Abstract
CG methylation is an epigenetically inherited chemical modification of DNA found in plants and animals. In mammals it is essential for accurate regulation of gene expression and normal development. Mammalian genomes are depleted for the CG dinucleotide, a result of the chemical deamination of methyl-cytosine in CG resulting in TpG. Most CG dinucleotides are methylated, but ~ 15% are unmethylated. Five percent of CGs cluster into ~20,000 regions termed CG islands (CGI) which are generally unmethylated. About half of CGIs are associated with housekeeping genes. In contrast, the gene body, repeats and transposable elements in which CGs are generally methylated. Unraveling the epigenetic machinery operating in normal cells is important for understanding the epigenetic aberrations that are involved in human diseases including cancer. With the advent of high-throughput sequencing technologies, it is possible to identify the CG methylation status of all 30 million unique CGs in the human genome, and monitor differences in distinct cell types during differentiation and development. Here we summarize the present understanding of DNA methylation in normal cells and discuss resent observations that CG methylation can have an effect on tissue specific gene expression. We also discuss how aberrant CG methylation can lead to cancer.
Keywords: Epigenetics, DNA methylation, transcription, CpG Island, Cancer, 5-azacytidine
1. Introduction
The great mystery in metazoan development is how different cells express different genes in spite of containing the same genetic material. Epigenetics was first defined by the developmental biologist Conrad H. Waddington in the early 1940’s as: “The interactions of genes with their environment that bring the phenotype into being”. But the modern definition of epigenetics has a more precise molecular basis, defined as the study of heritable changes in gene expression patterns that are not caused by changes in the nucleotide sequence of the genetic code itself. Epigenetic changes, frequently defined as “epigenome” involve chemically modified DNA (DNA methylation), and changes in DNA associate molecules, such as histones modifications, chromatin remodeling complexes and other small noncoding RNAs including miRNA and siRNAs.
After every cycle of DNA replication, fully methylated DNA becomes hemimethylated. DNA methyltransferase 1 (DNMT1), the maintenance methylase, binds the hemimethylated DNA and directs the addition of a methyl (CH3) group to the 5' carbon position of the cytosine ring. In mammals, three DNMTs have been identified: DNMT1, which is responsible for the maintenance of existing methylation patterns following DNA replication; DNMT3A and DNMT3B are considered to be de novo methyltransferases, which add methyl group to the previously unmodified DNA [1]. In mammals, most of the cytosine methylation occurs in the context of the CG dinucleotide, although other cytosine methylation is observed in the sequence context of CHG and CHH (H= A, C, T) in human embryonic stem cells and plants, but rarely in somatic mammalian cells [2, 3]. In this review, we examine the current understanding of the mammalian DNA methylation, the genomic distribution of CG methylation, and their predicted biological functions. Finally, we propose a model to explain an apparent contradiction surrounding the effects of 5-azacytidine that mediates demethylation and inhibits differentiation of normal cells whereas it induces differentiation in certain human cancers.
2. CG methylation in different parts of the genome
2.1. CG Islands
CGs are rare in the genome, probably, because of the higher mutation rate of the methylated CG that is prone to be chemically deaminated and converted to TG [4]. Figure 1 presents CG density at a megabase scale for some arbitrary chromosomal locations of both the human and Drosophila genome. The human genome has lower CG density across the genome compared to Drosophila, whereas CG rich clusters or CG islands (CGI) that occur ~20,000 times in mammalian genomes, are visible only in the human genome. CGI are generally 200 – 4,000 bp long, and often occur in the promoters and/or first exons of genes [5]. CGI were first detected using a 200 bp sliding window to identify regions that have CG content greater than 50% and an observed/expected CG dinucleotide ratio of greater than 0.6 [6] resulting in identification of 28,691 CGIs in the human genome, a definition used at the UCSC genome browser. This definition was subsequently modified by extending this window by 500 bp, increasing the CG content by greater than 55% and observed/expected ratio greater than 0.65 [7]. However, alteration of these thresholds significantly changed the number of predicted CGI and suggests that there may be different kinds of CGI [8].
