Abstract
PHLPP is a family of Ser/Thr protein phosphatases that serve as tumor suppressors by negatively regulating Akt. Our recent studies have demonstrated that the ubiquitin proteasome pathway plays an important role in the downregulation of PHLPP in colorectal cancer. In this study, we show that the deubiquitinase USP46 stabilizes the expression of both PHLPP isoforms by reducing the rate of PHLPP degradation. USP46 binds to PHLPP and directly removes the polyubiquitin chains from PHLPP in vitro and in cells. Increased USP46 expression correlates with decreased ubiquitination and upregulation of PHLPP proteins in colon cancer cells, whereas knockdown of USP46 has the opposite effect. Functionally, USP46-mediated stabilization of PHLPP and the subsequent inhibition of Akt result in a decrease in cell proliferation and tumorigenesis of colon cancer cells in vivo. Moreover, reduced USP46 protein level is found associated with poor PHLPP expression in colorectal cancer patient specimens. Taken together, these results indentify a tumor suppressor role of USP46 in promoting PHLPP expression and inhibiting Akt signaling in colon cancer.
Keywords: PHLPP, USP46, Akt, deubiquitination, tumor suppressor
INTRODUCTION
PHLPP (PH domain Leucine-rich-repeats Protein Phosphatase) belongs to a novel family of Ser/Thr protein phosphatases that play an important role in maintaining the balance in cell signaling (1). To date, PHLPP has been shown to negatively regulate signaling pathways activated by several critical protein kinases including Akt, protein kinase C (PKC), MAPK, and Mst1 in cells (2-5). Among them, Akt, conventional PKC isozymes, and Mst1 are identified as direct substrates of PHLPP. The functional outcome of PHLPP-mediated dephosphorylation of these substrates usually involves an increase in apoptosis and a decrease in proliferation in various cancer cell lines, thus indicating a tumor suppressor function of PHLPP. However, the contribution of each substrate to PHLPP-dependent inhibition of cell growth may be cell- and cancer-type specific (2, 5-7). Two isoforms of PHLPP, namely PHLPP1 and PHLPP2, are found in this phosphatase family (1). Although the two isoforms of PHLPP share their ability to dephosphorylate Akt, PKC, and Mst1 (3, 5, 6), different PHLPP isoforms have been shown to modulate a distinct subset of signaling events downstream of Akt, suggesting non-overlapping functions of the two isoforms (6).
Recently, an increasing number of studies have further confirmed the tumor suppressor role of PHLPP in vivo. For example, downregulation of both PHLPP isoforms occurrs at high frequency in human colorectal cancer specimens, and re-expression of PHLPP in colon cancer cells inhibits the growth of xenografted tumors (7). Moreover, knockout of the PHLPP1 gene on the partial PTEN-loss background promotes the development of prostate tumors in mouse models, and co-deletion of PHLPP1 and PTEN genes is associated with metastatic prostate cancer in patient samples (8). Given the importance of maintaining PHLPP levels in inhibiting tumor progression, it is of particular interest to investigate the mechanism controlling PHLPP protein expression.
Ubiquitination is a post-transcriptional modification that is commonly recognized by the proteasome pathway as a signal to degrade eukaryotic proteins (9). We have previously identified β-TrCP as the E3 ubiquitin ligase for PHLPP1, and this β-TrCP-mediated degradation requires GSK-3β-dependent phoshphorylation of PHLPP1 and is negatively regulated by Akt (10). Interestingly, increased Akt activity in glioblastoma cell lines is unable to stabilize PHLPP1 expression due to mislocalization of β-TrCP, thus resulting in unbalanced activation of Akt (11). The ubiquitin-dependent protein degradation is reversed by a family of proteases known as deubiquitinases (DUBs). By opposing the function of ubiquitin E3 ligases, DUBs play an important role in regulating protein stability and the homeostasis of ubiquitin (12). The functional importance of DUBs in tumorigenesis has been revealed by recent studies showing the expression of several oncogenes and tumor suppressors are controlled by specific DUBs (12). Specifically, deubiquitinase USP9X has been shown to stabilize the pro-survival protein MCL1, and increased USP9X expression correlated with increased MCL1 protein in human lymphomas (13). Similarly, increased expression of USP28 has been reported in colon and breast cancers that resulted in stabilization of MYC proteins (14).
In this study, we report the identification of USP46 as a deubiquitinase of both PHLPP isoforms. We show that the level of USP46 dictates the expression of PHLPP in cells and the tumor suppressor function of PHLPP is significantly potentiated in the presence of USP46. Furthermore, downregulation of both PHLPP1 and USP46 expression is highly associated in human colorectal cancer specimens.
RESULTS
USP46 interacts with PHLPP in cells
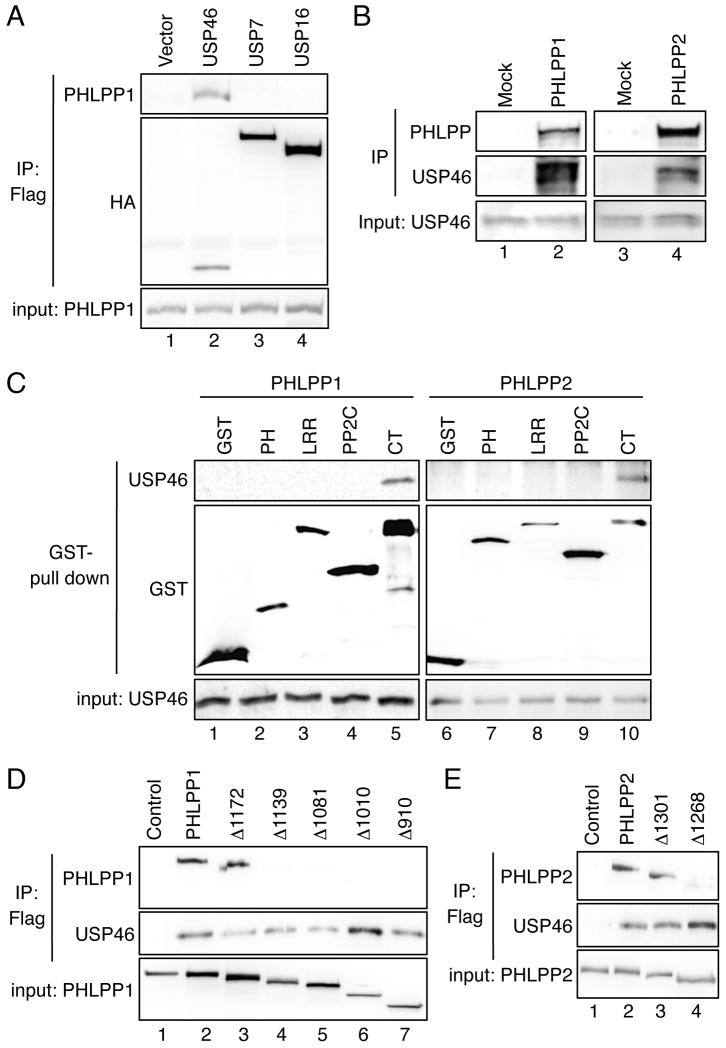
In a recent study to define the interaction landscape of human DUBs using the mass spectrometry analysis, Sowa et al. reported that both PHLPP isoforms formed complexes with three closely related DUBs including USP1, USP12, and USP46 (15). To determine the functional importance of these interactions, we first confirmed that both PHLPP isoform co-immunoprecipitated with all three USPs when co-expressed in cells (data not shown). Due to the availability of reagents for detecting endogenous USP proteins, we focused on investigating the regulation of PHLPP by USP46 in this study. Co-immunoprecipitation experiments showed that only USP46 specifically interacted with PHLPP1 in transfected 293T cells whereas other DUBs including USP7 and USP16 failed to binding PHLPP1, thus, demonstrating the specificity of USP46-mediated interaction (Fig. 1A). Similar results were obtained with PHLPP2 (data not shown). Furthermore, the interaction between both endogenous PHLPP isoforms and USP46 was readily detected in DLD1 cells (Figure 1B).
