Abstract
Structure-activity relationship (SAR) studies are essential in the generation of peptides with enhanced activity and efficacy as therapeutic agents. In this study we report a SAR study for a family of mimetic peptides derived from type IV collagen with potent anti-angiogenic properties. The SAR study was conducted using a number of validated in vitro assays including cell proliferation, adhesion, migration and tubule formation. We report a critical sequence (NINNV) within this peptide series which is required for the potent anti-angiogenic activity. Detailed amino acid substitutions resulted in peptides with superior efficacy. Specifically, substitutions with Isoleucine at positions twelve and eighteen along with the substitution of the Methionine at position ten with the non-natural amino acid d-Alanine led to an increase in potency by two orders of magnitude over the parent peptide. Several mimetic peptides in this series exhibit a significant improvement of activity over the parent peptide. This improved in vitro activity is expected to correlate with an increase in in vivo activity leading to effective peptides for anti-angiogenic therapy for different disease applications including cancer and age-related macular degeneration.
Keywords: angiogenesis, peptidomimetics, endothelial cell, cancer, age-related macular degeneration, lymphangiogenesis
Introduction
Peptides have been employed as therapeutics for multiple diseases (1) and recently have been investigated in clinical applications to target tumors either for imaging or therapy. (2),(3), (4–6) In general they are attractive tools as therapeutics due to their specific target binding, ability to penetrate cells and ease of modification giving flexibility for different applications. (6, 7) However, some of the properties that make peptides attractive candidates also contribute to their disadvantages such as reduced stability and bioavailability.(8) Thus, efforts in overcoming drawbacks such as these have been focused on increasing the stability of peptides by introducing non-natural amino acids (9) or conjugating them to larger and more stable carriers such as large proteins and antibodies.(10)
Peptides are sometimes characterized by having low binding affinities, usually in the micro-molar range. (11) Moreover, certain residues are known to be challenging in either synthesis or long term storage and stability; e.g. cysteines may oxidize or create intermolecular bridges, thus replacing them with more stable residues has been frequently explored. (12) In order to create more stable peptides a detailed understanding of the structure of the peptide is essential, so that non-critical amino acids can be replaced or eliminated to minimize the number of enzymatically labile residues. Systematic replacement of specific amino acid residues and evaluating the activity of the resulting peptides can result in enhanced peptides and thus decisions about the importance of each residue in the activity of the peptide can be made. (13) Another approach seeks to find shorter fragments (pharmacophores) within the peptide which are responsible for the activity, and this can usually be accomplished using systematic truncations. (14)
Identification of pharmacophores has been successfully used in the development of many therapeutic peptides and also in the development of anti-angiogenic peptides for treatment of cancer.(15) Angiogenesis, the process of blood vessel growth from preexisting vasculature, has been demonstrated to play a very important role in tumor growth and metastasis.(16, 17) Our laboratory has identified multiple anti-angiogenic peptides in several protein classes through a bioinformatics methodology. (18) These included several fragments from different fibrils of type IV collagen. One of the peptides in this series (referred to as SP2000, with the amino acid sequence LRRFSTMPFMFCNINNVCNF) is a 20 amino acid fragment found within the non-collagenous domain of the α5 fibril of type IV collagen. This peptide was shown to possess in vitro anti-angiogenic properties by inhibiting HUVECproliferation by 70% and migration by 60% at 20 µM (18, 19), and in vivo activity of inhibiting tumor growth by 50% after 14 days of peptide administration in both breast and lung xenograft models at a concentration of 5 mg/kg with daily intraperitoneal administration.(20, 21) Considerations of potential clinical applications lead us to substitute its two cysteines by L-α-amino-n-butyric acid (Abu) resulting in the peptide denoted SP2012 (sequence LRRFSTMPFMF-Abu-NINNV-Abu-NF), which serves as the parent peptide for this study. This peptide was also characterized in several in vitro and in vivo assays and demonstrated strong activity. (22) To facilitate translation of this peptide to clinical applications, a detailed understanding of its structure-activity relationship is essential. Thus, we constructed a series of truncated and modified peptides; truncations that eliminated residues from the C-terminus, N-terminus, both termini, and several substitutions within the body of the peptide. In this study we present the in vitro activities of these peptides on proliferation, adhesion, migration and tubule formation of human umbilical vein endothelial cells (HUVEC), microvascular endothelial cells (MEC), and lymphatic endothelial cells (LEC).
Methods and Materials
Cell culture
Human umbilical vein endothelial cells (HUVEC), microvascular endothelial cells (MEC), and lymphatic endothelial cells (LEC) were purchased from Lonza and maintained according to the manufacturer’s recommendation using Endothelial Basal Media (EBM-2) supplemented with the Bullet Kit (EGM-2, Lonza). The MEC and LEC were propagated in Microvascular Endothelial Cell Growth Medium-2 (EGM-2MV, Lonza). Breast cancer cells, MDA-MB-231 were supplied by Dr. Zaver Bhujwalla (JHMI, Radiology and Oncology). The cells were propagated in RPMI-1640 medium (Gibco, Carlsbad, CA) supplemented with 10% FBS and antibiotics (1% penicillin/streptomycin). Cells were maintained under standard conditions of 37°C and 5% CO2 and the passage number of the primary cells (HUVEC, LEC and MEC) was between 2 and 7.
Peptide synthesis
The peptides were synthesized using solid-phase synthesis and were supplied as TFA salts with an amidated C-terminus by two vendors (New England Peptide, Gardner, MA and American Peptide Company, Sunnyvale, CA). The purity of the peptides was > 95% and the suppliers provided product characterization (MALDI-TOF and HPLC traces) as proof of MW and purity accuracy. Peptides were solubilized in 5% DMSO and water due to their hydrophobic profile. The pH of solubilized peptides was checked and found to be around pH 7. For all experiments the DMSO % was maintained at non-toxic threshold (determined by toxicity curves of DMSO on cells) with a final DMSO percentage (< 0.2%) which was used as control in all experiments.
Proliferation assays
Colorimetric based proliferation assay using WST-1 (Roche, Basel Switzerland) proliferation reagent was carried using HUVEC. 2000 cells/well were plated in 96-well plates and allowed to adhere overnight. On the next day the media was exchanged with fully supplemented media containing peptides or equivalent DMSO vehicle for the controls. Three days later the media containing the peptides was replaced with serum-free EBM-2 media containing WST-1 reagent, and incubated for four hours as per manufacturer’s recommendations. Changes in color due to the formazan dye resulted from the cleavage of the tetrazolium salt WST-1 by the mitochondrial succinate-tetrazolium reductase, were read with a Victor V fluorescence plate reader (Perkin Elmer, MA) by measuring absorbance at 450 nm. Dose response curves of percent live cells (in comparison to untreated cells but incubated in complete media with 0.2% DMSO) were created. Assays were performed in at least two independent replicates and each replicate was performed using three experimental replicates.