Figure 1.
Frequency of CG dinucleotide across the human and drosophila genome shows the presence of CG clusters called CG Islands (CGI) only in the human genome.
Recently, a biochemical approach has been proposed to address this issue. CGIs are enriched based on an unmethylated CG affinity purification using the CXXC protein domain [8]. High-throughput DNA sequencing of this enriched DNA fragments identified a comprehensive set of ~25,000 unmethylated regions in both humans and mice, including the majority of CGIs plus an additional ~10,000 regions [9]. This approach identified the majority of predicated CGIs that are unmethylated but also other regions of the genome that are unmethylated. Unmethylated CGIs are present in the promoters or first exons of the housekeeping genes [5] and are transcriptionally active. However, in some circumstances, they become heavily methylated and correlate with silencing of the corresponding gene [10].
2.2. Promoter methylation
Approximately 50% of transcription start sites (TSS) [10], and ~70% of all genes are linked to CGIs in the human genome [11]. Promoters have been arbitrarily classified into three classes based on their CG density: low CG content promoters (LCP), high CG content promoters (HCP) and intermediate CG content promoters (ICP) [11]. When the methylation status of an entire promoter (−1,000 bp to + 500 bp) is determined using methylated DNA immunoprecipitation (MeDIP) followed by hybridization to promoter arrays, two distinct groups can be observed (Figure 2). Promoters that are CG dense tend to be unmethylated while low CG promoters tend to be methylated. Thus, majority of HCPs tends to be unmethylated and are associated with ubiquitously expressed housekeeping and tightly regulated developmental genes [1, 11–14]. However, many hypomethylated HCPs are also transcriptionally inactive or non-productive although bound by initiating form of RNA Polymerase II (RNAP) [15] and show a elevated levels of dimethylation of Lys4 of histone H3 [11]. These promoters are proposed to be transcriptionally permissive, and the productive transcription can be triggered by inducible transcription factor-dependent recruitment of P-TEFb followed by a second phosphorylation event on serine 2 (S2) of the CTD of RNAP [10, 15].
Figure 2.
CG methylation status of mouse promoters. Immunoprecipitated methylated DNA are hybridized to NimbleGen promoter arrays and the average methylation for each promoter is determined. Average methylation level (in log2) is plotted against number of CG dinucleotides (in log10) for each promoter. Two distinct clusters are formed, for low CG promoters, methylation increases with the CG density and they are methylated promoters while for high CG promoters, average methylation is low and are the unmethylated promoters.
In contrast, LCPs are hypermethylated in somatic cells and which does not preclude their activity [11]. Gene ontology analysis revealed that LCPs are associated with tissue specific genes. Recently, we reported that methylation of these LCPs are required for activation of some tissue specific genes [16]. CG methylation creates transcription factor binding sites (TFBS) for C/EBP family members, which are involved in differentiation of many cell types. C/EBP binds these methylated TFBS and is required for active transcription at tissue specific LCP. The effect of CG methylation on transcription factor function is vividly depicted in Figure 3: a reporter plasmid with no CGs in the backbone was used as a recipient for 4 copies of the consensus CRE and then the CG in each CRE was enzymatically methylated. The unmethylated plasmid is activated by CREB but not C/EBPα while the methylated plasmid is not activated by CREB but is activated by C/EBPα, in agreement with biochemical studies [16]. During keratinocyte and fibroblast differentiation, where C/EBPα is known to be an active player, 40% and 32% of the 452 and 655 differentiation specific genes, respectively, have methylated low CG promoters. Bisulfite treatment followed by cloning and sequencing of some of these promoters showed that they were methylated, both in undifferentiated cells and differentiated cells following gene activation [16]. Demethylation by either 5-azacytidine treatment or by inhibition of DNMT1 by siRNA treatment preferentially decreased C/EBPα binding to methylated promoters and resulted in inactivation of methylated promoters during differentiation suggesting that methylation is essential for C/EBPα binding and subsequent activation of gene expression. Overall, the general observation is that promoters with high CG density are generally unmethylated, whereas low CG containing promoters are hypermethylated and their methylation status does not preclude their activation. Indeed, in some cases, it is required for activation of transcription.