Figure 1.
PHLPP specifically interacts with USP46 in cells. (A) 293T cells were co-transfected with HA-PHLPP1 together with vector, USP46, USP7, or USP16 (lanes 1-4). Note that all USP proteins were tagged with both Flag and HA tags (15). The cell lysates were immunoprecipitated with the anti-Flag agarose and analyzed by immunoblotting. The presence of PHLPP1 in immunoprecipitates and cell lysates (5% input) was detected using the PHLPP antibody, while all USP proteins were detected by the HA antibody. (B) Equal amounts of DLD1 cell lysates were incubated with the protein A/G beads alone (Mock) or the beads plus the PHLPP1 or PHLPP2 antibody. The presence of PHLPP and USP46 in the immunoprecipitates and input were detected with the PHLPP and USP46 antibodies, respectively. (C) The GST vector or GST-tagged individual domains of PHLPP1 or PHLPP2 were co-expressed with USP46 in 293T cells. The cell lysates were pulled down with glutathione-Sepharose and analyzed using immunoblotting. (D) 293T cells were co-transfected with USP46 and WT PHLPP1 or one of the truncation mutants of PHLPP1 including Δ1172, Δ1139, Δ1081, Δ1010, and Δ910. The cell lysates were immunoprecipitated with the anti-Flag agarose and analyzed by immunoblotting. The WT and mutant PHLPP1 proteins were detected using the P1-PP2C antibody. (E) 293T cells were co-transfected with Flag-USP46 and WT PHLPP2 or one of the truncation mutants of PHLPP2 including Δ1301 and Δ1268. The WT and mutant PHLPP2 proteins were detected using the HA antibody.
We next determined the region in each PHLPP isoform responsible for binding USP46. The different domains of both PHLPPs were expressed as GST-tagged fusion proteins and GST pull-down experiments were performed using overexpressed USP46. We found that USP46 interacted with the C-terminus of PHLPP1 and PHLPP2 (Fig. 1C). To further narrow down the region in the C-terminus of PHLPP that is required for the interaction, sequential truncation mutants in the C-terminus of PHLPP1 were generated and used in the co-immunoprecipitation experiments with USP46. The results showed that PHLPP1 lost its ability to bind USP46 upon deletion of a small region residing between amino acid residues 1139 to 1172 in the C-terminus of PHLPP1 (Fig. 1D, Δ1172 vs. Δ1139). We defined this region as the minimal binding domain for USP46. Since the sequence in this the minimal binding domain is highly conserved between PHLPP1 and PHLPP2, two truncation mutants were constructed in the C-terminus of PHLPP2 representing the deletion of this domain. Indeed, binding of USP46 was abolished when this minimal binding domain was removed from PHLPP2 as well (Fig. 1E). Collectively, our data identified a minimal binding domain conserved in both PHLPP isoforms that is required for interacting with USP46.
USP46 controls the expression levels of PHLPP
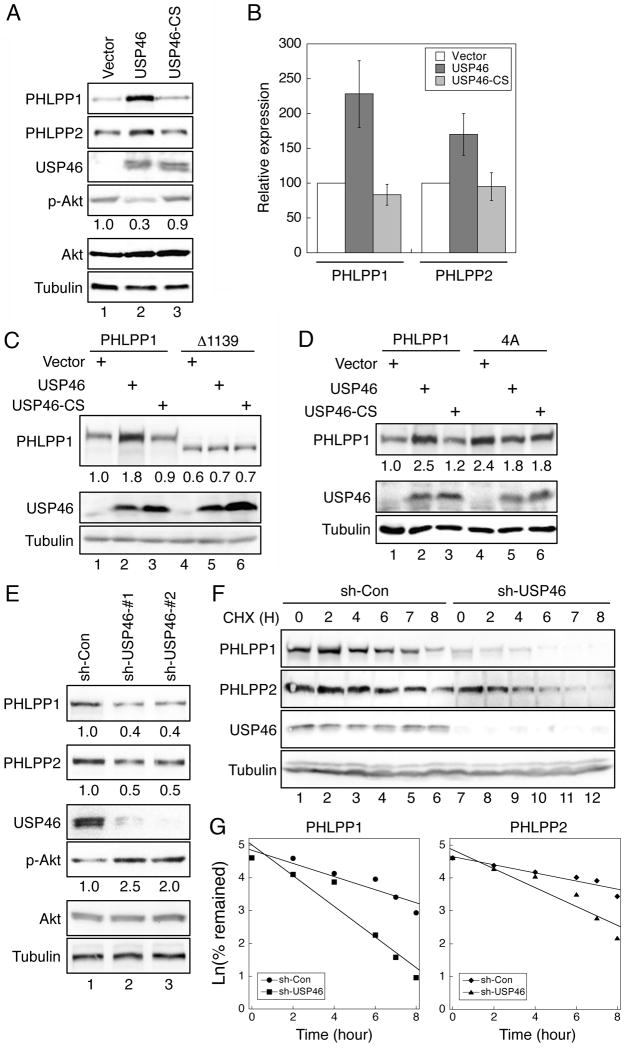
Since the main function of DUBs is to stabilize their target proteins, we next examined whether the expression of PHLPP is regulated by USP46. Overexpression of WT USP46 significantly increased expression of both PHLPP isoforms in HCT116 cells. As a result, this increased expression of PHLPP reduced the phosphorylation of Akt by 70% (Fig. 2A and 2B). To serve as a negative control, a catalytically inactive form of USP46 was generated by changing a conserved Cys residue in the catalytic center to Ser (USP46-C44S). This inactive form of USP46 was unable to regulate the expression of PHLPP or the phosphorylation of Akt, thus confirming the requirement for the deubiquitinases activity of USP46 (Fig. 2A and 2B). To determine if the interaction of PHLPP and USP46 is required for USP46-mediated increase of PHLPP expression, the full-length PHLPP1 or the USP46 binding-deficient Δ1139 mutant was co-expressed with USP46. While the expression level of the full-length PHLPP1 was readily increased upon co-expression of WT USP46, the expression of Δ1139 mutant remained constant regardless of USP46 levels (Fig. 2C). As a negative control, the inactive mutant USP46-C44S did not change the expression of either WT or mutant PHLPP1 (Fig. 2C).