Migration assay
The inhibitory potential of the peptides was measured using a real time migration assay system based on electrical impedance (RT-CIM, ACEA Biosciences, CA). CIM 16-well plates (Roche, Basel Switzerland) are composed of a top and bottom chamber separated by a microporous (8 µm) polycarbonate membrane. The membrane was coated with fibronectin (20µg/ml) and 45,000 cells/well in serum free media with or without peptides were added to the top compartment. Media with chemoattractant (i.e. fully supplemented EBM-2) was added to the bottom compartment of the chamber and the plate was incubated at 37°C for 20 hours. The sensors integrated on the bottom side of the membrane monitor and continuously record changes in impedance as the cells move through the membrane. The RT-CIM technology allows for easy quantification of cell migration by monitoring the cell index derived from the measured impedances. Assays were performed in at least two independent replicates and each replicate was performed using two experimental duplicates. Breast cancer cells MDA-MB-231 are not suitable for the RT-CIM type experiments due to their thin elongated phenotype resulting in low signal, thus the inhibition of migration was investigated using a wound healing type assay. This assay was performed using the Oris Pro Migration assay (Platypus Technologies, CMA 1.101). Briefly, 25,000 cells/well in full media were added to the 96-well plate containing stoppers to block migration of cells to the center region of the wells. Cells were allowed to adhere for 4 hours, after which the stoppers were removed. Cells were washed one time with PBS and fully supplemented media with or without compound was added to the wells. After 18 hours cells were stained with calcein AM (0.5µg/ml) (Invitrogen, CA) and the cells that migrated to the center of the well were quantified by fluorescence intensity measured using Victor V plate reader and imaged using a Nikon microscope (Eclipse T-100); images were acquired with the CCD Sensicam mounted on a Nikon microscope (Cooke Company, MI). The detection of the cells that migrated into the previously restricted region is possible due to the addition of a detection mask at the bottom of the plate, which obstructs from measurement cells that did not migrate.
Adhesion assay
Similar to the migration assays the inhibition activity of the peptide in cellular adhesion was assessed using the RT-CIM technology. In this case 25,000 cells/well were plated in 16-well E-plates (Roche, Basel Switzerland) in the presence or absence of the peptide. The adhesion was monitored over time (3 hours) by measuring changes in the electrical impedance, which is a direct measure of the cells adhering on the electrodes. For the most active peptides we calculate IC50 values from dose response curves. Assays were performed in at least two independent replicates and each replicate was performed using two experimental replicates.
Tubule formation assay
When cultured on gels of extracellular matrix endothelial cells form chord like structures indicative of their potential to rearrange in vascular structures, and these are referred to as tubules. The compounds were tested for their ability to inhibit tubuleformation, a process critical in angiogenesis. The protocol was described by Arnaoutova et. al. (23) and it consists of plating HUVEC on top of basement membrane extract and after incubation at 37°C the cells naturally rearrange themselves in a network of tubules. Thus 50 µL/well of Matrigel (BD Biosciences, San Jose, CA ) was plated in a cold 96-well plate and incubated at 37°C for 30 min for polymerization. 15,000 cells/well were added to the top of the gel and incubated in complete media in the presence or absence (which serves as the positive control) of peptide for 19 hours. Images were captured using the CCD Sensicam mounted on a Nikon microscope (Eclipse T-100) using a 10× objective to capture a large area of the wells (approximately 95% ) but a smaller area is presented in the composite images. Assays were performed in at least two independent replicates and each replicate was performed using three experimental replicates and one image of a randomly chosen field was acquired per well. Qualitative analysis was carried out by assessing the phenotype of the cells as the result of the peptide treatment; elongated cord like structures (tubules), cell clumps or short stubby elongations, and these were seen throughout the entirety of each well.
Data analysis
Statistical significance ( p < 0.05) within each experiment using the independent replicates was assessed using standard statistical assessments such as Student’s t-test and ANOVA accompanied by Dunnett’s test if we were comparing different sets of data to one group.
Results and Discussion
Inhibition of Proliferation
Truncations from the C-terminus
Table 1 lists the family of peptides that were generated by eliminating residues from the C-terminus of the parent peptide (SP2012). Truncations that eliminate the NINNV sequence show loss of inhibitory activity in proliferation of endothelial cells. Peptides that include representative residues from this critical sequence (i.e. SP2028, SP2029, SP2032 and SP2033) demonstrate potent activity in inhibiting proliferation, even improved activity as compared to the parent compound. The substitution of Asparagine, a hydrophilic residue, by Valine, a hydrophobic amino acid leads to loss in activity (SP2028 vs. SP2006). However, if in addition the Abu residue is replaced by Threonine, the new compound regains activity (SP2029 vs. SP2006). Substitution of Abu in SP2028 by a Threonine (SP2032), results in a reduction in activity while its replacement by Isoleucine maintains the same level of activity (SP2033) thus confirming that the nature of the amino acid residue at this position is important for the inhibition of proliferation.
Table 1.
C-terminus truncations and peptide proliferation activity
| Peptide Name | Peptide sequence | Number of residues | Modification from the SP2012 | IC50 ± 95%CI (µM) |
|---|---|---|---|---|
| SP2012 | LRRFSTMPFMFAbuNINNVAbuNF | 20 | Parent peptide for this study | 48.1 ± 23.1 |
| SP2004 | LRRFSTMPFMF | 11 | Truncated 9 residues | >100 |
| SP2006 | LRRFSTMPFMFAbuNINV | 16 | Truncated 4 residues | >100 |
| SP2007 | LRRFSTMPFMFAbu | 12 | Truncated 8 residues | >100 |
| SP2008 | LRRFSTMP | 8 | Truncated 12 residues | >100 |
| SP2013 | LNRFSTMPF | 9 | Truncated 12 residues & substituted one Arginine with Asparigine | >100 |
| SP2014 | LRRFSTXPFXF –X=NorLeu | 11 | Truncated 9 residues & substituted Methionine with NorLeucine | >100 |
| SP2028 | LRRFSTMPFMFAbuNINN | 16 | Truncated 4 residues | 5.1 ± 2.7 |
| SP2029 | LRRFSTMPFMFTNINV | 16 | Truncated 4 residues & substituted the Abu residues with Threonine | 4.8 ± 2.8 |
| SP2032 | LRRFSTMPFMFTNINN | 16 | Truncated 4 residues & substituted the Abu residues with Threonine | 10.3 ± 4.1 |
| SP2033 | LRRFSTMPFMFININN | 16 | Truncated 4 residues & substituted the Abu residues with Isolucine | 3.8 ± 1.7 |
Truncations from the N-terminus
The peptides generated by removing amino acids from the N-terminus are listed in Table 2. In contrast with the previous series of C-terminus truncations the peptides that lack residues from the N-terminus are virtually inactive at inhibiting proliferation of HUVEC.