Figure 3.

Relative luciferase activity is measured for the 4X CRE reporter plasmid, which has no CG in backbone. Unmethylated version of the 4X-CRE is transactivated by the CREB transcription factor while methylated version of 4X-CRE is transactivated by the C/EBPα transcription factor.
2.3. Gene body methylation
In mammalian genomes, gene bodies tend to be methylated with a positive correlation between the level of mRNA expression and CG methylation [17]. Gene body methylation is an ancient phenomenon, and has been observed in plants, invertebrates and vertebrates [3, 18, 19]. Gene-body methylation has been observed in the active human X chromosome when compared to its inactive counterpart [20]. Targeted genome scale CG methylation analysis revealed an association of gene body methylation with highly expressed genes in human B-lymphocytes, fibroblasts and induced pluripotent stem cells [21]. However, a recent report found that gene body methylation in rice has a parabolic correlation with gene expression, with modestly expressed genes having the highest amounts of gene body methylation [19]. The general observations are that exons tend to be more methylated than introns, and that the TSS proximal region and transcription termination sites are devoid of methylation [3, 22]. These findings suggest a role of DNA methylation in transcriptional elongation and termination, and perhaps alternative splicing [23]. One specific example is from different stages of lymphocyte development when these cells express different splice variants of CD45 transcripts with variable exclusions of exons 4–6. A recent report found that CCCTC-binding factor (CTCF) binds exon 5 of CD45 only when this exon is unmethylated [24]. This binding causes local pausing of RNA polymerase II and promotes inclusion of exon 5 in peripheral lymphocytes, a direct evidence of DNA methylation effect on transcriptional elongation and alternative splicing.
2.4. DNA methylation and nucleosome positioning
Nucleosomes are the fundamental unit of eukaryotic chromatin and the basic DNA packaging module. In nucleosomes, 146 bp of DNA are wrapped around a histone octamer which compacts and regulates the access to DNA in the nucleus. It plays two major roles: alters the accessibility of DNA to the cellular machinery, and regulates transcriptional activities by covalent modification of the tails of four core histones: H2A, H2B, H3 and H4 [25–29]. The nucleosome positioning in the genome is not random with several possibilities being suggested [30]. The recent publication of the in vitro binding of nucleosomes to human DNA highlight an enigma in mammalian gene regulation: CGI are bound well by nucleosomes in vitro, while in vivo, these same sequences are not bound by nucleosomes and represent regulatory regions [31]. This is in a contrast with yeast and fly promoters that are A&T rich [32] and do not bind nucleosomes well [33], suggesting that different mechanisms are operating in these metazoans to regulate gene expression.
Evidence of nucleosome repositioning by CG methylation was first found in vitro on the chicken adult β-globin gene promoter [34]. DNA methylation prevents the histone octamer from interacting with an otherwise high affinity positioning sequence in the promoter region of the β-globin gene. This exclusion is attributed to methylation-determined changes in DNA structure within a triplet of CG dinucleotides [34]. However, a recent genome-wide study on the role of methylation dependence in nucleosome positioning in Arabidopsis thaliana and human, came to the opposite conclusion. They found that methylated DNA was preferentially bound by nucleosomes in vivo [35] and nucleosomal DNA was more highly methylated than flanking DNA. This study also found that methyltransferases preferentially target nucleosome-bound DNA, thus maintaining the methylation of nucleosome bound DNA [35]. Nucleosome positioning has a striking effect on DNA methylation, as depletion of linker histone H1 induces hypomethylation of some CGIs, such as some imprinting control regions of the H19-Igf2 and Gtl2-Dlk1 loci [36]. Recent genome wide studies revealed that exonic regions are more enriched with nucleosomes than introns and promoters, in agreement with observations that exonic regions are more hypermethylated than the other regions in the genome [19, 37], again supporting the fact that nucleosomal DNA was more highly methylated than flanking DNA. Another study proposed that CGI promoters, which are generally devoid of methylation and have low nulceosome occupancy, does not require SWI/SNF nucleosome remodeling complex to facilitate the promiscuous induction, whereas CG poor promoters that assemble into stable nucleosomes require SWI/SNF complex and transcription factors to promote selective nucleosome remodeling [38]. Further studies will be needed to examine in more detail the effect of methylation on the competition between nucleosome binding and transcription factor binding.
