Abstract
Embryonic stem (ES) cells, like all cell types, are defined by their unique transcriptional signatures. The ability of ES cells to self-renew or exit the pluripotent state and enter differentiation requires extensive changes in their transcriptome and epigenome. Remarkably, transcriptional programs governing each cell fate must remain sufficiently malleable so that expression of only a handful of transcriptional activators can override the pre-existing state by collaborating with an unexpectedly elaborate collection of coactivators to specify, restrict and stabilize the new state. Here, we discuss recent advances in our understanding of how the same coactivator can interpret multiple lines of information encoded by different activators and integrate signals from diverse regulators into stem cell-specific transcriptional outputs.
Keywords: self-renewal, pluripotency, transcription activation, Mediator, TFIID, DNA repair
Functional interplay between activators and coactivators in embryonic stem cells
Embryonic stem (ES) cells are highly specialized cells with an unmatched ability to faithfully propagate themselves indefinitely (self-renewal) while retaining the capacity to undergo differentiation to generate every cell type in the body (pluripotency). During these processes, concerted, sweeping changes that involve the degradation, synthesis, and reorganization of transcription factors and their cofactors must occur to direct the rapid reprogramming of gene expression networks, remodeling and stabilization of the new transcriptional landscape.
The expression of only a handful of sequence-specific DNA-binding transcription factors is restricted to the pluripotent state (e.g. Oct4 and Nanog), while the majority of transcription factors that have been implicated in stem cell maintenance are expressed in many other cell types (e.g. Sox2, Klf4, c-Myc, Smads and Stats). How do ES cells utilize a limited number of cell type-specific transcription factors to define a stem cell-specific transcriptional signature? Although the combinatorial assembly of transcription factors directed by Oct4 on gene promoters offers some level of specificity [1, 2], their ability to recruit a wide array of cofactors adds to the necessary breadth and flexibility in their transcriptional repertoire to accommodate a wide range of transcriptional responses in ES cells.
Transcriptional cofactors are a large class of loosely grouped protein and multisubunit protein complexes that regulate gene expression through diverse mechanisms. Oct4 and Sox2 recruit histone modifiers (e.g. p300/CBP [3] and the Wdr5/trithorax complex [4]) and chromatin remodelers (e.g. esBAF [5]) to ensure active loci are maintained. Independent of chromatin, they also cooperate with multisubunit Mediator [6], TAFs/TFIID [7], and, unexpectedly, the nucleotide excision repair (NER) complex containing XPC, RAD23B and Centrin 2 (SCC [7]) to execute stem cell-specific transcriptional programs (Glossary and Figure 1).
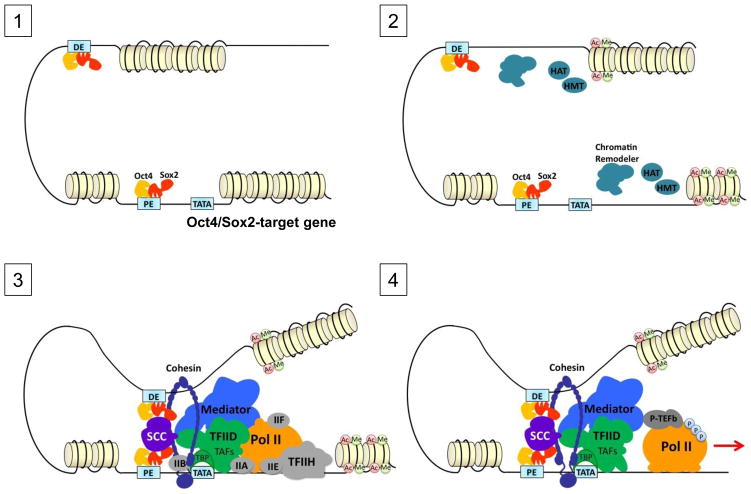
Figure 1.