Table 2.
N-terminus truncations and peptide proliferation activity
| Peptide Name |
Peptide sequence | Number of residues |
Modification from the SP2012 |
IC50 ± 95%CI (µM) |
|
|---|---|---|---|---|---|
| SP 2012 | LRRFSTMPFMFAbuNINNVAbuNF | 20 | Parent peptide for this study | 48.1 ± 23.1 | |
| SP 2009 | NINNVAbuNF | 8 | Truncated 12 residues | >100 | |
| SP 2010 | FMFAbuNINNVAbuNF | 12 | Truncated 8 residues | >100 | |
| SP 2011 | STMPFAbuNINNVAbuNF | 16 | Truncated 4 residues | >100 | |
Truncations from both termini
Peptide fragments generated by removing residues from both termini are listed in Table 3. The inhibitory activity suggests that shorter fragments are less active even if they retain most of the NINNV sequence (SP2021 and SP2030). However, addition of two more residues to the N-terminus of the NINNV fragment along with the replacement of Abu by Threonine (SP2031) restores activity to similar levels as the larger 16 amino acid C-terminus truncations. These results in conjunction with the fact that splitting the molecule in two almost equal pieces leads to an inactive and active compound, SP2004 and SP2020 respectively, indicates that the presence of particular residues is crucial. These results suggest that the NINNV sequence is important to the inhibitory activity of our peptides and especially the context in which is incorporated (the surrounding residues); thus, additional amino acid residues on either end (SP2031) or just at the C-terminus improve activity.
Table 3.
Truncations from both termini and peptide proliferation activity
| Peptide Name | Peptide sequence | Number of residues | Modification from the SP2012 | IC50 ± 95%CI (µM) |
|---|---|---|---|---|
| SP 2012 | LRRFSTMPFMFAbuNINNVAbuNF | 20 | Parent peptide for this study | 48.1 ± 23.1 |
| SP 2020 | FAbuNINNVAbuN | 9 | Truncated 11 residues | 9.8 ± 4.6 |
| SP 2021 | FAbuNIN | 5 | Truncated 15 residues | >100 |
| SP 2030 | FAbuNINV | 6 | Truncated 14 residues | >100 |
| SP 2031 | FTNINNVTN | 9 | Truncated 11 residues & substituted the Abu residues with Threonine | 4.7 ± 1.5 |
Proliferation activity of substitutions
The truncation studies demonstrated that the Abu positions (in SP2012) are important in maintaining strong activity in the inhibition of proliferation thus we set forth to investigate additional substitutions that could be introduced at these positions (12 and 18). These substitutions are presented in Table 4. The inhibitory activity of these peptides is not affected when the Abu is substituted by Serine (SP2022), Threonine (SP2025), AllyGly (SP2026) or Valine (SP2027) thus hydrophilicity at these locations is not crucial. Hydrophobic residues such as Alanine or Isoleucine increase the activity dramatically, 2.8 and 6.5 times, respectively.
Table 4.
Amino acid substitutions and peptide activity in proliferation
| Peptide Name | Peptide sequence | Modification from the SP2012 | IC50 ± 95%CI (µM) |
|---|---|---|---|
| SP 2012 | LRRFSTMPFMFAbuNINNVAbuNF | Parent peptide for this study | 48.1 ± 23.1 |
| SP 2022 | LRRFSTMPFMF S NINNV S NF | Abu residues replaced by Serine | 44.9 ± 29.4 |
| SP 2023 | LRRFSTMPFMF A NINNV A NF | Abu residues replaced by Alanine | 17.3 ± 6.0 |
| SP 2024 | LRRFSTMPFMF I NINNV I NF | Abu residues replaced by Isoleucine | 7.45 ± 5.7 |
| SP 2025 | LRRFSTMPFMF T NINNVT NF | Abu residues replaced by Threonine | 63.8 ± 35.0 |
| SP 2026 | LRRFSTMPFMF(AllyGly)NINNV (AllyGly)NF | Abu residues replaced by AllyGly | 47.3 ± 29.2 |
| SP 2027 | LRRFSTMPFMF V NINNV V NF | Abu residues replaced by Valine | 33.9 ± 17.9 |
Proliferation activity of 2nd generation substitutions
We modified one of the most active full length peptides, the SP2024 which contains Isoleucines instead of Abu residues present in the parent peptide (SP2012), to generate additional 2nd generation peptides. These compounds are illustrated in Table 5. Two of these peptides (SP2034 and SP2035) demonstrate submicromolar activity in proliferation (IC50 0.54±0.19 and 0.94±0.38µM respectively), a remarkable increase in activity from the parent peptide, SP2012 (IC50 48.1±23.1µM). It is interesting to note that substitution of the second methionine in position 10 (SP2034) with the non-natural amino acid d-Alanine or Alanine leads to considerable increases in activity, while the additional substitution of the methionine at position 7 results in a 10-fold loss of activity compared to substitution of methionine at position 10 (SP2036).
Table 5.
2nd Generation substitutions and peptide activity in proliferation
| Peptide Name | Peptide sequence | Modification from the SP2012 | IC50 ± 95%CI (µM) |
|---|---|---|---|
| SP 2024 | LRRFSTMPFM FI NINNVI NF | Abu residues replaced by Isoleucine this peptides becomes the parent peptide for this group | 7.45 ± 5.7 |
| SP 2034 | LRRFSTMPFdA FI NINNVINF | Second Methionine replaced by d-Alanine | 0.54 ± 0.19 |
| SP 2035 | LRRFSTMPFA FI NINNVINF | Second Methionine replaced by Alanine | 0.94 ± 0.38 |
| SP 2036 | LRRFSTAPFA FI NINNVINF | Both Methionine replaced by Alanine | 9.97 ± 8.3 |
Inhibition of Adhesion and Migration
Truncations from the N- or C-terminus
Figure 1 illustrates the inhibitory activity of peptides generated by deletions from the C-terminus of SP2012 in HUVEC adhesion and migration assays. The same four peptides that exhibited activity in the inhibition of proliferation are also very potent in inhibiting adhesion and migration, but do not display increased activity over the parent compound SP2012. In the proliferation assay they were more potent than SP2012. In addition SP2006, which did not inhibit proliferation, exhibited inhibitory activity in both the adhesion and migration assays. The significance of the NINNV sequence is supported by the absence of activity in the fragments which lack this region (SP2007, SP2008, SP2013 and SP2014). These results are also in accordance with the inhibitory activity in the proliferation assay.