3. Functional aspects of CG methylation
3.1. Development and Differentiation
Immediately after fertilization and prior to the first cell division, the paternal genome undergoes a genome-wide demethylation [39–42]. After the first cell cycle, the maternal genome also becomes demethylated, and this genome-wide demethylation continues, except for the imprinted genes, until the formation of the blastocyst [43, 44]. In contrast, in the primordial germ cells the parental imprinting is erased by DNA demethylation [45]. During blastocyst formation, DNA methylation levels of the pluripotent stem cells are restored by the de novo methyltransferases when the early cell fate decisions are established. Embryonic stem cells (ESC) derived from the inner cell mass have methylation at many regulatory regions except for the pluripotent promoters [46, 47], and have the ability to self renew and differentiate into all cell types. ESC deficient in either maintenance (Dnmt1) or in de novo methyltransferases (Dnmt3a/3b), lose their pluripotency and proper differentiation potential, but maintain their ability of self renewal [48]. A recent genome-wide study reported that cytosine methylation on non CG dinucleotides is prevalent in the ESC but not in fibroblasts expanding the complexity of this epigenetic mark during development [3, 49].
In the first stage of differentiation of pluripotent ESC, promoters of the several key plurioptent genes become methylated and undergo targeted repression, and remain methylated in somatic cells [50–52]. Differentiation of ESC into somatic cells does not change the global level of DNA methylation, but there is a redistribution of the methylation pattern throughout the genome during somatic cell formation [53]. DNA methylation controls a subset of critical tissue specific genes, and loss of methylation usually results in inhibition of proper differentiation [16, 54, 55].
Another important advancement is the discovery of 5-Hydroxymethylcytosine (5hmC). 5hmC is formed by modifying the cytosine base by addition of a methyl group and a hydroxy group. It was first found in bacteriophages in 1952 [56]. It has been recently found to be abundant in human and mouse brain [57], and in ESC [58]. The exact function of this modification is not fully understood, but it has been proposed to be involved in regulating gene expression and DNA demethylation. The idea that 5hmC is an intermediate product in promoting active DNA demethylation in mammalian cells has been supported by a recent finding that methylcytosine hydroxylase, TET1, converts 5-methyl cytosine to 5hmC and promotes DNA demethylation in mammalian cells through a process that requires the base excision repair pathway [59]. Another recent genome-wide study found that 5hmC is enriched at both gene bodies of actively transcribed genes and extended promoter regions of Polycomb-repressed developmental regulators, suggested a dual role of 5hmC in regulating gene expression [60].
3.2. Transcription
The 146 bp of DNA wrapped around histone octamers forms a nucleosome, and ~ 20 million nucleosomes are needed in each cell to package both copies of the human genome. Access to gene promoters is dependent upon the dynamic status of promoter-associated nucleosomes, which is controlled by a variety of factors, including covalent modifications of DNA, post-translational modifications of the histones and proteins of the transcription machinery. The covalent modification of DNA (DNA methylation, specifically at CG) can alter the recognition of the double helix by the transcriptional machinery. Two complementary phenomena are being proposed. First, the methylated DNA can prevent some transcription factors from binding their target. Second, DNA methylation can create binding sites for proteins that specifically recognize methylated DNA. DNMT1 knockout mice showed that expression of certain lineage specific genes is unaltered and methylation status of the 5' regions of a set of tissue-specific genes did not correlate with expression in tissues of fetal and newborn mice [61]. A global study of three human chromosomes (chromosomes 6, 20 and 22) found that methylation of one-third of the differentially methylated 5' UTRs are inversely correlated with transcriptional activation [13]. Another recent study showed that in human somatic cells low CG containing promoters are generally hypermethylated, and a set of genes with these hypermethylated promoters are transcriptionally active [11]. These genome-wide studies suggest that promoter CG density is the central player that determines how DNA methylation affects gene expression. Promoters with a high CG density are generally unmethylated in all tissues regardless of the expression of their linked gene [62]. When these promoters become methylated, they are strongly repressed [63].