Figure 2.
The expression of PHLPP is regulated by USP46. (A) HCT116 cells transfected with vector, USP46, or USP46/CS were lysed and analyzed using immunoblotting. The phosphorylation of Akt was determined using the phospho-S473 antibody. (B) Graphic representation of data shown in panel (A). The relative expression levels were obtained by normalizing the amount of PHLPP to tubulin, and the level of wild-type PHLPP in the vector-transfected cells was set to 1. Data represent the mean ± SEM (n=3). (C) HCT116 cells were transfected with WT (lanes 1-3) or Δ1139 mutant PHLPP1 (lanes 4-6) together with vector, USP46, or USP46/CS. The relative expression levels of PHLPP1 were quantified by normalizing the amount of PHLPP1 to tubulin and indicated below the PHLPP1 panel. (D) HCT116 cells were transfected with WT (lanes 1-3) and 4A mutant of PHLPP1 (lanes 4-6) together with vector, USP46, or USP46/CS. The expression of PHLPP1 and USP46 was analyzed in the cell lysates using the PHLPP1 and USP46 antibodies, respectively. The relative expression levels of PHLPP1 were quantified by normalizing the amount of PHLPP1 to tubulin and indicated below the PHLPP1 panel. (E) Cell lysates prepared from stable control (sh-Con) and two different USP46 knockdown (sh-USP46-#1 and -#2) DLD1 cell lines were analyzed using immunoblotting. The expression levels of PHLPP1 and PHLPP2 and the amount of Akt phosphorylation relative to the control samples were indicated below the corresponding panels. (F) Stable sh-Con (lanes 1-6) and sh-USP46 (lanes 7-12) DLD1 cells were treated with CHX, and the amount of PHLPP remaining at each time point was analyzed and quantified. (G) Graphs depict the turnover rates of PHLPP expressed as the natural log of the percentage of PHLPP remaining with respect to time. The data were fitted to a single exponential term by linear least-squares regression analysis. The half-life of PHLPP1 (t1/2) was calculated to be 4.6 h (r = 0.94) and 2.3 h (r = 0.96) whereas the half-life of PHLPP2 was 6.1 h (r = 0.95) and 3.3 h (r = 0.95) in sh-Con and sh-USP46 cells, respectively.
We have previously shown that a phosphorylation deficient mutant of PHLPP1, PHLPP1-4A, expresses at a much higher level compared to WT PHLPP1 in cells due to its resistance to ubiquitination-induced degradation (10). Consistent with our previous findings, the expression level of 4A mutant was higher compared to WT PHLPP1. However, USP46 was unable to further increase 4A expression suggesting that USP46 regulates PHLPP expression in an ubiquitination dependent manner (Fig. 2D).
To further confirm the functional effect of endogenous USP46 on PHLPP expression, we analyzed the level of PHLPP in control and USP46 knockdown DLD1 cells. Knockdown of USP46 using two different shRNA targeting constructs resulted in a marked decrease of PHLPP1 and PHLPP2 expression and a subsequent increase in Akt phosphorylation (Fig. 2E). Note that the stable USP46 knockdown cell line containing targeting sequence #1 was used in all subsequent experiments in our study. Similar results were obtained in the other knockdown cell line containing targeting sequence #2. In addition, knockdown of USP46 increased the insulin-induced phosphorylation of Akt at S473 site (Fig. S1). Loss of USP46-induced upregulation of Akt dephosphorylation is similar as that observed by knocking down of PHLPP directly (6, 7). Furthermore, the time course of PHLPP degradation in USP46 knockdown cells was significantly accelerated. The half-life of PHLPP1 and PHLPP2 was decreased from 4.6 h to 2.3 h and 6.1 h to 3.3 h, respectively (Fig. 2F and 2G). Taken together, we showed that the expression levels of PHLPP and USP46 are highly correlated in cells. Mutations in PHLPP1 that disrupt the interaction with USP46 or the ubiquitination of PHLPP1 become resistant to USP46-mediated stabilization.
USP46 directly regulates the ubiquitination of PHLPP1
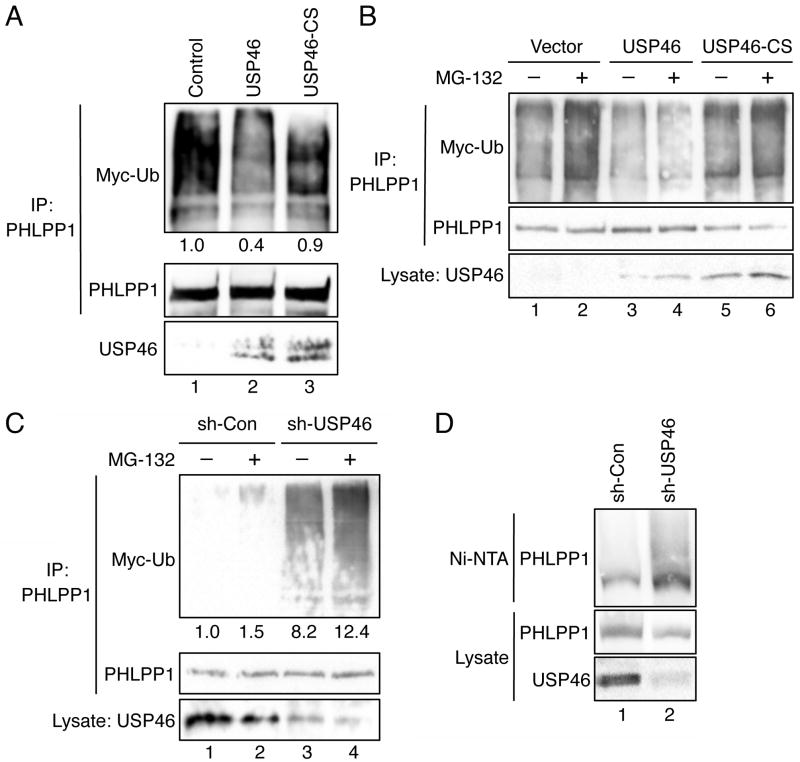
We have previously demonstrated that the ubiquitination of PHLPP1 is mediated by the E3 ligase β-TrCP (10). Here, we investigated whether USP46 directly regulates the level of PHLPP1 ubiquitination in vitro and in cells. The WT and mutant USP46 immunoprecipitated from transfected cells were used to treat ubiquitinated PHLPP1 in vitro. The level of PHLPP1 ubiquitination was decreased by incubating with WT USP46 but not with USP46-C44S mutant proteins confirming the deubiquitinase activity of USP46 against PHLPP1 in vitro (Fig. 3A). Furthermore, the WT or mutant USP46 was co-expressed with PHLPP1 in cells and the level of PHLPP1 ubiquitination was analyzed in the presence or absence of MG-132. Overexpression of WT USP46 significantly decreased the amount of ubiquitination of PHLPP1 both basally and after MG-132 treatment, while the catalytically inactive mutant USP46 had no effect (Figure 3B). In contrast, the ubiquination of PHLPP1 in USP46 knockdown cells was markedly increased (Fig. 3C). To further confirm the effect of USP46 on PHLPP1 ubiquitination, we analyzed the ubiquitination status of PHLPP1 under denaturing conditions in control and USP46 knockdown cells. Consistent with results shown in Fig. 3C, the amount of ubiquitinated PHLPP1 was increased in USP46 knockdown cells (Fig. 3D). Collectively, these results suggest that USP46 plays a positive role in stabilizing PHLPP1 expression by inhibiting the ubiquitination and protein degradation.
