Abstract
Copy number variants (CNVs) are widely distributed throughout the human genome, where they contribute to genetic variation and phenotypic diversity. De novo CNVs are also a major cause of numerous genetic and developmental disorders. However, unlike many other types of mutations, little is known about the genetic and environmental risk factors for new and deleterious CNVs. DNA replication errors have been implicated in the generation of a major class of CNVs, the nonrecurrent CNVs. We have found that agents that perturb normal replication and create conditions of replication stress, including hydroxyurea and aphidicolin, are potent inducers of nonrecurrent CNVs in cultured human cells. These findings have broad implications for identifying CNV risk factors and for hydroxyurea-related therapies in humans.
Introduction
In recent years, copy number variants (CNVs), defined as deletions or duplications of 50 bp to over a megabase, have been found to be widely distributed throughout the human genome [1-7]. The discovery of CNVs is tied to the advent of new genomic technologies that have enabled high-resolution analysis, including oligonucleotide microarrays and next generation sequencing approaches. With over 25,000 polymorphic CNVs, including nearly 1000 large CNVs greater than 50 kb now described in normal individuals [8], it is clear that human genetic variation is profoundly influenced by large-scale structural changes. It is also clear that many CNVs have deleterious consequences. Spontaneous or de novo CNVs are an important and frequent cause of genetic and developmental disorders, including severe intellectual disability, autism, schizophrenia, heart defects and many others [9-13], and they arise frequently in cancer cells. The frequency at which they arise suggests a high de novo mutation rate.
Despite their importance, there is limited understanding of how many CNVs arise, and little knowledge of risk factors involved. Like all mutation classes, it is certain that risk for new and deleterious CNVs will be increased by exposures to precipitating environmental mutagens as well as by inherited genetic predisposition. A key to predicting and identifying these factors is a clear understanding of the underlying mechanisms by which CNVs are formed. At least two distinct pathways are involved in the formation of most disease-associated CNVs: unequal meiotic recombination and replication errors. We have found that agents that perturb replication induce a high frequency of CNVs in normal human cells that resemble non-recurrent CNVs in humans in all aspects [14-16]. These agents include the polymerase inhibitor aphidicolin and the ribonucleotide reductase inhibitor, hydroxyurea, which is commonly used in the treatment of sickle cell disease and other disorders. These data provide experimental support for replication error models for the origins of CNVs and further suggest that many agents or conditions that lead to replication stress have the potential to induce deleterious CNVs.
Classes of CNVs
As with all mutation types, the risk for new and deleterious CNVs will undoubtedly be increased by inherited genetic predisposition and by exposure to precipitating environmental mutagens. Understanding the mechanisms involved in their formation is key to defining genetic and environmental risk factors for new and deleterious CNV mutations. However, we know little about the molecular mechanisms involved in the formation of this important class of CNVs. Most human CNV research to date has focused on cataloguing their occurrence and association with various disease states [17-20], with few experimental studies aimed at defining molecular mechanisms of formation. Mechanisms giving rise to CNVs have therefore largely been inferred from the observed CNV breakpoint junction sequences of normal and disease-associated CNVs and from the genetic architecture in the vicinity of breakpoints. In addition to the large class of smaller CNVs created by retrotransposition events or VNTR rearrangements, this approach has revealed two major categories of both polymorphic and de novo, pathogenic CNVs with distinctly different structures and cellular origins, frequently termed “recurrent” and “non-recurrent” CNVs, respectively.
Recurrent CNVs
Approximately 20-40% of normal polymorphic CNVs and many de novo, disease-related CNVs show recurrent breakpoints in low-copy repeats or segmental duplications [3,5,6,8,17,19]. As one would expect, hotspots for these CNVs exist in regions containing large segmental duplications. These variants include a growing number of recurrent CNVs associated with distinct clinical phenotypes, such as those identified on chromosomes 16p11.2 and 17q11.2 in individuals with neurological disorders, including severe intellectual disability, autism and schizophrenia [21-24].