Low CG promoters are typically associated with tissue specific genes, which are methylated and not active, reinforcing the positive correlation between methylation and the lack of promoter activity [11]. Recently, we observed that when tissue specific genes become active, their promoters are still methylated [16]. We identified that the CG methylation in the promoters of tissue specific genes create transcription factor binding sites (TFBS) for C/EBPα that are needed for tissue specific gene activation [16]. When primary newborn mouse keratinocytes are differentiated with calcium treatment for two days, many genes with methylated proximal promoters become activated [16]. Similar observation was observed when primary dermal fibroblasts were differentiated into adipocytes by adipogenic media containing IBMX, dexamethasone, insulin and rosiglitazone. The role of C/EBPα in adipocyte differentiation is well established in the literature, and we found that CG methylation marks in the promoters of tissue specific genes create TFBS for C/EBPα needed for tissue specific gene activation [16]. So, the general conclusion from this study is that the differentiation system where C/EBPα is involved might show similar effects in regulating tissue specific gene expression with low CG containing promoters. However, in both, keratinocyte and adipocyte differentiation studies, promoter methylation for differentiation specific low CG containing genes did not change with differentiation, which raises a question of finding the molecular switch that triggers the tissue specific gene expression and is discussed in the next section. Methylation of the E2BS1 and Zta response elements (ZRE) TFBS results in strong activation instead of inhibition of the promoter, phenomenon also observed in HPV and EBV viral genomes [64, 65], indicating a possibility of being a context depend general phenomena of this activation mark.
The importance of CG methylation for promoter function was examined by global demethylation using either 5-azacytidine or by transient DNMT1 depletion. Demethylation preferentially inhibited C/EBPα binding to methylated promoters and their subsequent activation during differentiation in both primary keratinocyte and adipocyte differentiation system suggesting a link between methylation and differentiation. These tissue specific low CG promoters are methylated irrespective of their expression [11, 16]. This conundrum can be explained by several mechanisms. For examples, low CG promoters sometimes are associated with some distal enhancer that contains high CG regions. Differential methylation of these enhancer regions might be the regulator for active transcription of these genes (Figure 4). Other possibility is that bivalent histone marks might present to maintain their inactive state, and alteration of histone modifications causes the activation of these genes. Posttranslational modification of the bound transcription factors might also be responsible for their activation [16]. However, it is likely that these mechanisms act in concert for proper activation of tissue specific promoters. Overall, it can be concluded that, although DNA methylation is typically considered to be involved in silencing mechanism, methylation of CG poor promoters is linked to activation of some tissue specific gene expression.
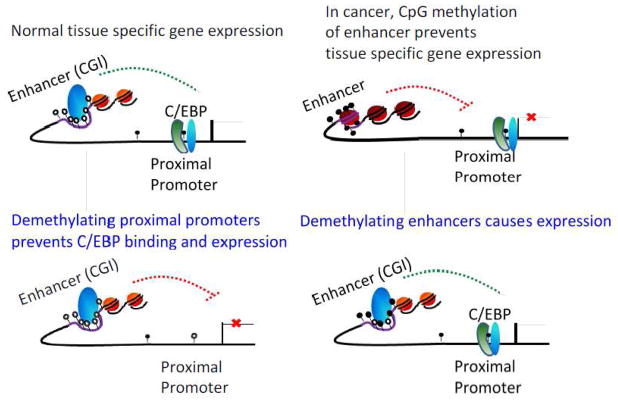
Figure 4.