Figure 3.
Ubiquitination of PHLPP1 is negatively controlled by USP46. (A) In vitro deubiquitination experiments were carried out as described in the Materials and Methods. The level of ubiquitination was detected using the Myc antibody. The amount of PHLPP1 (the substrate) and USP46 (the deubiquitinase) in the reaction were detected using the HA and USP46 antibodies, respectively. The relative ubiquitination level detected on PHLPP1 was quantified by normalizing ECL signals of Myc-Ub to those of HA and indicated below the Myc-Ub panel. (B) 293T cells were co-transfected with HA-P1 and Myc-ubiquitin (Myc-Ub) together with vector (lanes 1 and 2), USP46 (lanes 3 and 4), or USP46/CS (lanes 5 and 6). The cells were treated with DMSO or MG-132 for 30 minutes, and the cell lysates were immunoprecipitated with the PHLPP1 antibody. The ubiquitination of PHLPP1 was detected using the Myc antibody. The blot was stripped and reprobed with the HA antibody to visualize total PHLPP1 in the immunoprecipitates. The expression of wild-type and mutant USP46 in cell lysates were detected using the USP46 antibody. (C) Stable sh-Con and sh-USP46 HCT-P1 cells transfected with Myc-Ub were treated with DMSO or MG-132 for 30 minutes, and the cell lysates were immunoprecipitated with the PHLPP1 antibody. The ubiquitination of PHLPP1 was detected using the Myc antibody. The amount of total PHLPP1 in the immunoprecipitates and USP46 in cell lysates were detected using the HA and USP46 antibodies, respectively. The relative ubiquitination level detected on PHLPP1 was quantified by normalizing ECL signals of Myc-Ub to those of HA and indicated below the Myc-Ub panel. (D) Detection of PHLPP1 ubiquitination under denatured conditions. Stable sh-Con and sh-USP46 HCT-P1 cells transfected with His-Ub were lyzed in denature buffer and incubated with Ni-NTA. The ubiquitinated PHLPP1 associated with beads was detected using the PHLPP1 antibody. The expression of PHLPP1 and USP46 in cell lysates was detected using the PHLPP1 and USP46 antibodies, respectively.
USP46 enhances PHLPP-mediated negative regulation of Akt in cells
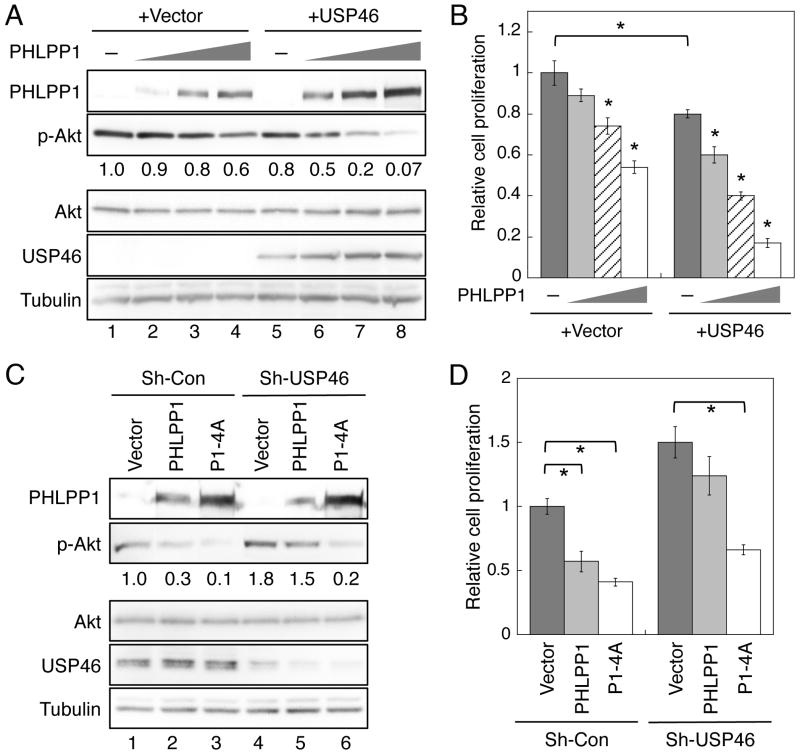
Our previous studies have shown that overexpression of PHLPP inhibits cell proliferation via dephosphorylation of Akt in colon cancer cells (7, 10). To determine if PHLPP and USP46 function together to negatively regulate cell proliferation, increasing amount of PHLPP1 plasmids was transfected into HCT116 cells alone or in combination with USP46. When PHLPP1 was expressed alone, dose-dependent dephosphorylation of Akt was observed as the expression of PHLPP1 increased. However, the degree of Akt dephosphorylation was relatively small (Fig. 4A, lanes 1-4). Similarly, overexpression of USP46 by itself resulted in a small degrease in Akt phosphorylation, likely due to the fact that the endogenous expression of PHLPP1 is very low in this colon cancer cell line (7). In contrast, co-expression of USP46 with PHLPP1 markedly potentiated PHLPP1-mediated dephosphorylation of Akt as the expression of PHLPP1 was stablized by USP46 (Fig. 4A, lanes 5-6). In the experiments to determine the effect of PHLPP1 and USP46 on the proliferation of HCT116 cells, we found that the level of Akt phosphorylation tightly correlated with the rate of cell proliferation in the transfected cells (Fig. 4B). Increased expression of PHLPP1 by itself resulted in a dose-dependent inhition of cell growth, however, the anti-proliferative effect of PHLPP1 was significantly enhanced when USP46 was co-expressed (Fig. 4B). When a low concentration of PHLPP1 plasmid was transfected into cells alone, the level of PHLPP1 was not sufficient to significantly reduce the rate of cell proliferation, whereas co-expression of USP46 allowed the expression of PHLPP1 to surpass the threshold needed to induce the significant effect (Fig. 4B). As a negative control, the inactive form of USP46 had no effect alone or in combination with PHLPP1 on Akt dephosphorylation and cell proliferation (Fig. S2). Furthermore, knockdown of USP46 attenuated the ability of PHLPP1 to dephosphorylate Akt and to inhibit cell proliferation (Fig. 4C and 4D). Specifically, overexpression of WT or 4A mutant of PHLPP1 decreased the phosphorylation of Akt by 70% and 90% in control DLD1 cells, respectively. Knockdown of USP46 resulted in an increase of Akt phosphorylation basally, however, WT PHLPP1 became much less effective to antagonize Akt whereas the degradation-resistent 4A mutant maintained its potency at dephosphorylating Akt (Fig. 4C). The rate of cell proliferation followed the pattern of Akt phosphorylation that WT PHLPP1 was unable to significantly inhibit cell growth in USP46 knockdown cells (Fig. 4D).