Multi-step activation model of an Oct4/Sox2-target gene in embryonic stem cells. The transcription apparatus is assembled on an Oct4/Sox2-target gene promoter in a spatially and temporally regulated manner to initiate gene expression. The stepwise assembly is depicted here in the order proposed in [68]. (1) ‘Orchestration’: sequence-specific transcription factors, Oct4 and Sox2, bind to both the proximal (PE) and distal (DE) enhancers. (2) ‘Access’: Chromatin remodelers and histone modifying enzymes such as histone acetyltransferases (HATs) and histone methyltransferases (HMTs) act coordinately to remodel chromatin around the gene loci by altering nucleosome positioning and histone modifications. This allows access of other transcription factors and cofactors to the gene promoter. (3) ‘Initiation’: Preinitiation complex (PIC) and various coactivators (e.g. TAFs/TFIID, Mediator and SCC) are assembled onto the core promoter via interaction with activators bound on PE and DE (e.g. TAFs-Oct4/Sox2 or SCC-Oct4/Sox2), promoter DNA elements (e.g. TBP-TATA), and modified nucleosomes (e.g. TAF3-H3K4me3). DNA looping by cohesin and Mediator can further stabilize the long range enhancer-promoter DNA interaction. (4) ‘Elongation’: phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) by TFIIH facilitates promoter escape while subsequent phosphorylation by P-TEFb stimulates the transition of Pol II into productive elongation.
It is worth noting though, that many, if not all, of these cofactors are also involved in transcriptional activation in cell types other than ES cells. Therefore, the challenge is how this expansive but rather universal set of coregulators can coordinate diverse transcriptional outputs that must be precisely tuned in a cell type-specific manner. The multisubunit nature of many of these cofactors and variations in the subunit composition provide unique contact surfaces for select transcription factors. The ability of cofactors (e.g. Mediator) to adopt distinct conformations induced by different activators permits specific activator-dependent readouts sensed by the transcription apparatus [8]. Furthermore, multisubunit coactivator complexes are often made up of functional submodules that allow them to participate in various facets of transcriptional regulation, and in some cases, processes beyond transcription. In ES cells, for example, using a DNA repair complex as a transcriptional coactivator for Oct4 and Sox2 represents yet another strategy to further diversify their transcriptional repertoire by coordinating with other cellular processes.
In this review, we will focus on the roles of TAFs, Mediator, and the XPC/SCC complex in stem cell-specific gene activation. Although they represent only a slice of the known transcriptional network in ES cells, they serve to highlight a recurring theme in cell type-specific transcriptional control: functional diversity, structural flexibility and compositional changes lend coactivators the unprecedented ability to interact and coordinate with a wide array of transcription factors and cofactors to execute stem cell-specific gene regulatory programs (Figure 2).
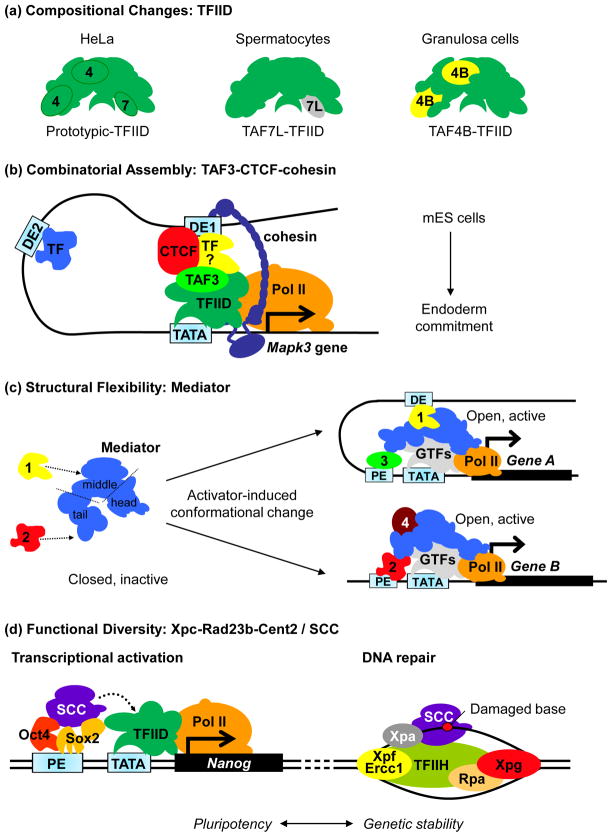
Figure 2.