5-azacytidine can prevent or cause tissue specific gene expression: A proposed model of methylation mediated tissue specific gene expression in normal and cancer cells. A) In normal cells low CG promoters are methylated and bound by C/EB;, while the high CG enhancers are unmethylated and activating the transcription. Demethylation by 5-azacytidine causes demethylation at the proximal promoters and inhibits binding of C/EBP and thus represses gene expression. B) In cancer cells, proximal promoters are methylated and bound by C/EB; while high CG enhancers become methylated, which causes formation of heterochromatin and thus tissue specific gene silencing. After 5- azacytidine treatment, demethylation begins stochastically at the high CG enhancers and allows transcription factor binding to activate differentiation specific genes.
4. DNA Methylation and Cancer
Proper epigenetic states are essential for the regulation of normal cells, and it is expected that deregulation could result in aberrant biological processes and lead to human diseases including cancer [66–71]. Since the 1980s, when the reduction of methylation was first observed in cancer tissues compared to the normal tissues [72], the role of DNA methylation in cancer has been studied extensively [71]. In cancers, global hypomethylation occurs at gene bodies, transposable elements and repetitive sequences, and hypermethylation is observed at promoters, which leads to aberrant transcription initiation and genome instability. DNA hypomethylation also contribute to the development of cancer by activating the transposable elements, generating the chromosomal instability and loss of imprinting. Demethylation of transposable elements could help them to transcribe or translocate to other genomic regions, to inactivate tumor suppressor genes and to facilitate genomic rearrangements. Hypomethylation can also lead to loss of imprinting. An example of a gene activated by loss of imprinting is the insulin-like growth factor gene (IGF2). Loss of imprinting of IGF2 is reported to be a risk factor for colorectal cancer [73, 74] and also contribute to the development of Wilms’ tumor [75, 76].
Most cancer cells predominantly produce energy by a high rate of glycolysis followed by lactic acid fermentation in the cytosol even in the presence of oxygen, rather than by a comparatively low rate of glycolysis followed by mitochondrial pyruvate oxidation as occurs in normal cells [77]. In a recent publication by Chi et al. it was proposed that environmental toxins affect the global epigenetic pattern by interfering with the metabolism by activating the TET proteins and thus by regulating the 5-hmC and 5-mC levels [78]. They speculated that the altered methylation pattern might be a consequence of oxidative stress mediated alteration in metabolism due to activation of TET and other chromatin modifying proteins [78]. Another recent study also proposed an epigenetic mechanism of the Warburg effect [79]. They found the NF-kappaB pathway mediates down-regulation of fructose-1,6-bisphosphatase-1 (FBP1), which functions to antagonize glycolysis, while NF-kappaB inhibition restored FBP1 expression, which is partially through demethylation of FBP1 promoter. NF-kappaB could interact with co-repressors like HDAC1 and HDAC2 to negatively regulate the gene expression [80, 81], and intern HDACs could interact with the DNMT1 and might cause the formation of localized promoter hypermethylation to promote stable gene silencing [82–85].
Hypermethylation of the CGI associated promoters and transcriptional repression or loss of corresponding gene function is the most studied epigenetic alteration in cancer [70, 71]. Promoter hymermethylation associated silencing of many tumor suppressor genes, such as, retinoblastoma (Rb) in retinoblastoma cancer, cyclin-dependent kinase inhibitor 2A (CDKN2A) in several tumors, Von Hippel–Lindau (VHL) in renal cancer, breast cancer–associated-1 (BRCA1) in breast and ovarian cancer, adenomatous polyposis coli (APC) in several tumors, GATA4 and GATA5 in colorectal and gastric cancer [86], and glutathione s-transferase P1 (GSTP1) in prostate cancer [87] are observed [67, 69–71]. In recent studies, several somatic mutations are also reported in genes involved in epigenetic pathways. For example, somatic mutations are observed in DNMT3A and TET2 in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) [88–92], mutation in DNMT1 in colorectal cancer [93], and mutations in isocitrate dehydrogenase genes, IDH1 and IDH2, in gliomas and AMLs [94–96], which strongly supports the role of altered DNA methylation in cancers.