Figure 1. Adhesion and migration of C-terminus fragments.
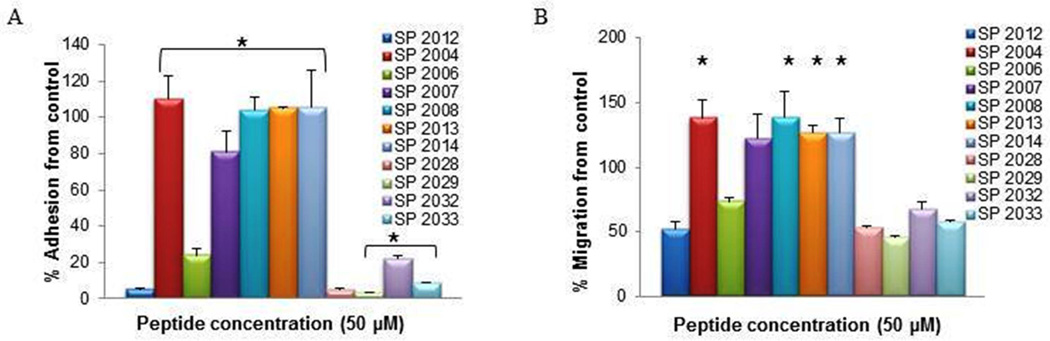
The activity in adhesion (A) and migration (B) of peptides generated by truncation of residues from the C-terminus. * indicate statistical difference (p<0.05) in activity in comparison to the parent peptide (SP2012).
Figure 2 illustrates the inhibitory activity of compounds generated by truncations from the N-terminus in adhesion and migration. Unlike the proliferation assay where all the N-terminus truncations were inactive, these compounds display a range of inhibitory activity in the adhesion and migration assays. For the purpose of comparison we have established a theoretical threshold of activity: any compound displaying an inhibitory potential of greater than 30% (in the graphs a % inhibition from control lower than 70%) will be considered active. Thus in the group of truncations from the N-terminus, peptides SP2010 and SP2011 show low inhibitory activity in the adhesion assay and SP2009 shows inhibitory activity in the migration assay. Since all these compounds include the critical NINNV fragment we asked what other features distinguish them in their pattern of activity. The peptides active in adhesion inhibition contain residues which extend in both directions from the NINNV sequence while SP2009 which only inhibits migration includes additional residues to the C-terminus of the NINNV sequence. Thus the context in which the NINNV region is presented seems to play a role in influencing the type of activity that a compound will demonstrate.
Figure 2. Adhesion and migration of N-terminus fragments.
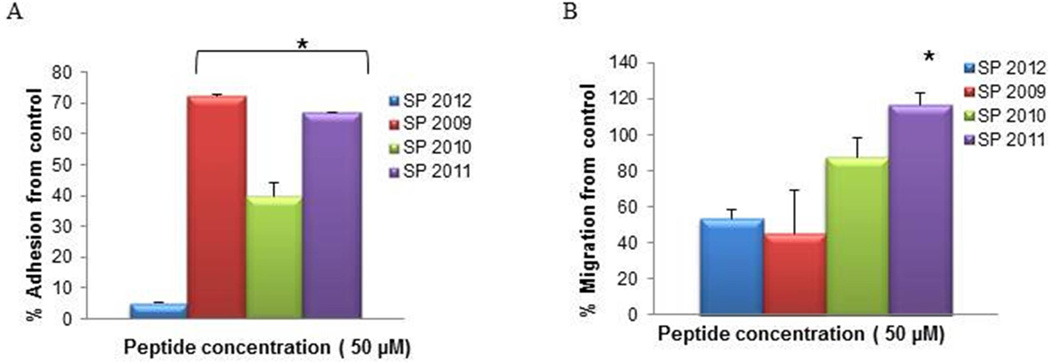
The activity in adhesion (A) and migration (B) of peptides generated by truncation of residues from the N-terminus. * indicate statistical difference (p<0.05) in activity in comparison to the parent peptide (SP2012).
Truncations from both termini
The pattern of activity in adhesion and migration inhibition by fragments generated by truncations from both termini is presented in Fig. 3. The SAR is similar to the one observed in inhibitory activity of proliferation; the absence of the NINNV sequence leads to inactivation of these compounds (SP2021 and SP2030). SP2031 which includes the NINNV sequence along with substitution of Threonine in lieu of the Abu residue displays robust activity across all assays; proliferation inhibition (IC50 4.6 µM), adhesion (>80% inhibition), and migration ( >50 % inhibition).
Figure 3. Adhesion and migration of both termini fragments.
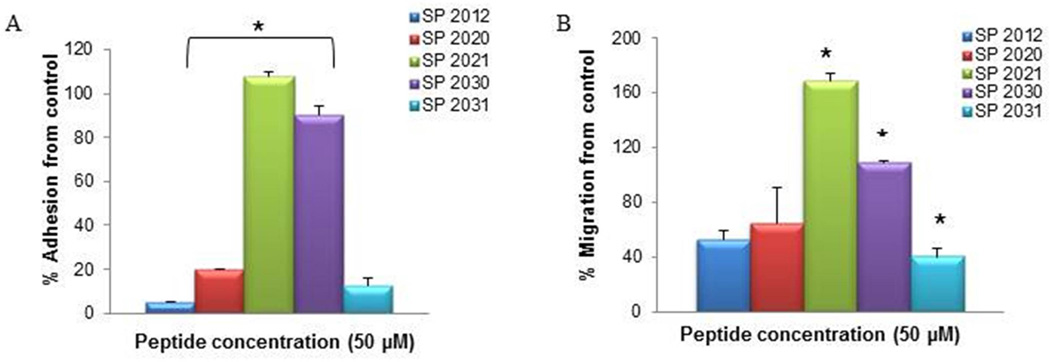
The activity in adhesion (A) and migration (B) of peptides generated by truncation of residues from both termini. * indicate statistical difference (p<0.05) in activity in comparison to the parent peptide (SP2012).