Mechanisms of cell type-specific transcriptional activation by coactivators. (a) Prototypic TFIID purified from HeLa cells is composed of the TATA-binding protein (TBP) and 13–14 TBP-associated factors (TAFs). Canonical TAFs are replaced with gonad-selective TAFs in both male and female germ cells. TAF7 is substituted with its paralog TAF7L in spermatocytes while TAF4 is replaced with TAF4B in granulosa cells of the ovary. Genetic inactivation of TAF7L and TAF4B in mice led to sterility. Incorporation of non-canonical TAFs like TAF7L and TAF4B in TFIID may provide unique contact surfaces for germ cell-specific transcription factors to mediate transcriptional activation [9]. (b) In mouse embryonic stem (ES) cells, the combinatorial assembly of TAF3, CTCF (CCCTC-binding factor), cohesin and presumably an endoderm lineage-specific TF (yellow) permits selective long distance enhancer-promoter DNA interaction (via DE1 but not DE2) that leads to activation of the Mapk3 (mitogen-activated kinase 3) locus and endoderm commitment during ES cell differentiation. (c) Binding of transcription factors (TFs) to Mediator induces conformational changes that convert Mediator from an inactive, closed conformation to an open, transcriptionally competent one, which in turns, facilitates additional contacts with other TFs occupied at the proximal (PE) or distal (DE) enhancers as well as the general transcription machinery (general transcription factors, GTFs, and RNA polymerase II, Pol II) assembled on a gene promoter (TATA). The modular nature of Mediator provides unique contact surfaces for diverse TFs. In the hypothetical example depicted above, recruitment of Mediator to the DE of Gene A by TF1 via the middle module causes a conformational change in Mediator that exposes unique, TF1-dependent interacting surfaces for Pol II and PE-bound TF3. On the other hand, activation of Gene B involves the binding of TF2 to Mediator tail at PE. TF2-induced conformation change in Mediator exposes new docking sites for another coactivator (“4”) and the general transcription machinery [69]. (d) In ES cells, the Xpc-Rad23b-Cetn2 complex (SCC) functions both as a coactivator for Oct4 and Sox2 at the Nanog gene, and as an initiator of global genome-nucleotide excision repair (GG-NER) by binding to specific DNA lesion and recruiting other NER factors (Xpa, Rpa, TFIIH, Xpg and Xpf-Ercc1). SCC may function to couple stem cell-specific transcription with genome surveillance.
TAFs
The recruitment of transcription factor TFIID to protein-coding gene promoters represents a defining step in the ordered assembly of the general transcription factors (GTFs) into a functional preinitiation complex (PIC) that culminates in transcription initiation [9] (Figure 1). TFIID is a multiprotein complex comprising the TATA binding protein (TBP) and 13–14 TBP-associated factors (TAFs). TAFs are an eclectic collection of proteins that possess enzymatic activity, recognize core promoter elements, anchor TFIID to nucleosomes, and function as transcriptional coactivators [9–11]. Given the multitude of essential protein-protein and protein-DNA transactions that TFIID mediates, the composition of TFIID has traditionally been thought to be invariant. However, with the discovery of tissue-specific TAFs, TAF paralogs, and TBP-related factors (TRFs) comes the appreciation that TFIID is just as capable as any other coactivators for imparting gene selectivity and functional flexibility to the PIC [9, 12] (Figure 2a).