Global DNA hypomethylation and CGI associated hypermethylation are also observed in early stage tumors. Thus, determination of aberrant DNA methylation pattern could be used as an epigenetic biomarker for early detection of different types and subtypes of cancer. It can also be used for the determination of tumor prognosis and personalized therapy. Another important aspect of epigenetic changes is that as these are reversible in nature, so inhibiting the enzymes of epigenetic machinery can be used to develop epigenetic drugs. For example, the US FDA has approved DNA methyltransferase (DNMT) inhibitors, azacytidine and decitabine (5-aza- and 5-aza-2'-deoxycytidine), for therapy of patients with myelodysplastic syndrome, which can lead to development of acute leukemia; and several similar drugs are undergoing clinical trials [70, 71]. While high doses of these drugs can inhibit DNA synthesis and eventually lead to cell death by cytotoxicity, administration of low doses of these drugs over a prolonged period resulted in a therapeutic efficacy [97–100].
How does one reconcile the observation that DNA methyltransferase inhibitor, 5-azacytidine inhibits differentiation of both epithelial and mesenchymal cell types whereas it induces differentiation of cancer cells? We suggest that differentiation involves activation of tissue specific genes that are regulated by unmethylated CG rich enhancers which activate methylated CG poor promoters (Figure 4). Some methylated sequences in tissue specific gene promoters are recognized by transcription factors that activate tissue specific gene expression [16]. CG methylations of CRE and C/EBP like sequences (NNNCGTCA and NNNCGCAA) create a binding site that is recognized by C/EBPα, a transcription factor that regulates tissue specific gene expression in many tissues [16]. In normal cells, 5-azacytidine treatment demethylates the promoters and prevents C/EBPα binding, resulting in gene inactivation. In the cancer cells, differentiation is inhibited because of hypermethylation of the CG rich enhancer. One potential effect of hypermethylation is preferential nucleosome binding that result in the repression of gene expression. In certain cancers, both the CG rich enhancer and CG poor promoter are methylated. With 5-azacytidine treatments, the enhancer stochastically begins demethylation before the promoter because of large number of methylated CGs. With demethylation, nucleosome binding is lost at the enhancer and TF binding is increased, which drives enhancer activity and thus differentiation via tissue specific gene expression. Thus, CG methylation drives a molecular switch between nucleosomes preferentially binding to the methylated enhancer, and transcription factors preferentially binding to the unmethylated enhancer. These transcription factors could be the same molecules that drive housekeeping function in the unmethylated promoters of the mammalian genome.
5. Concluding remarks
DNA methylation plays an essential role in normal mammalian development by maintaining the accurate epigenetic environment. DNA methylation is involved in many processes including gene regulation, imprinting, dosage compensation, cellular identity, differentiation and genomic integrity. CG methylation was initially thought to be a general repressive epigenetic mark in vertebrate genomes. But recent studies revealed a new role of CG methylation, as an activation mark that creates transcription factor binding sites, suggesting a more complex role for CG methylation. With the advent of high-throughput technologies, additional functions of epigenetic regulations are being unraveled, although many more are yet to be discovered. With the discoveries of new modification (e.g., 5hmC) and diverse functional implications, it is becoming more complex and challenging to fully uncover the mechanisms of epigenetic regulations leading to changes in DNA methylation. Understanding DNA methylation landscape in normal cells will help to identify the genomic context and functional implications of methylation, and moreover to determine the biomarker of epigenetically mediated human diseases.
Highlights.
Present understanding of DNA methylation in normal cells
A new function of CpG methylation: positive effect on tissue specific gene expression.
Aberrant CpG methylation and cancer.