Figure 4.
USP46 and PHLPP1 function cooperatively to inhibit Akt phosphorylation and cell proliferation. HCT116 cells were transfected with increasing concentrations of PHLPP1 (0, 0.2μg, 1 μg, and 2 μg of plasmid DNA in lanes 1-4 and 5-8, respectively) in combination with vector or USP46. (A) Cell lysates were prepared from transfected cells and analyzed for Akt phosphorylation using the phospho-S473 antibody. The amount of relative Akt phosphorylation was quantified by normalizing the amount of p-Akt to total Akt and indicated below the p-Akt panel. (B) The transfected HCT116 cells were subjected to cell proliferation analysis. Equal numbers of cells transfected with different constructs were seeded into regular growth medium and allowed to grow for additional 48 hours. The rate of cell proliferation was set to 1 for cells transfected with the vector alone, and the cells in all other settings were normalized accordingly. Each experimental point was done in duplicate and the experiments were repeated three times. Data shown in the graph represent the mean ± SEM (n=3, * indicates p<0.01 by Student t-test when compared to cells transfected with the vector alone). (C) Stable sh-Con and sh-USP46 DLD1 cells were infected with retrovirus encoding vector, PHLPP1, or P1-4A, and the phosphorylation of Akt was analyzed in cell lysates using the phospho-S473 antibody. The amount of relative Akt phosphorylation is indicated below the p-Akt panel. (D) Graph showing the relative rate of cell proliferation of stable DLD1 cells as described in panel (C). The rate of cell proliferation was set to 1 for sh-Con cells infected with vector, and the cells in all other settings were normalized accordingly. Each experimental point was done in duplicate and the experiments were repeated three times. Data shown in the graph represent the mean ± SEM (n=3, * indicates p<0.05 by Student t-test).
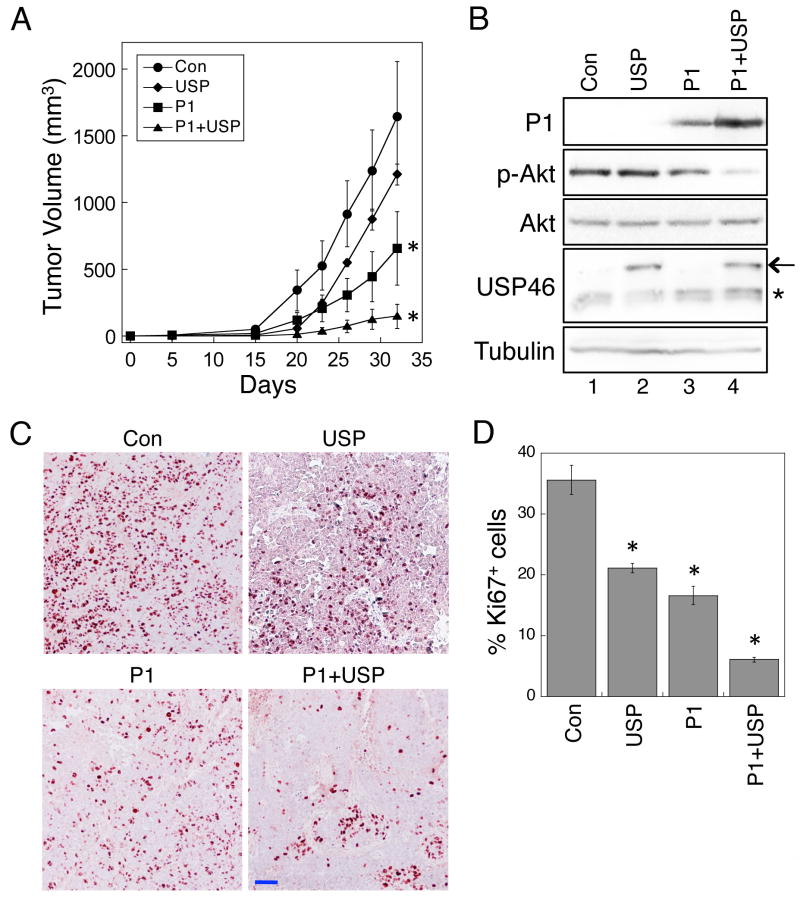
USP46 potentiates the tumor suppressor function of PHLPP1 in vivo
To further determine the functional interaction between USP46 and PHLPP, we generated four stable HCT116 cell lines expressing control vector, USP46 alone, PHLPP1 alone, or PHLPP1 plus USP46. The cells were subcutaneously injected into nude mice and the tumorigenesis process was monitored over a period of 32 days. As shown in Fig. 5A, the control cells formed tumors effectively, and the tumor size reached approximately 1600 mm3 at the end of the experiment. Overexpression of USP46 by itself decreased the size of tumors, however, the difference was not statistically significant. Consistent with our previous findings (7), overexpression of PHLPP1 in HCT116 cells significantly reduced the tumor size. More importantly, co-expression of USP46 markedly enhanced the ability of PHLPP1 to suppress tumor formation (Fig. 5A). The expression of PHLPP1 and USP46 was detected in the tumor tissues isolated from mice. The expression of PHLPP1 was increased in cells co-expressed with USP46, while the phosphorylation of Akt was substantially decreased (Fig. 5B). Similar results were obtained using IHC staining of the tumor tissues (Fig. S3). Furthermore, the rate of proliferation in the tumor tissues was determined by IHC staining of Ki67 (Fig. 5C). The numbers of Ki-67 positive cells in the tumor sections were significantly reduced in tumors expressing USP46, PHLPP1, or USP46 plus PHLPP1, and co-expression of USP46 and PHLPP1 had the largest inhibitory effect (Fig. 5D). Taken together, we demonstrated that USP46 and PHLPP1 function together to suppress tumorigenesis in vivo.