Non-recurrent CNVs
These CNVs have unique breakpoints that are not dependent on segmental duplications. In most cases, they are characterized by microhomologies with a smaller number having blunt ends or short insertions at the breakpoint junctions. The majority of normal CNVs and a large percentage of pathogenic CNVs fall into this class [19]. Many of these CNVs are unique, though overlapping CNVs with widely variable breakpoints can be clustered in regions. These CNV-prone regions are often ascertained by disease association, such as the MBD5 gene region, which harbors deletions in humans with autism and other neurological abnormalities [25]. Most non-recurrent CNVs are simple deletions or tandem duplications, but some are more complex and are interrupted by normal sequences or inversions and can contain both deleted and duplicated segments within the same interval. Some non-recurrent CNVs are highly complex, with dozens of events clustered in a single genomic region [26], similar to a phenomenon termed chromothripsis (for “chromosome shattering”), recently described in cancer cell genomes [27]. It is likely that the observed incidence of these complex events is currently underestimated because of the difficulty in obtaining accurate sequence data at the breakpoints of such events.
Mechanisms of CNV formation
Recurrent CNVs with common breakpoints are deletions and reciprocal duplications that are thought to arise by meiotic unequal or non-allelic homologous recombination (NAHR), typically mediated by misalignment of large flanking segmental duplications or repeated sequences. These CNVs therefore arise in the same manner as was first elegantly described two decades ago for human microdeletion/micorduplication syndromes such as Charcot-Marie Tooth and Prader-Willi syndromes [28], and recurrent CNV disorders can therefore be considered an extension of this class of syndromes.
Less is known about the origins and molecular mechanisms leading to non-recurrent CNVs. Interestingly, data from breakpoint sequences and experimental systems all point to a mitotic, rather than meiotic, cell origin. Errors in DNA replication, rather than meiotic homologous recombination are predicted to give rise to the observed breakpoint junctions (Figure 1). In addition, inhibitors of replication that create conditions of “replication stress” can induce similar CNVs experimentally [14-16], as discussed in detail below. The simplest model that is consistent with both breakpoint sequence data and induction by replication stress is one whereby there is aberrant restoration of stalled replication forks. The molecular mechanisms involved thus undoubtedly include pathways such as cell cycle checkpoints to restore normal replication and prevent CNV formation, as well as DNA replication and repair factors that create the CNV lesion. A number of pathways have been suggested for the latter. These possible mechanisms include simple rejoining of two or more double strand breaks by nonhomologous end-joining (NHEJ) or the related microhomology-mediated end-joining (MMEJ) pathway, and mechanisms involving template switching events [14,15,18,29-31]. Most notable of the latter are the models of “Fork Stalling and Template Switching” (FoSTeS) of Lee et al. [30] and a modification termed microhomology-mediated break-induced replication (MMBIR) proposed by Hastings et al. [29]. These models are based on template switching mechanisms proposed for stress-induced amplifications in E. coli [32] and at sites of stalled replication in yeast [33-35] and BIR events principally described in yeast [36]. In the FoSTeS model, replication forks encountering low-copy repeats or areas that are difficult to replicate are prone to stalling, leading to a switch to another active fork to bypass the DNA lesion or to resume replication. The MMBIR model invokes template switching repair of single-sided DSBs formed at collapsed replication forks into regions of microhomology rather than the longer homology typically observed at BIR-mediated events [36]. These models provide an appealing explanation of the molecular mechanisms involved in non-recurrent CNV formation. However, most of the data to support these models come from observations of breakpoint junction sequences of CNVs found in normal genomes and arising in patients. There is a clear need for rigorous experimental testing of the possible models explaining CNV formation.
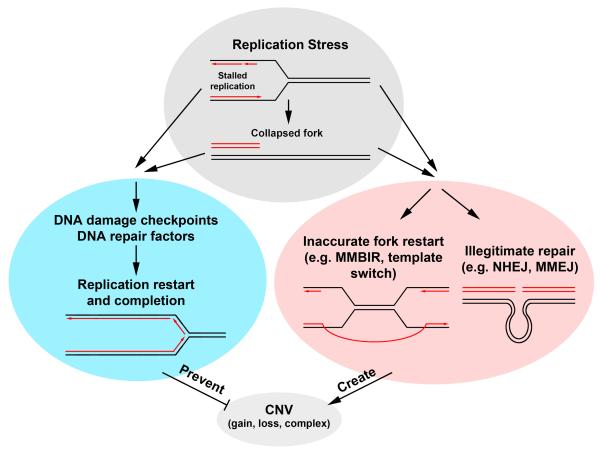
Figure 1.