TFIID levels tend to be elevated in highly proliferative cell types (e.g. cancer cells and ES cells) and are down-regulated in terminally differentiated cells (e.g. muscle and adult hepatocytes) [9, 13]. This is consistent with the observation that transcription of cell cycle genes often requires TFIID [14]. Therefore, it is difficult at times to decipher the role of TAFs in ES cells, that is, to separate stem cell-specific functions of TAFs/TFIID from their general role in proliferation. Nevertheless, several unbiased genome-wide RNA interference (RNAi) screens identified multiple TAFs as important regulators of Oct4 expression in both human (TAF2, 7, and 12 [15]) and mouse (TAF1, 5, 6L, 10, 11 and 12 [6]) ES cells. We surmise that the partial depletion of TAFs by RNAi retains enough intact TAFs/TFIID to sustain transcription of housekeeping genes but falls below a critical threshold level to support the transcription of Oct4 and other ES cell genes. It remains possible that certain TAFs can function as coactivators for stem cell-specific activators (e.g. Oct4, Sox2 and Nanog) to activate ES cell genes. Indeed, TAF1 and TBP were identified in a proteomic study as Oct4-associated proteins [16]. Furthermore, Oct4/Sox2-dependent activation of the Nanog gene in vitro requires TAFs/TFIID, whereas purified TBP alone failed to substitute for TFIID [7]. Since TFIID and most of the GTFs used in our transcription assays were purified from HeLa cells, it is unlikely that Oct4 and Sox2 require an ES cell-specific TAF(s) or component(s) in the general transcription machinery to support Nanog transcription. However, a full Oct4/Sox2-dependent activation requires at least two additional coactivator activities derived from ES cells [7], one of which, XPC/SCC, will be discussed in detail later in this review. Although it doesn’t appear that a cell type-restricted cofactor is required to activate ES cell genes, transcriptional specificity can be achieved through the combinatorial assembly of cofactors and activators that are highly enriched in ES cells (e.g. TAFs/TFIID, XPC/SCC, Oct4 and Sox2).
There is growing evidence that TAFs may function beyond transcription initiation. For example, TAF7 has been implicated in transcriptional elongation control by association with transcription elongation factors TFIIH and P-TEFb [17]. Although still controversial, TAF12 has been shown to promote DNA demethylation at ribosomal RNA genes by recruiting the DNA damage inducible protein Gadd45a [18]. Recently, we reported an unexpected role for TAF3 in mediating long distance enhancer-promoter DNA interaction in mouse ES cells [19]. Even though TAF3 is selectively enriched in ES cells (but not in a variety of other highly proliferative cell types such as C2C12 cells), knockdown (KD) of TAF3 has minimal effect on the expression of pluripotency genes. However, a striking defect in the gene expression program specifying endodermal lineage was observed in TAF3 KD ES cells upon differentiation into embryoid bodies in vitro and teratoma formation in vivo. Genome-wide studies on TAF3 binding sites revealed a significant overlap with those of CTCF-cohesin, a complex with a well documented role in mediating long distance DNA looping and enhancer insulation [20]. TAF3 directly interacts with CTCF and is required to establish proper DNA looping around the Mapk3 locus, a key signaling inducer for endoderm differentiation, by bridging a distal enhancer to the core promoter [19] (Figure 2b).
An outstanding question is what drives TAF3 to search, select and isolate a subset of enhancers for looping during a highly dynamic process like ES cell differentiation. The PHD domain of TAF3 can direct TFIID to actively transcribing and developmentally-poised gene promoters through specific interaction with H3K4 trimethylated (H3K4me3) histones [21, 22]. However, this alone cannot account for the selective defect in endoderm specification observed in TAF3 KD ES cells, as H3K4me3 marks are also present in ‘poised’ ectodermal and mesodermal genes [21]. We hypothesize that TAF3 in TFIID may interact with endoderm-specific activators (e.g. GATA4) at distal enhancers and promote DNA looping through formation of a higher order protein ensemble containing TAF3-CTCF-cohesin. Although none of these proteins alone contains the necessary information to carry out ES cell-specific function, the multiple contacts between lineage-specific activators, TAF3, CTCF and cohesin may act as ‘check points’ to ensure activation of the right gene at the right time (Figure 2b).