Acknowledgments
We thank our lab members for their encouragement and support. This work is supported by the intramural project of National Cancer Institute, NIH, USA. We thank guest editor and two anonymous reviewers for their thoughtful comments and suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird AP. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Illingworth RS, Bird AP. CpG islands--'a rough guide'. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 7.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 12.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 19.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 20.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 21.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, McCombie WR, Wigler M, Hannon GJ, Hicks JB. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeltsch A. Molecular biology. Phylogeny of methylomes. Science. 2010;328:837–838. doi: 10.1126/science.1190738. [DOI] [PubMed] [Google Scholar]
- 24.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011 doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 26.Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 29.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davey C, Pennings S, Allan J. CpG methylation remodels chromatin structure in vitro. J Mol Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 35.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 40.Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 42.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 43.Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- 44.Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 48.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 49.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 52.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 55.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170:1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- 57.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 63.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 64.Vinokurova S, von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS One. 2011;6:e24451. doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flower K, Thomas D, Heather J, Ramasubramanyan S, Jones S, Sinclair AJ. Epigenetic Control of Viral Life-Cycle by a DNA-Methylation Dependent Transcription Factor. PLoS One. 2011;6:e25922. doi: 10.1371/journal.pone.0025922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 67.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 69.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 70.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 71.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 73.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 74.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 75.Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 76.Feinberg AP. Imprinting of a genomic domain of 11p15 and loss of imprinting in cancer: an introduction. Cancer Res. 1999;59:1743s–1746s. [PubMed] [Google Scholar]
- 77.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 78.Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics. 2011;6:853–856. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- 79.Liu X, Wang X, Zhang J, Lam EK, Shin VY, Cheng AS, Yu J, Chan FK, Sung JJ, Jin HC. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442–450. doi: 10.1038/onc.2009.332. [DOI] [PubMed] [Google Scholar]
- 80.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhat KP, Pelloski CE, Zhang Y, Kim SH, deLaCruz C, Rehli M, Aldape KD. Selective repression of YKL-40 by NF-kappaB in glioma cell lines involves recruitment of histone deacetylase-1 and -2. FEBS Lett. 2008;582:3193–3200. doi: 10.1016/j.febslet.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 82.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 83.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 84.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Jin H. The epigenetic basis of the Warburg effect. Epigenetics. 2010;5:566–568. doi: 10.4161/epi.5.7.12662. [DOI] [PubMed] [Google Scholar]
- 86.Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, Baylin SB. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, Fulton R, Schmidt H, Kalicki-Veizer J, O'Laughlin M, Kandoth C, Baty J, Westervelt P, DiPersio JF, Mardis ER, Wilson RK, Ley TJ, Graubert TA. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 90.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, Fulton R, Schmidt H, Kalicki-Veizer J, O'Laughlin M, Kandoth C, Baty J, Westervelt P, DiPersio JF, Mardis ER, Wilson RK, Ley TJ, Graubert TA. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 92.Langemeijer SM, Jansen JH, Hooijer J, van Hoogen P, Stevens-Linders E, Massop M, Waanders E, van Reijmersdal SV, Stevens-Kroef MJ, Zwaan CM, van den Heuvel-Eibrink MM, Sonneveld E, Hoogerbrugge PM, van Kessel AG, Kuiper RP. TET2 mutations in childhood leukemia. Leukemia. 2011;25:189–192. doi: 10.1038/leu.2010.243. [DOI] [PubMed] [Google Scholar]
- 93.Kanai Y, Ushijima S, Nakanishi Y, Sakamoto M, Hirohashi S. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003;192:75–82. doi: 10.1016/s0304-3835(02)00689-4. [DOI] [PubMed] [Google Scholar]
- 94.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 97.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, Schoch R, Gattermann N, Sanz G, List A, Gore SD, Seymour JF, Bennett JM, Byrd J, Backstrom J, Zimmerman L, McKenzie D, Beach C, Silverman LR. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vigil CE, Martin-Santos T, Garcia-Manero G. Safety and efficacy of azacitidine in myelodysplastic syndromes. Drug Des Devel Ther. 2010;4:221–229. doi: 10.2147/dddt.s3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia JS, Jain N, Godley LA. An update on the safety and efficacy of decitabine in the treatment of myelodysplastic syndromes. Onco Targets Ther. 2010;3:1–13. doi: 10.2147/ott.s7222. [DOI] [PMC free article] [PubMed] [Google Scholar]