Possible mechanisms involved in the prevention and formation of nonrecurrent CNVs. Exposure of cells to conditions that cause replication stress will result in stalled replication, which in turn might lead to fork collapse and result in a single-ended double strand break (Top). Either of these structures will activate a number of DNA damage checkpoint and repair pathways, which should faithfully restore replication at the site of the stalled or collapsed fork (Left). These checkpoint and repair pathways serve to protect the integrity of the genome, preventing the formation of CNVs and other structural variants. However, stalled replication or collapsed replication forks could also lead to restart or repair via alternate pathways. A stalled fork may be inaccurately restarted at a distant site, using a template switching or MMBIR pathway, giving rise to a CNV. Long range end-joining of two distant DNA breaks could also lead to deletions of large amounts of intervening sequence, resulting in a CNV (Right). It is expected that mutations that inhibit a cell’s ability to properly respond to a stalled or collapsed fork will result in an increased CNV frequency
Replication stress induces CNVs
Our laboratory has published direct experimental evidence that aberrant replication can induce a high frequency of CNVs in cultured mammalian somatic cells. We found that inhibiting replication with the DNA polymerase inhibitor aphidicolin (APH) induces a high frequency of de novo CNVs that mimic non-recurrent human CNVs in size, distribution and breakpoint structures [14,16]. This finding arose from studies of aberrations induced by replication stress at chromosome fragile sites. Treatment of cells with low doses of APH is highly effective in inducing expression of common fragile sites. We found that treatment of a human chromosome 3 somatic cell hybrid cell line with doses of APH used to induce fragile sites gave rise to a high frequency of CNV-like deletions of tens to hundreds of kb in the FRA3B fragile site that mimic those frequently found in tumor cells [16]. The breakpoints of these APH-induced CNVs all showed microhomologies, blunt ends or short insertions, which were concurrently being found in human non-recurrent CNVs. When this approach was applied to normal human fibroblasts, we found that APH induced CNVs across the human genome that resembled human non-recurrent CNVs in size, structure and breakpoint sequences. These CNVs were distributed throughout the genome with most (81%) found in regions containing genes [14].
These results strongly suggested that replication fork stalling is mechanistically responsible for these CNVs and, furthermore, that any agent that leads to replication stress could be a risk factor for their induction. To begin to test this, we performed a series of experiments using the mechanistically-distinct and clinically-relevant replication inhibitor, hydroxyurea (HU). HU leads to replication stress via inhibition of ribonucleotide reductase and perturbation of nucleotide pools [37], resulting in stalled replication and DNA double strand breaks. In addition to its well-studied properties as a replication inhibitor, HU is an important drug, especially for treatment of sickle-cell disease. Chronic HU treatment leads to increased expression of fetal hemoglobin, possibly through direct stimulation of cellular nitric oxide and cGMP signaling in erythroid progenitors [37-41], which results in reduced erythrocyte sickling and amelioration of disease severity [42]. As a result, HU-treated patients have fewer vaso-occlusive events and require fewer transfusions and hospitalizations [42]. HU treatment is therefore an effective drug for long-term management of sickle cell disease and it is effective for a number of other disorders, including certain cancers, myeloproliferative disorders, thalassemias and HIV infection. Thus, in addition to allowing a test of our replication stress hypothesis, HU is an important drug for many thousands of individuals.
These experiments showed that HU, at doses equivalent to the peak serum levels achieved in sickle cell patients, is also a potent inducer of CNVs in cultured normal human fibroblasts [15]. The sizes, structures and breakpoint junction sequences of HU-induced CNVs were consistent with APH-induced CNVs [14,16,43] and the non-recurrent class of normal and pathogenic CNVs [5,8,17-19,30,44-46]. It is notable that the sizes of CNVs induced by APH and HU are the same as those that arise spontaneously during culture, indicating that exogenous replication stress is not inducing a new type of event, but rather increasing the incidence of events that occur at a measurable frequency during normal cellular growth. While replication stress induced CNVs throughout the genome in these experiments, there were also hotspots where distinct overlapping CNVs were found. These include hotspots at 3q13.31 near the LSAMP gene, a deletion hotspot in cancers and cancer cell lines [47-49], at 16q23.3 in the WWOX locus, and at 7q11.2 spanning AUTS2, a gene deleted in some cases of autism and other neurological disorders [50-52]. These hotspots coincide with the location of chromosomal fragile sites [15,53,54], strongly suggesting a mechanistic link between the events leading to fragile site expression and these CNVs. Notably, the most significant CNV hotspot we observed in human fibroblasts, at 3q13.31, corresponds to a fragile site that was recently shown by Le Tallec et al. [55] to be highly expressed in fibroblasts, but not lymphoblasts, in a manner that correlated with reduced levels of replication origin firing in this region and cell type.