Mediator
The Mediator complex exemplifies the versatility and adaptability coactivators must possess in order to coordinate a full range of transcriptional programs and integrate them into a functional biological response. The 30 or so subunits in Mediator endow this megadalton protein complex with the ability to interact with a wide array of transcriptional activators and RNA polymerase II (Pol II) and to mediate the synergistic response by these activators [8]. The MED26 subunit of Mediator has been shown to function as a docking site for both TFIID and transcriptional elongation factors and is thought to act as a molecular sling to facilitate promoter escape of Pol II and its entrance into productive elongation [23]. In the context of chromatin, Mediator collaborates with TAFs/TFIID and PBAF to overcome the transcriptional block imposed by nucleosomes and to generate a robust activator-dependent transcriptional response [24, 25]. Recent studies have identified expanded roles for Mediator in alternative splicing [26] as well as in termination of transcription [27]. Given the generality of Mediator action in transcription, it has been proposed that Mediator should be considered as part of the general transcription machinery [8].
The critical role of Mediator in mouse ES cell transcription was revealed in an RNAi screen for candidates required for the proper expression of Oct4 [6]. Although this may not come as a surprise considering Mediator’s general function in transcription, what was unanticipated was the ability of Mediator to directly interact with the cohesin complex, thereby stabilizing long distance enhancer-promoter DNA interactions at ES cell genes. Intriguingly, Mediator occupies only a subset of cohesin-bound sites that contain the cohesin loading factor Nipbl but are largely devoid of CTCF. This may indicate a gene-selective function of Mediator in directing the activation of ES cell genes. The recent finding that TAF3 cooperates with cohesin-CTCF to form long distance chromatin loops to control the expression of early developmental genes (but not pluripotency genes, i.e. Oct4, Sox2 and Nanog) during ES cell differentiation [19] underscores how starkly different transcriptional outcomes can be achieved through assembly of distinct protein ensembles (e.g. Mediator-cohesin-Nipbl versus TAF3-cohesin-CTCF) that mediate surprisingly similar processes (i.e. DNA looping).
It is worth pointing out that Mediator-dependent DNA looping is not unique to ES cells [6]. MED1 has been shown to facilitate chromatin loop formation during nuclear receptor-activated transcription [28, 29]. Extensive colocalization between Mediator and cohesin was also observed in mouse embryonic fibroblasts (MEFs) and adult thymocytes [30]. However, comparison of Mediator and cohesin bound sites in ES cells and MEFs revealed only a limited overlap of their targets [6]. These observations suggest that Mediator plays an integral part, in conjunction with cell type-specific activators, in forming functional long distance DNA loops in a cell type-specific manner.
In addition to acting as a molecular adaptor linking activators to the general transcription machinery and enhancers to gene promoters, Mediator also functions as a molecular hub onto which various signaling pathways relevant to ES cell maintenance and homeostasis converge. For example, the CDK8/cyclin C kinase submodule helps recruit β-catenin, a key player in the Wnt signaling cascade, to its target genes (e.g. c-Myc) [31] and antagonizes the repressive function of E2F1 on β-catenin-dependent transcription [32]. CDK8 can also impact BMP and TGFβ pathways by activating Smad proteins through phosphorylation at the linker region [33]. In addition, the Mediator tail subunit MED15, through interaction with the nucleocytoplasmic shuttling/transcriptional regulator TAZ (WWTR1), specifically recruits Smad2/3–4 complexes, but not Smad1–4, to TGFβ response elements [34].
Perhaps a testament to the multifunctional nature of Mediator in transcription, myriad cofactors not mentioned in this review (histone acetyltransferase p300 [35], chromatin remodeler Chd1 [36], elongation factor Paf1C [37] and many more) have also been reported to interact with Mediator in ES cells. Accordingly, disruption of these factors often results in spontaneous differentiation and/or skewed developmental potential. How all these fit together to generate a cell type- and gene-specific transcriptional response is less well understood. Perhaps the many activator-induced conformations adopted by Mediator can uniquely orient Mediator with respect to other transcription factors assembled on a gene promoter. This may allow Mediator to selectively present (or obscure) one or more of its submodules (head, middle, tail and kinase) to effect downstream events in transcription in an activator-dependent manner, while enabling crosstalk with various cell signaling pathways to establish a cell type-specific transcriptional signature (Figure 2c).