Substitutions and 2nd generation substitutions
The inhibition of adhesion and migration by peptides generated by specific substitutions are shown in Fig. 4. The inhibitory activity in the adhesion assay is quite similar to that of the parent compound (SP2012), resulting in almost complete inhibition at 50 µM. However, the activity in migration is significantly increased from 60% in the parent compound (SP2012) to > 90% in the modified peptides. Thus we investigated the activity at 5 µM, a much lower concentration, and observed that SP2022 shows similar activity to the parent compound (SP2012) while the other compounds SP2023 through SP2027 show much greater activity.
Figure 4. Adhesion and migration of peptides generated by specific substitutions.
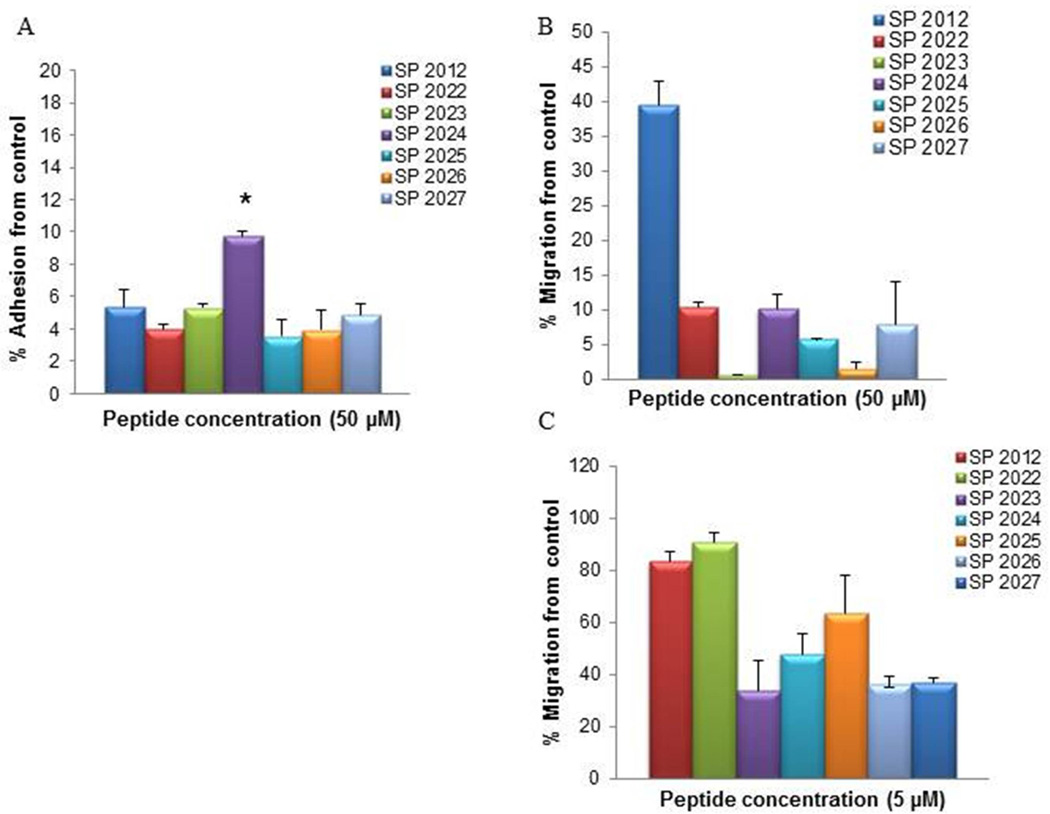
The activity in adhesion at 50 µM (A) and migration at 50 µM (B) and migration at 5 µM of peptides generated by specific substitution of the Abu residues. * indicate statistical difference (p<0.05) in activity in comparison to the parent peptide (SP2012). Note that the y-axis range is reduced to 0–20% for panel A and 0–50% for panel B.
Further introduction of methionine substitutions leads to significant increases in activity in adhesion inhibition as illustrated in Fig. 5 and 6. The parent compound (SP2012) exhibits an IC50 in adhesion inhibition of 2.39±1.55 µM, which was not affected by the introduction of the Isoleucine in lieu of the Abu residues at positions 12 and 18 (SP2024 IC50 2.35±1.3 µM). The IC50 was not affected by further substitutions of either one Methionine (position 10) or both (position 7 and 10) with Alanine (SP2035 IC50 2.51±0.88 µM and SP2036 IC50 2.75±1.22 µM). However, this activity was increased four times by the introduction of D-Alanine in lieu of the Methionine at position 7 (SP2034 IC50 0.55±0.37 µM). Similarly the inhibition of proliferation is increased from the parent SP2012 (IC50 48.1±23.1µM) by one order of magnitude in SP2024 (IC50 7.45±5.7µM) and even further by two order of magnitudes from SP2012 to SP2034 and SP2035 with IC50 0.54±0.19µM and IC50 0.98±0.38µM respectively.
Figure 5. Adhesion and migration of 2nd generation peptides.
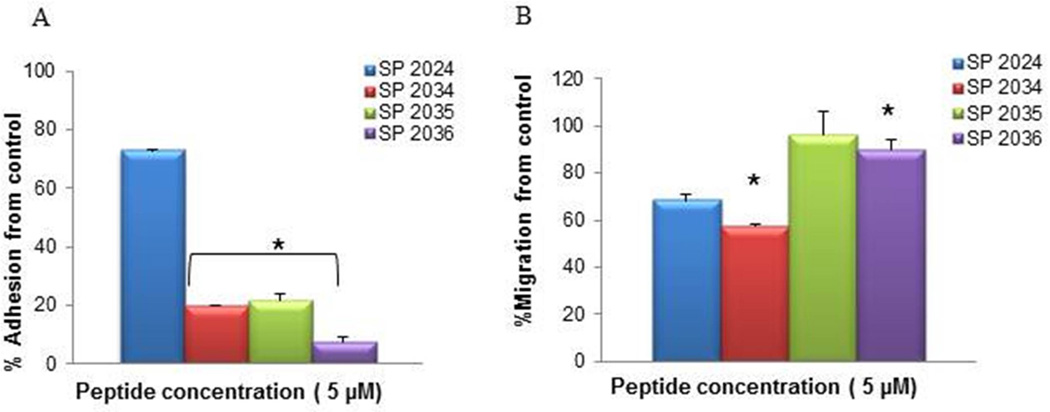
The activity in adhesion in adhesion (A) and migration (B) of the family of peptides that include the isoleucine substitution at positions 9 and 11 along with additional substitutions. * indicate statistical difference (p<0.05) in activity in comparison to the 1st generation peptide, SP2024 which contains the isoleucine substitutions.