HU is now the second agent experimentally shown to induce CNVs. Although HU and APH impair DNA replication via different mechanisms, both agents induce CNVs with similar frequencies and size distributions that are identical to many normal and pathogenic CNVs. These results strongly support a common mechanism mediated by replication stress for the formation of the non-recurrent class of CNVs found in vivo and those induced experimentally. They also suggest that any agent or condition that leads to replication stress has the potential to induce deleterious, de novo CNVs. They also have direct implications for HU therapy. HU is FDA-approved for the treatment of sickle cell disease and has clear benefits for the treatment of sickle cell patients, as well as those with some types of cancer, myeloproliferative disorders, thalassemias and HIV infection [56]. While HU is well-tolerated and has low toxicity in patients, reproductive studies are limited and the long-term effects of HU on the genomes of subsequent generations have not been evaluated. The observation that HU induces CNVs in cultured cells, at concentrations equivalent to the peak serum levels achieved in sickle cell patients [57,58] and does so in one or two cell divisions, strongly suggests that further studies are necessary. In particular, the intergenerational, germline effects of HU and other replication inhibitors should be determined to directly test the replication stress hypothesis for CNV formation in vivo and to further assess the potential risk for submicroscopic genomic structural changes in the genomes of HU-treated patients and their future generations.
Risk Factors for CNV formation
Unlike other types of mutations studied for decades, our current knowledge of the mechanisms involved in CNV formation only allow us to begin to identify potential genetic and environmental risk factors (Table 1). For recurrent CNVs formed by NAHR during meiosis I, the greatest risk factor thus far identified is variation in genomic architecture, including the orientation and size of segmental duplications. Such structural polymorphisms impact the likelihood that NAHR will create a de novo CNV in these regions [59,60]. There does not appear to be a parental origin bias for recurrent CNVs [61] and the importance of variation in genes involved in meiotic recombination is unknown. In addition, nothing is known about environmental factors that could influence meiotic NAHR that could lead to CNVs.
For non-recurrent CNVs, the mitotic cell origin hypothesis has important implications for the genetic and environmental factors involved in their formation. For example, males complete ongoing mitotic divisions leading to mature germ cells throughout adulthood while females do so during fetal development. We thus predicted a male sex bias in risk for de novo, non-recurrent CNVs, coupled with a possible age effect [14,15]. The recent studies of Hehir-Kwa et al. [62] and Sibbons et al. [61] support this prediction. These two groups determined the parent of origin of rare CNVs associated with intellectual disability and found that the majority of non-recurrent CNVs, or CNVs not mediated by segmental duplications, originated on the paternal allele. In addition, agents that perturb replication may be a factor in producing CNVs in the maternal grandchildren of females exposed during pregnancy. The mitotic origin hypothesis also predicts that CNVs will arise frequently in post-zygotic somatic cells, leading to somatic mosaicism within or between tissues. Indeed, substantial evidence exists for somatic mosaicism of pathogenic CNVs, such as in the NF1 and DMD genes [44,63,64], and for apparently benign CNVs in identical twins [65], and different tissues within individuals [66,67].
Because the precise molecular mechanisms involved in producing non-recurrent CNVs are not well understood, it is more difficult to precisely predict genetic risk factors, other than parental origin. If NHEJ is mechanistically involved, the genetic factors are well known and variation in those genes could be tested. However, the genetic factors involved in MMEJ and template switching in mammalian cells have not been identified. Thus, tests for the influence of variation in these genes are not currently possible. We can speculate that variation in genes in DNA damage checkpoint pathways that respond to replication stress to prevent CNV formation can potentially influence non-recurrent CNV risk. Based on our current knowledge, we can also predict from our findings in a cell culture model that agents or conditions leading to replication stress are good candidates for risk factors for inducing non-recurrent CNVs in both the germline and somatic cells in vivo. Nevertheless, aside from a handful of well-characterized laboratory reagents, we currently have a very poor understanding of what these agents are and the scope of the risk for de novo CNVs resulting from environmental agents since, unlike direct DNA damaging agents, comprehensive genomic studies defining agents that cause replication stress and their effects are lacking. These results demonstrate the importance of identifying and studying the effects of such agents on our genomes, both in experimental models and directly in human populations in order to better understand the risks of replication stress for de novo CNVs.