XPC/SCC
Interactome studies of Oct4 and Sox2 revealed an extended network of protein-protein interactions [16, 38–40]. With these extensive data sets comes the difficulty of sorting out functionally relevant interactors that are required for Oct4 and Sox2 to activate transcription. Compounding this challenge is that many functional coactivator-activator interactions are often weak and transient. Using an unbiased and defined in vitro transcription assay to systematically screen for factors that are required for Oct4 and Sox2 to activate the transcription of Nanog, we unexpectedly identified the XPC/SCC complex as an Oct4/Sox2 coactivator in ES cells [7]. Consistent with our in vitro results, XPC/SCC binds Oct4 and Sox2 and co-occupies the majority of their target genes in mouse ES cells. Surprising as it may seem, these data underscore the ability of Oct4 and Sox2 to recruit a wide range of multisubunit and multifunctional protein complexes to establish a transcriptional network that can crosstalk with other cellular processes to generate a coordinated biological response. It has been shown that ES cells are particularly sensitive to DNA damage induced by UV as they undergo rapid differentiation followed by apoptosis [41]. This is due in part to the activation of a p53-dependent DNA damage response that coordinately represses key pluripotency genes while activating those that promote differentiation [42–44]. Given the unique position of XPC/SCC in both transcriptional activation and DNA repair in ES cells, the XPC/SCC complex may also serve as a nexus between the DNA damage response and the transcriptional machinery at gene promoters (Figure 2d). Perturbation of the Oct4/Sox2 transcriptional circuitries by redirecting XPC/SCC from transcription duties to damaged DNA sites could provide an efficient sensing mechanism to eliminate damaged ES cells by destabilizing the stem cell-specific gene regulatory network.
Although best known as an initiator of global genome-NER pathway, XPC/SCC has also been shown to play a role in base excision repair (BER) by activating DNA glycosylases that excise damaged DNA bases [45]. In mammals, BER is also thought to be one of the major DNA demethylation pathways from which deaminated (5mC to T) or highly oxidized derivatives of methylated cytosine (5mC to 5hmC/5fC/5caC) can be recognized by DNA glycosylases as “damaged” bases and subsequently removed [46]. Consistent with the notion that XPC/SCC regulates DNA demethylation, the global DNA methylation level was dramatically increased in XPC-depleted HeLa cells [47]. DNA methylation and demethylation are essential for the proper execution of differentiation programs in ES cells as well as the reacquisition of a pluripotent state during somatic cell reprogramming [48]. Aberrant DNA methylation/demethylation or 5mC/5hmC turnover in XPC/SCC-depleted MEFs may therefore partially account for the strong block in induced pluripotent stem (iPS) cell derivation [7]. More controversial is the proposed role of NER factors including XPC in recruiting Gadd45a to aid DNA demethylation at gene promoters [47]. Regardless of the exact mechanism, XPC/SCC is likely to participate in various aspects of transcriptional control in ES cells that includes regulating DNA methylation homeostasis genome-wide or at gene promoters.
Further highlighting the multifunctional nature of XPC/SCC, the RAD23B subunit of the XPC/SCC complex is an unusual protein with independent domains that interact with the proteasome and ubiquitinated proteins. It chaperones ubiquitinated substrates to the proteasome for degradation [49], and in some instances, protects proteins like XPC from proteolysis [50, 51]. The recruitment of RAD23B/SCC by Oct4 and Sox2 to enhancers and gene promoters in ES cells [7] may permit XPC/SCC to modulate transcription through regulating transcription factor turnover. This is consistent with the observation in yeast that Rad23 deletion affects transcription of many genes [52]. Since the proteasome itself has also been implicated in transcriptional elongation control [53, 54], SCC may impact transcription at multiple steps by acting as a classical transcriptional coactivator, a regulator of DNA methylation and possibly by recruiting the proteasome to regulate protein turnover and transcriptional elongation.