Figure 6. Dose response curves.
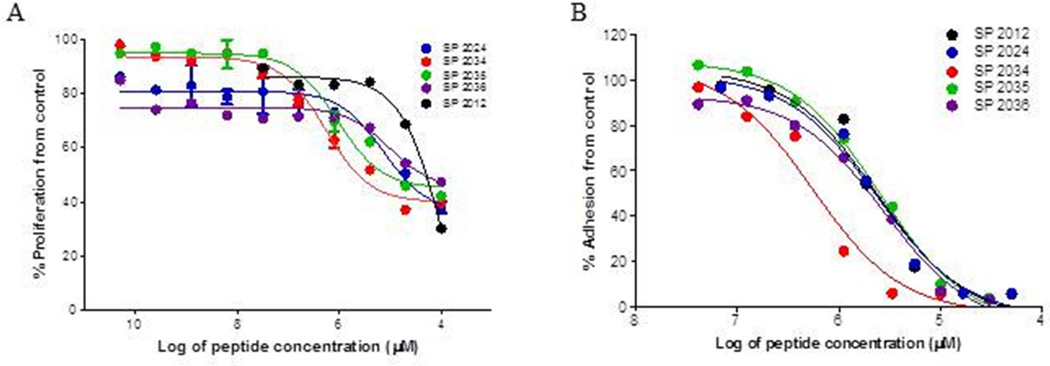
Dose response curves used to determine IC50 values for (A) proliferation activity, (B) adhesion inhibition for the most active compounds. Compound SP2034 demonstrates a significant increase (p < 0.05) in potency in comparison to the parent compound and even the 1st generation substitution SP2024.
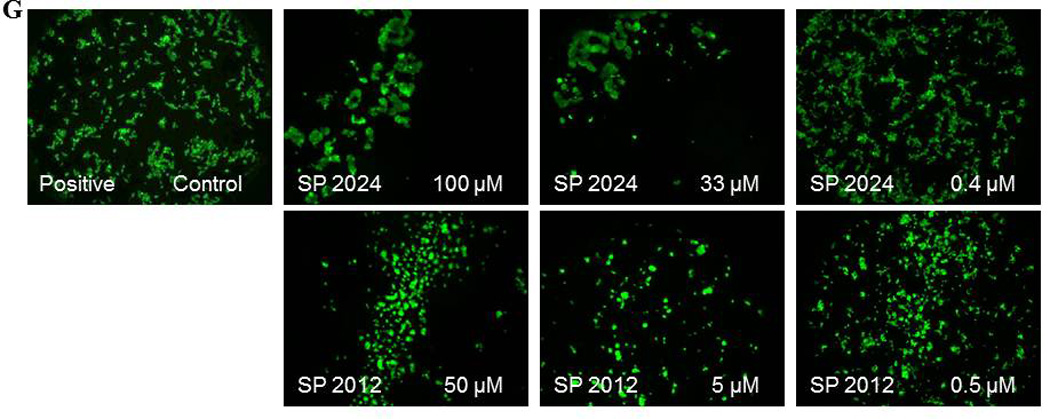
Inhibition of tubules formation
Endothelial cells spontaneously form a network of tubules when plated on extracts of extracellular matrix. This in vitro assay combines aspects of adhesion and migration and it is routinely used in angiogenesis research.(23) The ability to inhibit tubules formation is a comprehensive assessment of the anti-angiogenic potential. Figure 7 illustrates the activity of these peptides on HUVEC tubules formation. We chose to test the inhibition of tubules formation at a high dose, 100 µM so that the inhibition will be obvious thus eliminating the need for quantification. Based on the results from adhesion and migration it was expected that in the group of C-terminus truncation peptides (Figure 7, panel A) SP2028, SP2029, SP2032 and SP2033 will have activity in the inhibition of tubules formation. Surprisingly SP2006 is also active at inhibiting tubules formation, even though it was inactive in the migration assay. The compound has 18% activity in inhibiting migration which is below our threshold of at least 70% activity. However SP2006 has strong activity in adhesion assay thus it supports its activity in inhibition of tubules formation. On the other hand the N-terminus truncated SP2011 (Figure 7, panel B) shows no activity in tubules formation as expected because it is inactive in the migration assay and it only has minimal activity in adhesion. Both SP2009 and SP2010 display activity in inhibiting tubules formation as expected and are active at inhibiting adhesion or migration, respectively.
Figure 7. Inhibition of tubules formation.
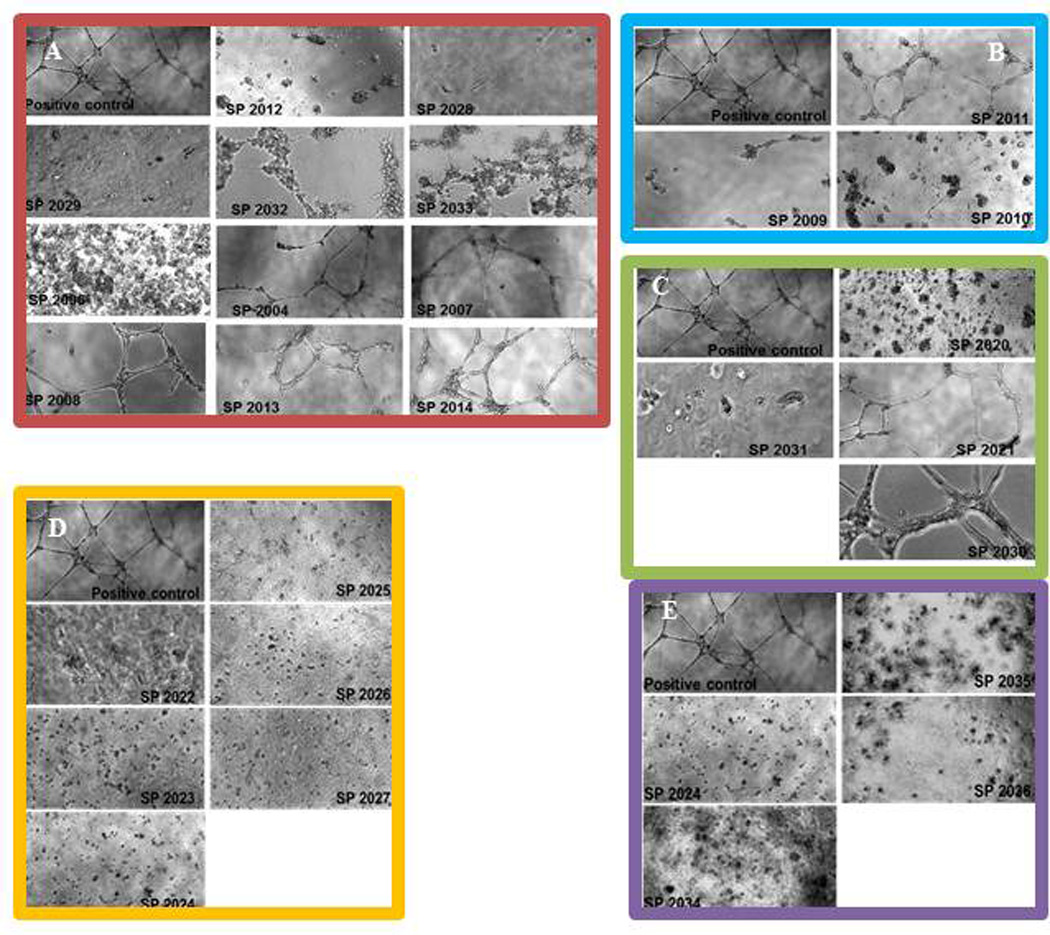
(A) activity of C-terminus truncations; (B) inhibition of the peptides generated by substitutions; (C) inhibition activity of peptides generated by truncation from both termini; (D) activity of the 2nd generation peptides; (E) inhibition activity of the peptides generated by truncations of the N-terminus residues. The positive control images are HUVEC cultured on matrigel extract in complete media without any peptide addition.