Conclusions and Perspectives
The development and implementation of high resolution, genome-wide analyses over the past decade has revealed that the human genome contains much more structural variation than previously realized. Major progress has been made showing that CNVs are important factors in human genomic variation and in genetic disease. From these studies, we have learned that there are distinct classes of CNVs, as defined by the molecular mechanisms responsible for their formation, and which therefore dictate the risk factors involved for new mutation. Recurrent CNVs have been shown to be the result of NAHR, a well-understood mechanism resulting in rearrangements during meiosis. Surprisingly, the large class of non-recurrent CNVs appears to have a mitotic cell origin, associated with replication stress. While several models have been proposed to explain this class of CNV, there is little experimental evidence to explain how they are formed. Current challenges include experimentally defining these mechanisms and beginning to identify the risk factors for all classes of CNVs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* Of special interest
** Of outstanding interest
- *1.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Phenotypic variability and genetic susceptibility to genomic disorders Detection of large-scale variation in the human genome. Nature Genetics. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- *2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Måner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- *3.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews D, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:428–249. doi: 10.1038/nature05329. Three of the earliest high-density, whole-genome analysis studies demonstrating the extent of of stuctural variants in the human geneome
- 4.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **8.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. This paper analyzed whole genome sequence data from the 1000 Genomes Project, mapping over 25,000 CNVs and other structural variants, including nucleotide resolution breakpoint data for over half the events. These data provided key insights into the molecular mechanisms that give rise to CNVs in the human genome, and also revealed that CNVs arising via the same mechanism tend to cluster in the genome.
- 9.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valduga M, Philippe C, Bach Segura P, Thiebaugeorges O, Miton A, Beri M, Bonnet C, Nemos C, Foliguet B, Jonveaux P. A retrospective study by oligonucleotide array-CGH analysis in 50 fetuses with multiple malformations. Prenat Diagn. 2009;30:333–341. doi: 10.1002/pd.2460. [DOI] [PubMed] [Google Scholar]
- 11.Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 13.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. This paper demonstrated that replication stress caused by the polymerase inhibitor, aphidicolin induced CNVs across the entire human genome in cultured primary fibroblasts. This experimental observation led to the replication stress model of CNV formation.
- *15.Arlt MF, Ozdemir AC, Birkeland SR, Wilson TE, Glover TW. Hydroxyurea induces de novo copy number variants in human cells. Proceedings of the National Academy of Sciences, USA. 2011;108:17360–17365. doi: 10.1073/pnas.1109272108. This paper reported that hydroxyurea, an important drug in the treatment of sickle cell disease induces de novo CNVs in cultured human cells. These results supported a generalized replication stress model for non-recurrent CNV formation and also demonstrated the need to further assess the potential risk for submicroscopic genomic structural changes in the genomes of HU-treated patients and their future generations
- *16.Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proceedings of the National Academy of Sciences, USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. This paper was the first description of replication stress induction of CNV-like microdeletions in the fragile site, FRA3B with microhomologies at breakpoint junctions in mammalian cells.
- 17.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, de Vries BB, Gilissen C, Hoischen A, et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- *19.Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, Lee C, Turner DJ, Hurles ME. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet. 2010;42:385–391. doi: 10.1038/ng.564. These two papers demonstrated that non-recurrent, pathogenic CNVs are primarily characterized by microhomologous sequences at their breakpoint junctions, supporting template switching or end-joining models of CNV formation.