Concluding remarks
Most, if not all, of the cofactors discussed here are utilized by a variety of cell types (ES cells or not) to regulate the expression of a large number of genes. It is therefore somewhat perplexing that disruption of these cofactors in ES cells seems to preferentially affect pluripotency gene expression without severely impacting the transcription of genes important for housekeeping and cell viability. Although this may in part be due to the nature of the experiments (partial knockdown vs. knockout), it does highlight potential differences in the mechanisms governing the transcription of pluripotency genes (e.g. Oct4 and Nanog) versus housekeeping genes (e.g. Gapdh and β-actin). We surmise that the many transcription factors and cofactors required to activate Oct4 and Nanog, for example, allow transcription to occur rapidly and over a large dynamic range [55]. This is particularly important in ES cells as many of the key pluripotency and lineage specification genes must be turned on and off or precisely tuned in a developmental stage-specific manner. The expression of housekeeping genes, on the other hand, is largely sustained at some basal level and not as dynamically regulated, which may explain why their levels tend to be less reliant on cofactor abundance [56–58].
We showed that XPC/SCC-depleted MEFs failed to be fully reprogrammed to a pluripotent state [7]. The role of TAFs and Mediator in this process remains less clear. Given that the transcriptome of iPS cells closely resembles that of bona fide ES cells, it is likely that similar gene regulatory mechanisms govern both cell types. Indeed, a partially purified protein fraction enriched in BAFs and TAFs is competent for reprogramming [59]. Although ectopic expression of several BAF subunits in MEFs has been shown to facilitate somatic cell reprogramming, the role of TAFs, if any, in iPS cell derivation remains an open question.
Although this review primarily focused on coactivators, it is not our intention to de-emphasize the profound influence cell-type specific transcriptional activators have on cell fate choice. The reacquisition of pluripotency in differentiated cell types by ectopic expression of only a handful of transcription factors (e.g. Oct4, Sox2, Klf4 and c-Myc [60]), and the ability of ‘master’ regulators like Oct4 and MyoD to ‘hijack’ other sequence-specific transcription factors (e.g. Smad proteins) to their respective cognate binding sites [2] and to directly convert fibroblasts to a variety of functional differentiated cell types [61–65], underscore the ability of sequence-specific DNA binding transcription factors to build basic frameworks for cell type-specific transcriptional programs. However, execution of such drastic reorganization and stabilization of the transcriptional landscape also requires an unexpectedly diverse list of coactivators, corepressors, and epigenetic regulators. Furthermore, the potential role of non-coding RNAs in transcription implicates even more complex regulatory networks involving protein-protein and protein-nucleic acid-mediated gene control mechanisms [66, 67]. The emerging picture reveals an elaborate mechanism of gene regulation with no single protein or protein complex standing out as more important than the rest as they appear to be physically and functionally intertwined.
Acknowledgments
The authors wish to thank E. Zhang, C. Inouye, Z. Liu and members of our laboratory for critical reading of the manuscript. C.C. is a postdoctoral fellow of California Institute for Regenerative Medicine (CIRM training program TG2-01164). T.Y. is a Jane Coffin Childs fellow.