The group of peptides generated by truncations from both ends (Figure 7, panel C) includes two active compounds (SP2020 and SP2031) and two inactive compounds (SP2021 and SP2030). All compounds in the substitution and 2nd generation substitution group (Figure 7, panel D and E) are very active at inhibiting tubules formation which is expected as their potency in migration and adhesion was either increased or not affected by the substitutions.
Multimodal inhibitory activity: effects on MEC, LEC and cancer cells
Even though HUVEC remain the most common cells used in angiogenesis assays, they are derived from veins, and thus there are questions whether they are the most appropriate model for studying angiogenesis. We and others have shown that many of the in vitro assays with HUVEC and microvascular endothelial cells (MEC) exhibit qualitatively similar behaviors. (22) Here we repeated some of the experiments described above on MEC using one of the 1st generation peptide, SP2024 along with the parent peptide SP2012. These results are shown in Fig. 8 panels C and D, and they demonstrate that these peptides are quite active at inhibiting adhesion and migration of MEC. Furthermore, we tested the activity of these two peptides at inhibiting the adhesion and migration of lymphatic endothelial cells (LEC) and the MDA-MB-231 breast cancer cells. These results are depicted in Fig. 8 panels E, F, and G indicating that these peptides exhibit a strong inhibitory activity on the adhesion and migration of these cell types. Thus these peptides possess inhibitory activity towards multiple cell types thus potentially inhibiting the growth of blood and lymphatic vessels and also tumor growth and metastasis.
Figure 8. Inhibitory activity of the peptides on multiple cell types.
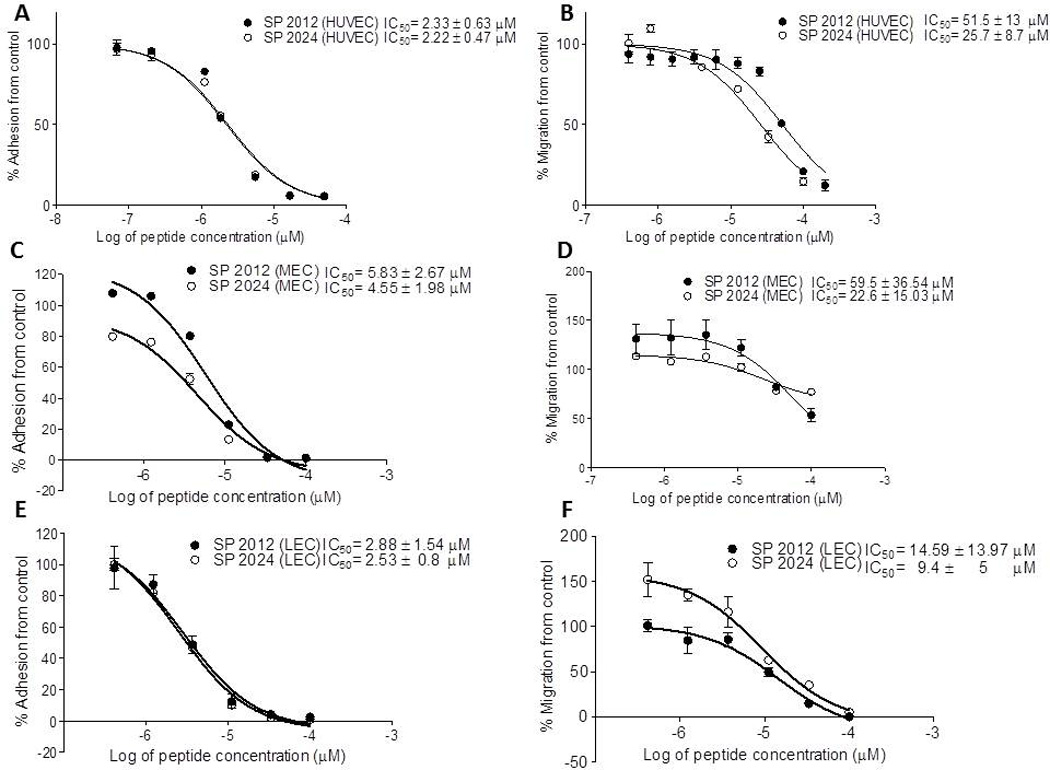
(A) and (B) inhibition of adhesion and migration on human vascular endothelial cells (HUVEC); (C) and (D) inhibition of adhesion and migration on microvascular cells (MEC); (E) and (F) inhibition of adhesion and migration on lymphatic endothelial cells (LEC); (G) migration inhibition on breast cancer cells MDA-MB-231
Conclusions and Future Directions
Peptides have emerged as therapeutic agents and many are used clinically or in clinical trials for multiple diseases (1). Although peptides can interact specifically with cellular receptors, sometimes these interactions may be of low affinity and their application is limited due to the short half-life and reduced bioavailability. Thus substitutions or introduction of non-natural amino acids are used to increase peptides activity and prolong their stability.