Figure 5.
Overexpression of USP46 enhances the tumor suppressor function of PHLPP1 in vivo, (A) Nude mice of BALB/c background were inoculated subcutaneously with stable HCT116 cells as described in the Experimental Procedures. The size of the tumors was measured every 3-5 days. Five mice were used in each group. Data in the graph represent the mean ± SEM, and the analysis was based on a longitudinal model to account for repeated measurements with each mouse (the asterisk indicates p<0.001 for HCT-P1 and HCT-P1+USP compared to HCT-Con). (B) Protein lysates prepared from tumor samples isolated from mice were analyzed by immunoblotting. Note that the overexpressed Flag-USP46 and the endogenous USP46 were marked by an arrow and an asterisk, respectively. (C) The rate of cell proliferation was determined in the fixed tumor tissues isolated from mice using IHC staining with the anti-Ki67 antibody. The tissue slides were viewed using 10x objective. The scale bar = 200 μm. D, The percentage of Ki67 positive cells were quantified using the Leica Application Suite EZ software. The numbers from 3 random viewing fields were averaged, and representative images are shown. Data in the graph represent the mean ± SEM (n=3, and the asterisk indicates p<0.05 compared with HCT-Con by two-sample t-tests).
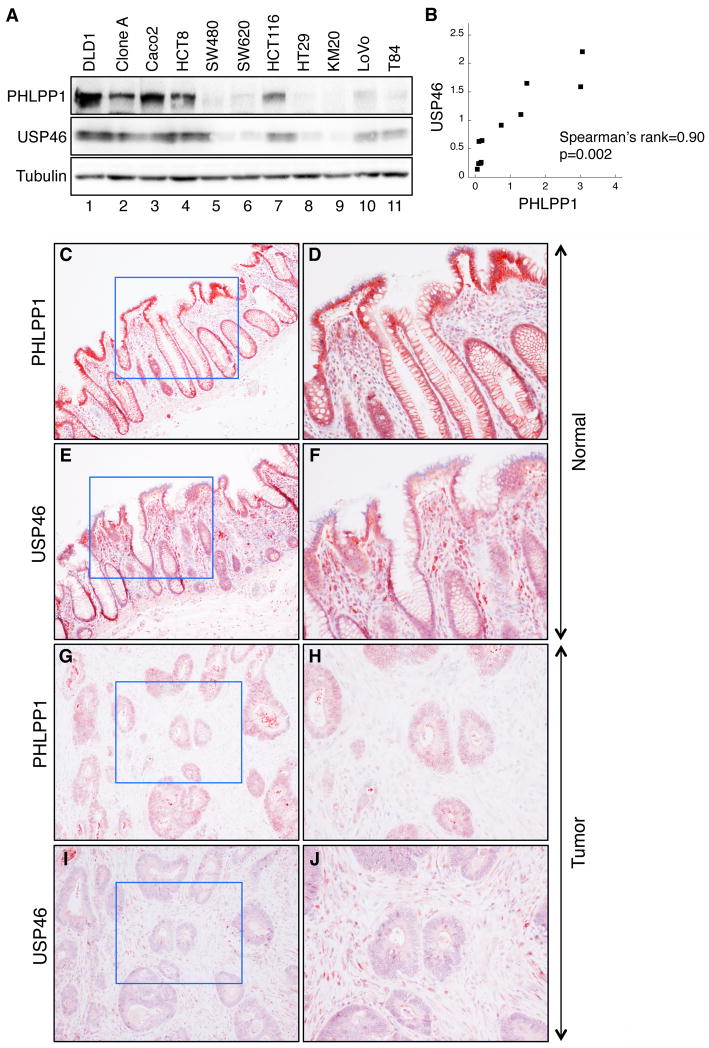
Expression of USP46 and PHLPP1 in human colon cancers
We next investigated the correlation between USP46 and PHLPP1 expression in colon cancer cell lines and colorectal cancer specimens. If USP46 enhances the stability of PHLPP1, then human tumors with downregulation of PHLPP1 protein might also have decreased expression of USP46. To test this, we analyzed the expression of PHLPP1 and USP46 in 11 colon cancer cell lines (Fig. 6A), and the results revealed that the expression of these two proteins correlated significantly (P = 0.002, Spearman’s rank correlation) (Fig. 6B). Furthermore, samples from colorectal cancer and normal mucosa were analyzed for expression of both PHLPP1 and USP46 using IHC staining. Our previous studies showed that PHLPP1 is localized to the membrane of normal colonic epithelia in human tissues and its expression is frequently lost in colorectal cancers (7). Consistent with these findings, the PHLPP1 expression was readily detected in normal mucosa and localized to the membrane (Fig. 6C and 6D). Similarly, USP46 showed strong mixed cytoplasmic and membrane expression in normal tissues (Fig. 6E and 6F). In contrast, the expression of PHLPP1 or USP46 was largely decreased in the colorectal cancer specimens (Fig. 6G-6J). Pearson correlation analyses of quantitative IHC staining results based on a four-tier scoring system revealed that there is a significant positive correlation between PHLPP1 and USP46 in both normal and tumor tissue samples (P < 0.001 and P < 0.05, respectively, Table S1). In addition, the expression of USP46 was significantly decreased in 60% of colorectal cancer samples (P < 0.01) indicating a potential tumor suppressor role of USP46.
Figure 6.
Downregulation of PHLPP1 and USP46 expression of in colorectal cancer tissues. (A) Western blots of PHLPP1 and USP46 in colon cancer cell lines. (B) The expression of PHLPP1 and USP46 is highly correlated in the colon cancer cell lines analyzed in (A). Spearman’s rank correlation coefficient = 0.90; P = 0.002. (C-J) Representative images taken from the normal and tumor specimens stained with the PHLPP1 or USP46 antibody. Images shown in the left panels were obtained with a 10X objective, and the enlarged images of areas enclosed by the blue boxes are shown in the panels directly on the right.
In summary, our findings demonstrate that USP46 stabilizes PHLPP expression by promoting deubiquitination. USP46 and PHLPP1 function together to suppress tumorigenesis in vivo, and the expression of PHLPP1 correlates with USP46 in human colorectal cancers.