- 20.Park CH, Rha SY, Jeung HC, Kang SH, Ki DH, Lee WS, Noh SH, Chung HC. Identification of novel gastric cancer-associated CNVs by integrated analysis of microarray. J Surg Oncol. 2010;102:454–461. doi: 10.1002/jso.21585. [DOI] [PubMed] [Google Scholar]
- 21.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 22.Brunetti-Pierri N, Grange DK, Ou Z, Peiffer DA, Peacock SK, Cooper ML, Eng PA, Lalani SR, Chinault AC, Gunderson KL, et al. Characterization of de novo microdeletions involving 17q11.2q12 identified through chromosomal comparative genomic hybridization. Clin Genet. 2007;72:411–419. doi: 10.1111/j.1399-0004.2007.00896.x. [DOI] [PubMed] [Google Scholar]
- 23.Bedoyan JK, Kumar RA, Sudi J, Silverstein F, Ackley T, Iyer RK, Christian SL, Martin DM. Duplication 16p11.2 in a child with infantile seizure disorder. Am J Med Genet A. 152A:1567–1574. doi: 10.1002/ajmg.a.33415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ, Graham BH, Pursley A, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, Repnikova EA, Gastier-Foster J, Thrush DL, Kathiresan S, Ruderfer DM, et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stankiewicz P, Inoue K, Bi W, Walz K, Park SS, Kurotaki N, Shaw CJ, Fonseca P, Yan J, Lee JA, et al. Genomic disorders: genomic architecture results in susceptibility to DNA rearrangements causing common human traits. Cold Spring Harbor Symposia on Quantitative Biology. 2003;68:445–454. doi: 10.1101/sqb.2003.68.445. [DOI] [PubMed] [Google Scholar]
- **29.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. In this paper, Hastings et al. describe MMBIR, a model to explain CNVs that form via non-homologous mechanisms. MMBIR is a modification of FoSTeS, which used a template switching mechanism at stalled replication forks to explain CNV formation.
- 30.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Hermetz KE, Jackson JM, Mulle JG, Dodd A, Tsuchiya KD, Ballif BC, Shaffer LG, Cody JD, Ledbetter DH, et al. Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Hum Mol Genet. 2011;20:3769–3778. doi: 10.1093/hmg/ddr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplication induced under stress in Escherichia coli. Public Library of Science Genetics. 2006;2:385–398. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. Elegant studies in yeast showing that template switching during BIR can occur by repeated rounds of strand invasion, DNA synthesis, and dissociation. The authors demonstrate that such events can give rise to chromosome rearrangements.
- 34.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol. 2009;29:1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proceedings of the National Academy of Sciences, USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- 38.Atweh GF. Hydroxyurea in sickle cell disease: What will it take to change practice? Am J Hematol. 2010;85:401–402. doi: 10.1002/ajh.21733. [DOI] [PubMed] [Google Scholar]
- 39.Ware RE, Aygun B. Advances in the use of hydroxyurea. Hematology Am Soc Hematol Educ Program. 2009:62–69. doi: 10.1182/asheducation-2009.1.62. [DOI] [PubMed] [Google Scholar]
- 40.Scott JP. Hydroxurea and sickle cell disease: Its been a long, long time coming. Pediatr Blood Cancer. 2010;54:185–186. doi: 10.1002/pbc.22340. [DOI] [PubMed] [Google Scholar]
- 41.Lou TF, Singh M, Mackie A, Li W, Pace BS. Hydroxyurea generates nitric oxide in human erythroid cells: mechanisms for gamma-globin gene activation. Exp Biol Med (Maywood) 2009;234:1374–1382. doi: 10.3181/0811-RM-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charache S, Terrin ML, Moore RD, Dover GJ, Barton FB, Eckert SV, McMahon RP, Bonds DR. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. Investigators of the Multicenter Study of Hydroxyurea in Sickle Cell Anemia. N Engl J Med. 1995;332:1317–1322. doi: 10.1056/NEJM199505183322001. [DOI] [PubMed] [Google Scholar]
- 43.Arlt MF, Ozdemir AC, Birkeland SR, Lyons RH, Glover TW, Wilson TE. Comparison of Constitutional and Replication Stress-induced Genome Structural Variation by SNP Array and Mate-pair Sequencing. Genetics. 2011 doi: 10.1534/genetics.110.124776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers ALJ, Lalic T, Janson AAM, Ginjaar HB, Bruening MH, den Dunnen JT. Duplications in the DMD gene. Human Mutation. 2006;27:938–945. doi: 10.1002/humu.20367. [DOI] [PubMed] [Google Scholar]
- 45.Inoue K, Osaka H, Thurston VC, Clarke JT, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kresse SH, Ohnstad HO, Paulsen EB, Bjerkehagen B, Szuhai K, Serra M, Schaefer KL, Myklebost O, Meza-Zepeda LA. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2009;48:679–693. doi: 10.1002/gcc.20675. [DOI] [PubMed] [Google Scholar]
- 48.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- *49.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. Bignell and colleagues demonstrated unexplained hotspot regions of recurrent deletions in cancer cell lines, including a region near LSAMP at 3q13.31. This region corresponds to a major hotspot for de novo CNV formation in primary human fibroblasts after treatment with replication inhibitors.
