghe B, Wu X, Libbrecht L, De Vos R, Quondamatteo F, Klempt M, Brakebusch C, van Roy F. Continuous cell injury promotes hepatic tumorigenesis in cdc42-deficient mouse liver. Gastroenterology. 2008;134(3):781–92. doi: 10.1053/j.gastro.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Walters KA, Smith MW, Pal S, Thompson JC, Thomas MJ, Yeh MM, Thomas DL, Fitzgibbon M, Proll S, Fausto N, Gretch DR, Carithers RL, Jr, Shuhart MC, Katze MG. Identification of a specific gene expression pattern associated with HCV-induced pathogenesis in HCV- and HCV/HIV-infected individuals. Virology. 2006;350(2):453–64. doi: 10.1016/j.virol.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Wanchu A, Rana SV, Pallikkuth S, Sachdeva RK. Short communication: oxidative stress in HIV-infected individuals: a cross-sectional study. AIDS Res Hum Retroviruses. 2009;25(12):1307–11. doi: 10.1089/aid.2009.0062. [DOI] [PubMed] [Google Scholar]
- Wang J, Koyota S, Zhou X, Ueno Y, Ma L, Kawagoe M, Koizumi Y, Okamoto H, Sugiyama T. Expression and localization of regenerating gene I in a rat liver regeneration model. Biochem Biophys Res Commun. 2009;380(3):472–7. doi: 10.1016/j.bbrc.2009.01.126. [DOI] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, Karlsson J, Castellani LW, Mumick S, Wang K, Xie T, Coon M, Zhang C, Estrada-Smith D, Farber CR, Wang SS, van Nas A, Ghazalpour A, Zhang B, Macneil DJ, Lamb JR, Dipple KM, Reitman ML, Mehrabian M, Lum PY, Schadt EE, Lusis AJ, Drake TA. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 2009;41(4):415–23. doi: 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonkers NL, Rodriguez B, Asaad R, Lederman MM, Anthony DD. Systemic immune activation in HIV infection is associated with decreased MDC responsiveness to TLR ligand and inability to activate naive CD4 T-cells. PLoS One. 2011a;6(9):e23884. doi: 10.1371/journal.pone.0023884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonkers NL, Sieg S, Rodriguez B, Anthony DD. Reduced naive CD4 T cell numbers and impaired induction of CD27 in response to T cell receptor stimulation reflect a state of immune activation in chronic hepatitis C virus infection. J Infect Dis. 2011b;203(5):635–45. doi: 10.1093/infdis/jiq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan RH, Jeng YM, Chen HL, Hsieh FJ, Yang CY, Lee PH, Hsu HC. Opposite roles of human pancreatitis-associated protein and REG1A expression in hepatocellular carcinoma: association of pancreatitis-associated protein expression with low-stage hepatocellular carcinoma, beta-catenin mutation, and favorable prognosis. Clin Cancer Res. 2005;11(7):2568–75. doi: 10.1158/1078-0432.CCR-04-2039. [DOI] [PubMed] [Google Scholar]
- Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, Hao K, Woo S, Fabre-Suver C, Qian S, Tota MR, Keller MP, Kendziorski CM, Yandell BS, Castro V, Attie AD, Kaplan LM, Schadt EE. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6(5):e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lum PY, Lamb J, GuhaThakurta D, Edwards SW, Thieringer R, Berger JP, Wu MS, Thompson J, Sachs AB, Schadt EE. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105(2–4):363–74. doi: 10.1159/000078209. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Schadt EE. A systems biology approach to drug discovery. Adv Genet. 2008;60:603–35. doi: 10.1016/S0065-2660(07)00421-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using the same approach as that used for the liver network, the PBMC network containing 1,117 genes was derived from inputting 2,479 signature sequences to deCODE blood Bayesian network built from about 1,000 samples, and retaining 131 genes with at least 3 connected nodes to grow out.