DISCUSSION
The funtional importance of PHLPP as a tumor suppressor has been entensively investigated by serval recent studies. To date, loss of PHLPP expression has been found associated with human colon, brain, and prostate cancers (7, 8, 16). Downregulation of PHLPP proteins may result from decreased gene expression (8). However, our previous studies have also shown that PHLPP expression can be regulated at the post-transcriptional level. For example, the degradation of PHLPP is tightly regulated by E3 ubiquitin ligase β-TrCP, and PHLPP is a translational target of mTOR (10, 17). Here, we identified USP46 as a novel regulator of PHLPP expression. USP46 functions as a deubiquitinase to remove ubiquitin from PHLPP and stabilizes the protein. This USP46-mediated regulation of PHLPP requires the interaction between the two proteins and the emzymatic activity of USP46. Futhermore, a positive corelation of PHLPP1 and USP46 expression has been observed in normal and colon cancer patient samples, thus suggesting a novel tumor suppressor function of USP46. It is interesting to determine whether loss of USP46 expression provides additional proliferative advantages to tumor cells with mutations in the PI3K/Akt pathway in future studies.
It has been shown recently that USP46 plays a role in controlling the abundance of glutamate receptor GLR-1 in nerve systems (18). Our study here identifies a potential tumor suppressor role of USP46. By stabilizing the expression of PHLPP, USP46 negatively regulates Akt activity in cells and in vivo. Here, we show that USP46 and PHLPP function together to suppress Akt signaling in a mutually-dependent manner. There are several lines of evidence indicating that USP46-mediated regulation of Akt depends on PHLPP: (i) since the endogenous expression of PHLPP is relatively low in the colon cancer cells used in our study, overexpression of USP46 by itself has relatively small impact on Akt phosphorylation and cell proliferation; (ii) the effect of USP46 knockdown on Akt phosphorylation and cell proliferation is reversed by overexpressing a degradation-resistant form of PHLPP1 (the 4A mutant); and (iii) co-expression of USP46 with PHLPP significantly potentiates the antagonistic effect of both proteins on Akt phosphorylation and tumorigenesis. Although the expression of both PHLPP isoforms are regulated by USP46, alteration of USP46 levels usually has larger effect on PHLPP1. This is likely due to the intrinsic protein turnover rate of PHLPP2 is at least three times slower than that of PHLPP1 (Fig. 2). Future studies are needed to dissect the functional contribution of each PHLPP isoform in response to the alteration of USP46 expression. In summary, we have identified USP46 as a novel regulator of PHLPP-dependent tumor suppressor function. The dose-dependent effect of PHLPP1 on inhibiting Akt phosphorylation and cell proliferation highlights the importance of maintaining sufficient levels of PHLPP1 and USP46 in order to prevent the hyperactivation of oncogenic signaling in cells. Futher understanding of how USP46 expression is regulated will help to develop new therapeutic strategies targeting PHLPP and USP46 in cancer.
MATERIALS AND METHODS
Antibodies and reagents
The PHLPP1 and PHLPP2 antibodies were obtained from Bethyl Laboratories. The phospho-Akt (p-Akt for Ser473) antibody was from Cell Signaling. The Akt1/2/3 (pan-Akt) polyclonal antibody and mouse monoclonal antibodies (mAb) for Akt1 and Akt2 were from Santa Cruz Biotechnology. The USP46 and anti-γ tubulin antibody was from Sigma-Aldrich. The P1-PP2C antibody was prepared in rabbits against the recombinant PP2C domain of human PHLPP1 (residues 642-910) and described previously (19).
Cells
Human colon cancer cells HCT116 and HCT-P1 [HCT116 cells overexpressing wild-type PHLPP1(7)] were cultured in McCoy’s 5A medium, and DLD1 and 293T cells were cultured in DMEM (Cellgro). All media were supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals) and 1% penicillin/streptomycin. Transient transfection of all cell types was carried out using polyethylenimine (PEI, Polysciences).
The shRNA for human USP46 and Akt2 gene was constructed in pLKO.1-puro vector and purchased from Sigma-Aldrich, and the targeting sequence is as the following: 5’-CAGCTATCCATCCTAAAGGTA-3’ (#1) and 5’-CGCTTACCAATGAAACTCGAT-3’ (#2) f o r U S P 4 6 ; a n d 5’-ACGGGCTAAAGTGACCATGAA-3’ (# 1) a n d 5’-ACGGGCTAAAGTGACCATGAA-3’ (#2) for Akt2. The shRNA targeting sequence for human Akt1 gene has been described previously (10). The lentivirus-mediated delivery of shRNA and selection for stable knockdown cells were carried out as previously described (7). To generate stable cells overexpressing USP46, retrovirus encoding USP46 was produced using the pCL-Ampho packaging plasmid (20). HCT116 cells infected with the control or USP46 retroviruses were subjected to selection with puromycin (2 μg/ml).
Expression constructs
The following mammalian expression plasmids have been described previously: HA-PHLPP1, HA-PHLPP2, HA-P1/4A, GST-P1/PH, GST-P1/LRR, GST-P1/PP2C, and GST-P1/CT (2, 6, 10); and Flag-HA-USP46 (15). The C44S mutant in USP46 was created using the Quickchange site-directed mutagenesis kit (Stratagene). To generate C-terminal truncation mutants in PHLPP1, including Δ910, Δ1010, Δ1081, and Δ1139, Δ1172, stop codons were incorporated into the corresponding amino acid residues by site-directed mutagenesis using pcDNA3-HA-PHLPP1 as the template. Similarly, C-terminal truncation mutants of PHLPP2, including P2-Δ1268, and P2-Δ1301, were generated using pcDNA3-HA-PHLPP2 as the template.
Cycloheximide (CHX) chase assay
Equal numbers of stable control and USP46 knockdown DLD1 cells were seeded onto 12-well plates and allow to attached for ~18 hours. The cells were treated with CHX (40 μg/ml) and harvested at indicated time points. The cell lysates were prepared in SDS sample buffer directly, and the equal amount of lysates was analyzed by Western blotting.
Immunoprecipitation and GST-pull down assays
Immunoprecipitation and GST-pull down experiments were performed following procedures described previously (2). Briefly, the detergent-solubilized cell lysates were incubated with one of the following beads including anti-Flag agarose (Sigma-Aldrich), indicated antibodies coupled to protein A/G agarose, or glutathione-Sepharose at 4°C for 3 hours. The precipitated proteins were analyzed by SDS-PAGE and Western blotting.