- 50.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’Arcy M, de Berardinis R, Frackelton E, Kim C, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase a inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Human Genetics. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 54.Mrasek K, Schoder C, Teichmann AC, Behr K, Franze B, Wilhelm K, Blaurock N, Claussen U, Liehr T, Weise A. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int J Oncol. 2010;36:929–940. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- 55.Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- 56.Kovacic P. Hydroxyurea (therapeutics and mechanism): Metabolism, carbamoyl nitroso, nitroxyl, radicals, cell signaling and clinical applications. Med Hypotheses. 2010 doi: 10.1016/j.mehy.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 57.Zimmerman SA, Schultz WH, Davis JS, Pickens CV, Mortier NA, Howard TA, Ware RE. Sustained long-term hematologic efficacy of hydroxyurea at maximum tolerated dose in children with sickle cell disease. Blood. 2004;103:2039–2045. doi: 10.1182/blood-2003-07-2475. [DOI] [PubMed] [Google Scholar]
- 58.Flanagan JM, Howard TA, Mortier N, Avlasevich SL, Smeltzer MP, Wu S, Dertinger SD, Ware RE. Assessment of genotoxicity associated with hydroxyurea therapy in children with sickle cell anemia. Mutat Res. 2010;698:38–42. doi: 10.1016/j.mrgentox.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–187. doi: 10.1093/hmg/ddq366. This paper examined phenotypic variability and CNV content in patients, revealing that disease phenotypic variability may be due to the inheritance of additional rare events in families. They show that variability in the number, size, and orientation of segmental duplication blocks in the genome influences the formation of CNVs in those regions.
- 60.Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, Girirajan S, Alkan C, Campbell CD, Vives L, Malig M, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sibbons C, Morris JK, Crolla JA, Jacobs PA, Thomas NS. De novo deletions and duplications detected by array CGH: a study of parental origin in relation to mechanisms of formation and size of imbalance. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *62.Hehir-Kwa JY, Rodriguez-Santiago B, Vissers LE, de Leeuw N, Pfundt R, Buitelaar JK, Perez-Jurado LA, Veltman JA. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J Med Genet. 2011;48:776–778. doi: 10.1136/jmedgenet-2011-100147. These two papers examined patients with neurological abnormalities that were characterized by de novo CNVs. They found a paternal bias in de novo CNV formation, consistent with the hypothesis that mitotic replication stress in the germline can create deleterious CNVs that can be passed on to offspring, and that such stress is more likely to create CNVs in the male germline, which undergoes mitotic divisions leading to germ cells throughout an individuals life.
- 63.Kehrer-Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne DE, Hansmann I, Mautner VF. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet. 2004;75:410–423. doi: 10.1086/423624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White SJ, den Dunnen JT. Copy number variation in the genome; the human DMD gene as an example. Cytogenetic and Genome Research. 2006;115:240–246. doi: 10.1159/000095920. [DOI] [PubMed] [Google Scholar]
- 65.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Stahl TD, Menzel U, Sandgren J, von Tell D, Poplawski A, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Piotrowski A, Bruder CE, Andersson R, de Stahl TD, Menzel U, Sandgren J, Poplawski A, von Tell D, Crasto C, Bogdan A, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 67.Mkrtchyan H, Gross M, Hinreiner S, Polytiko A, Manvelyan M, Mrasek K, Kosyakova N, Ewers E, Nelle H, Liehr T, et al. Early embryonic chromosome instability results in stable mosaic pattern in human tissues. PLoS One. 2010;5:e9591. doi: 10.1371/journal.pone.0009591. [DOI] [PMC free article] [PubMed] [Google Scholar]