Glossary
- General transcription factors
an evolutionarily conserved set of protein or protein complexes (TFIIA, B, D, E, F and H) that, together with RNA polymerase II, form the preinitiation complex (PIC) that is responsible for the transcription of virtually all eukaryotic mRNA and miRNA genes
- Sequence-specific transcription factor
protein that recognizes and binds specific DNA motif in the promoter and/or enhancer region of a given gene, and can mediate gene activation or repression
- Transcriptional cofactor
protein or protein complex that mediates transcriptional activation or repression via interaction with general and/or sequence-specific transcription factors
- Transcriptional coactivator
a transcriptional cofactor that enhances transcription of a target gene
- Transcriptional corepressor
a transcriptional cofactor that represses transcription of a target gene
- Histone modifier
protein or protein complex that deposits or removes post-translational modifications on histone cores and/or tails. Known modifications of histones include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, citrullination, glycosylation, propionylation, butyrylation, proline cis-trans isomerization, ADP-ribosylation, and proteolysis
- Chromatin remodeler
ATP-dependent protein complexes that disrupt and reform histone-DNA contacts to facilitate nucleosome shifting along a piece of DNA, re-orient DNA around histone octamers, position nucleosomes on the DNA, and/or exchange histones
- PHD finger domain (Plant Homeo Domain)
zinc-coordinating domain of 50–80 amino acids that anchors PHD-containing proteins to unmodified or various forms of modified lysines on histone H3 tail (H3K4me3, H3K9me3, H3K4me0)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 4.Ang YS, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong YW, et al. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cler E, et al. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell Mol Life Sci. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller F, et al. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev. 2010;20:533–540. doi: 10.1016/j.gde.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 13.D’Alessio JA, et al. Core promoter recognition complex changes accompany liver development. Proc Natl Acad Sci USA. 2011;108:3906–3911. doi: 10.1073/pnas.1100640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson I, et al. New insights into TAFs as regulators of cell cycle and signaling pathways. Cell Cycle. 2005;4:1486–1490. doi: 10.4161/cc.4.11.2120. [DOI] [PubMed] [Google Scholar]
- 15.Chia NY, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 16.Ding J, et al. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–167. doi: 10.1038/cr.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gegonne A, et al. TFIID component TAF7 functionally interacts with both TFIIH and P-TEFb. Proc Natl Acad Sci USA. 2008;105:5367–5372. doi: 10.1073/pnas.0801637105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz KM, et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, et al. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorsett D. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21:199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 22.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi H, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemon B, et al. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 25.Naar AM, et al. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, et al. Mediator Complex Regulates Alternative mRNA Processing via the MED23 Subunit. Mol Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukundan B, Ansari A. Novel role for mediator complex subunit Srb5/Med18 in termination of transcription. J Biol Chem. 2011;286:37053–37057. doi: 10.1074/jbc.C111.295915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, et al. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Park SW, et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris EJ, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varelas X, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 35.Black JC, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 36.Gaspar-Maia A, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding L, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 40.van den Berg DL, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Waard H, et al. Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts. DNA Repair. 2008;7:1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 43.Li M, et al. Distinct Regulatory Mechanisms and Functions for p53-Activated and p53-Repressed DNA Damage Response Genes in Embryonic Stem Cells. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain AK, et al. p53 Regulates Cell Cycle and MicroRNAs to Promote Differentiation of Human Embryonic Stem Cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y, et al. Stimulation of DNA Glycosylase Activities by XPC Protein Complex: Roles of Protein-Protein Interactions. J Nucleic Acids. 2010:2010. doi: 10.4061/2010/805698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branco MR, et al. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 47.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade SL, Auble DT. The Rad23 ubiquitin receptor, the proteasome and functional specificity in transcriptional control. Transcription. 2010;1:22–26. doi: 10.4161/trns.1.1.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okuda Y, et al. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair. 2004;3:1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Ng JM, et al. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade SL, et al. The Snf1 kinase and proteasome-associated Rad23 regulate UV-responsive gene expression. Embo J. 2009;28:2919–2931. doi: 10.1038/emboj.2009.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Szutorisz H, et al. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 55.Darzacq X, Singer RH. The dynamic range of transcription. Mol Cell. 2008;30:545–546. doi: 10.1016/j.molcel.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen-Gihon I, et al. Modular genes with metazoan-specific domains have increased tissue specificity. Trends Genet. 2005;21:210–213. doi: 10.1016/j.tig.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 57.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singhal N, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 60.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 64.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 65.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 66.Wilusz JE, et al. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Cri Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci USA. 2010;107:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]