Ubiquitination and deubiquitination of PHLPP1
To examine the ubiquitination of PHLPP1 in cells, stable control or USP46 knockdown HCT-P1 cells were transfected with Myc-Ub. The transfected cells were pretreated with DMSO or MG-132 (10 μM) for 30 minutes and lysed in Buffer A (50 mM Tris, pH 7.4, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, 1 mM DTT, 200 μM benzamidine, 40 μg ml-1 leupeptin, 200 μM PMSF, and 10 mM N-Ethylmaleimide). The detergent-solubilized cell lysates were incubated with the PHLPP1 antibody coupled to protein A/G agarose at 4°C for 3 hours. The beads were washed three times in Buffer B (Buffer A plus 250 mM NaCl) and once in Buffer A. The immunoprecipitated proteins were analyzed by SDS-PAGE and Western blotting. To detect the ubiquitination or PHLPP1 under denature conditions, stable sh-Con and sh-USP46 HCT-P1 cells were transfected with His-tagged ubiquitin (21, 22). Cell lysates were prepared in denatured buffer (6M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole) and incubated with Ni-NTA beads to pull down all ubiquitinated proteins.
In vitro deubiquitination assays were performed as described previously (23). Briefly, 293T cells transfected with Myc-Ub and HA-PHLPP1 proteins were immunoprecipitated using the PHLPP1 antibody coupled to protein A/G agarose. Similarly, lysates obtained from vector (for the control treatment), WT USP46, or USP46-C44S transfected 293T cells were immunoprecipitated using anti-HA Affinity Matrix (Roche Applied Science). The USP46 proteins were eluded from the beads by incubating with HA peptide, dialyzed, and subsequently used as the deubiquitinating enzyme by incubating with the immunoprecipitated PHLPP1. The ubiquitination status of immunoprecipitated PHLPP1 after the treatment was analyzed using SDS-PAGE and Western blotting.
Cell proliferation assay
To determine the rate of cell proliferation, HCT116 cells were co-transfected with HA-PHLPP1 and USP46. Approximately 24 hours post-transfection, equal numbers of transfected cells were seeded onto 12-well plates and allowed to grow for additional 48 hours. Subsequently, the cells were fixed and stained with 0.5% crystal violet in 20% methanol. The stained cells were dissolved in 1% SDS and absorbance at 570 nm was measured (7).
Immunohistochemical (IHC) staining
Paraffin embedded normal and colorectal cancer tissue section slides were obtained from US Biomax. IHC staining was performed as previously described (7). The stained sections were visualized using a Leica DM750 microscope. Scoring of PHLPP1 and USP46 staining was done blindly according to the intensity of staining with a four-tier system (levels 0-3) (24). Comparisons were performed using the Wilcoxon rank-sum test for intensity scores of USP46 expression levels as detected by IHC staining. Pearson coefficient was calculated to assess the correlation in intensity scores between PHLPP1 and USP46.
Tumorigenesis analysis in xenografted nude mice
The HCT-Con and HCT-P1 cells (7) were infected with retrovirus encoding a control vector or Flag-USP46. The resulting stable cells, including HCT-Con, HCT-USP46, HCT-P1, and HCT-P1+USP46, were collected in PBS and inoculated subcutaneously into 6-week-old male BALB/c-nu/nu mice at 1×106 cells/injection site. The tumor size was measured every 3-5 days with a caliper, and the tumor volumes was defined as (longest diameter) × (shortest diameter)2/2. At the end of the experiments, the mice were sacrificed and the tumors dissected from the individual mouse were fixed in 10% buffered formalin. The paraffin embedded samples were prepared, and 5 mm sections were used for IHC staining. For Western blot analysis, fresh tumor tissues were flash frozen in liquid N2 and detergent-soluble protein lysates were extracted and analyzed as previously described (2). All animal procedures were done in the nude mouse facility using protocols approved by the University of Kentucky Animal Care and Use Committee. Tumor growth in nude mice studies were summarized at each time point of follow-up and analysis was performed using longitudinal models to account for repeatedly-measured tumor volume over time within each mouse.
Supplementary Material
Acknowledgments
We thank Dr. Wade Harper (Harvard Medical School) for providing USP46, USP7, and USP16 expression plasmids, and Dr. Hui-Kuan Lin (University of Texas M. D. Anderson Cancer Center) for providing His-tagged ubiquitin construct. The Biostatistic Core at the Markey Cancer Center (University of Kentucky) provided assistant with the statistical analysis in our study. This work was supported by NIH R01CA133429 (T.G.), American Cancer Society RSG0822001TBE (T.G.), P20CA1530343 (UK GI SPORE), and R01DK48498 (BME).
References
- 1.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008 Aug;19(6):223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005 Apr 1;18(1):13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Gao T, Brognard J, Newton AC. The Phosphatase PHLPP Controls the Cellular Levels of Protein Kinase C. J Biol Chem. 2008 Mar 7;283(10):6300–11. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 4.Shimizu K, Mackenzie SM, Storm DR. SCOP/PHLPP and its functional role in the brain. Mol Biosyst. 2010 Jan;6(1):38–43. doi: 10.1039/b911410f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiao M, Wang Y, Xu X, Lu J, Dong Y, Tao W, et al. Mst1 is an interacting protein that mediates PHLPPs’ induced apoptosis. Mol Cell. 2010 May 28;38(4):512–23. doi: 10.1016/j.molcel.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007 Mar 23;25(6):917–31. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009 Feb 19;28(7):994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O’Neill A, et al. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011 Aug 16;20(2):173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23(11):2028–36. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Liu J, Gao T. b-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol. 2009;29(23):6192–205. doi: 10.1128/MCB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warfel NA, Niederst M, Stevens MW, Brennan PM, Frame MC, Newton AC. Mislocalization of the E3 ligase, beta-transducin repeat-containing protein 1 (beta-TrCP1), in the pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) and Akt. J Biol Chem. 2011 Jun 3;286(22):19777–88. doi: 10.1074/jbc.M111.237081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009 Aug;10(8):550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 13.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010 Jan 7;463(7277):103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 14.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007 Jul;9(7):765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009 Jul 23;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, et al. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2011 Aug 1; doi: 10.1038/onc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Stevens PD, Gao T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem. 2011 Feb 25;286(8):6510–20. doi: 10.1074/jbc.M110.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans. J Neurosci. 2011 Jan 26;31(4):1341–54. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Yang H, Liu J, Schmidt MD, Gao T. Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. Embo Rep. 2011;12(8):818–24. doi: 10.1038/embor.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucl Acids Res. 1990 Jun 25;18(12):3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003 May;11(5):1177–88. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 22.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009 Aug 28;325(5944):1134–8. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009 Jan 9;136(1):123–35. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Reiner A, Neumeister B, Spona J, Reiner G, Schemper M, Jakesz R. Immunocytochemical localization of estrogen and progesterone receptor and prognosis in human primary breast cancer. Cancer Res. 1990 Nov 1;50(21):7057–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.