In this study we report the structure-activity relationship analysis of anti-angiogenic peptides whose original peptide (SP2000) was derived from the α5 fibril of type IV collagen. Our potent cysteine-free peptide (SP2012), the parent peptide for this study, was tested in vitro and in vivo xenograft mouse model of triple negative breast cancer, a form of cancer which is not amendable to conventional chemotherapeutic and hormonal treatments. (22) We have developed a small family of peptides (using truncations and substitutions of amino acids) which we screened in a series of in vitro assays: proliferation, adhesion, migration and tubules formation with macrovascular endothelial cells (HUVEC) commonly used in angiogenesis studies. We also tested selected peptides using microvascular endothelial cells (MEC), lymphatic endothelial cells (LEC) and MDA-MB-231 breast cancer cells and showed similarity of responses for these cell types. We found that the proliferation inhibition activity was retained when most of the sequence was preserved (SP2028) or when specific substitutions were introduced. Shorter fragments that still maintained activity must include the NINNV sequence which has to be coupled with additional amino acids on both sides (SP2009 vs. SP2020). Work by Eikesdal et al. investigated important residues in tumstatin, another short peptide derived from collagen IV, and identified critical residues responsible for the activity but not a contiguous region as in this case. (24) The in vitro activity was improved by two orders of magnitude in comparison to that of the parent peptide, when the length was maintained and certain substitutions were introduced (SP2034). Overall adhesion and migration profiles are similar; generally a compound active at inhibiting adhesion would also be active at inhibiting migration. Similarly the activity in the inhibition of adhesion and migration is maintained if the NINNV sequence is conserved (SP2004 vs. SP2028). However, the adjacent residues which could be flanking this region also appear to play a role in the activity of the peptide. For instance when the NINNV region is flanked with residues from the C-terminus (SP2009) the compound is active in migration and not adhesion. However, when it is flanked with residues from the N-terminus (SP2006) the activity is reversed, i.e. the compound is active in adhesion but not migration. Despite the similarities in the activity profiles of adhesion and migration there are a few exceptions, e.g. SP2006, SP2010, SP2011 compounds which are active in adhesion but not in migration and compound SP2009 which is active in migration but not adhesion. This could be due to the activity of particular residues (i.e. Valine) or perhaps to the threshold levels that were chosen to define what constitutes an active compound. As expected, full length compounds that contain specific substitutions are active in both assays and actually display a significant increase in activity in both assays. These point substitutions were guided by in silico prediction of proteolytic cleavage sites from NetChop 3.1 (http://www.cbs.dtu.dk/services/NetChop) or ExPASy Proteomics (http://expasy.org) with the intention of creating a more stable compound which should translate to an increase in potency in vivo; another quantitative SAR approach guiding the design of the peptides was based on a computational method called the least absolute shrinkage and selection operator (Lasso). (25) Experiments investigating the stability and the bioavailability of these compounds are currently underway.
Inhibition of tubules formation correlates very well with activity in either inhibition of adhesion or migration; if a compound is active in either of those two assays it consistently demonstrated activity in the inhibition of tubules formation. We have used high concentration of peptides to assess the activity in this assay so that the inhibition is complete and there is no need for quantification of the effects. Also, we observe two different phenotypes in the inhibition by these compounds; one in which the cells are clumped (e.g., Fig. 7, panel D, SP 2023-SP 2027 ), and the other in which the cells are less clumped and form fragile and short tubule (e.g., Fig.7, panel A - SP 2032 and panel E- SP2034). The clumped profile is seen with all the compounds that inhibit adhesion while the short tubule profile is associated with compounds which are active in migration. In the compounds which are active in both, adhesion and migration, the clump profile predominates however. Live-dead staining of cells treated with the compounds (results not shown) have indicated that cells which are at the bottom of the clump are dead while cells which are in contact with other cells are still alive but cannot rearrange due to their inability to migrate.
In conclusion, we presented a structure-activity relationship study for a family of anti-angiogenic peptides. We found a number of important amino acids and amino acid sequences which are important for in vitro activity of these compounds. We report that a short sequence, NINNV, is very important in the activity of this peptide in inhibiting proliferation, adhesion, migration and tubules formation in HUVEC. The results are qualitatively similar to those in MEC and LEC. We have recently demonstrated that another family of short peptides, derived from the somatotropin domain-containing proteins, inhibit angiogenic activity in LEC. (26) To the best of our knowledge, these are the only known peptides that exhibit anti-lymphangiogenic activity. Through specific and directed substitutions we were able to create compounds (SP2034 and SP2035) which exhibited a significant (two orders of magnitude) increase in activity over the activity of parent peptide. We postulate that these compounds, especially SP2034, due to the d-Alanine residue will be more stable and thus also will be more active in vivo at inhibiting tumor growth; this prediction will be tested in vivo.
Table 6.
Summary of activity for all compounds
| Peptide Name |
Peptide sequence | Activity in Proliferation |
Activity in Adhesion |
Activity in Migration |
Activity in Tube formation |
|
|---|---|---|---|---|---|---|
| SP 2012 | LRRFSTMPFMFAbuNINNVAbuNF | Yes | Yes | Yes | Yes | |
| SP 2004 | LRRFSTMPFMF | No | No | No | No | |
| SP 2006 | LRRFSTMPFMFAbuNINV | No | Yes | No | Yes | |
| SP 2007 | LRRFSTMPFMFAbu | No | No | No | No | |
| SP 2008 | LRRFSTMP | No | No | No | No | |
| SP 2013 | LNRFSTMPF | No | No | No | No | |
| SP 2014 | LRRFSTXPFXF–X= NorLeu | No | No | No | No | |
| SP 2028 | LRRFSTMPFMFAbuNINN | Yes | Yes | Yes | Yes | |
| SP 2029 | LRRFSTMPFMFTNINV | Yes | Yes | Yes | Yes | |
| SP 2032 | LRRFSTMPFMFTNINN | Yes | Yes | Yes | Yes | |
| SP 2033 | LRRFSTMPFMFININN | Yes | Yes | Yes | Yes | |
| SP 2009 | NINNVAbuNF | No | No | Yes | Yes | |
| SP 2010 | FMFAbuNINNVAbuNF | No | Yes | No | Yes | |
| SP 2011 | STMPFAbuNINNVAbuNF | No | Yes | No | No | |
| SP 2020 | FAbuNINNVAbuN | Yes | Yes | Yes | Yes | |
| SP 2021 | FAbuNIN | No | No | No | No | |
| SP 2030 | FAbuNINV | No | No | No | No | |
| SP 2031 | FTNINNVTN | Yes | Yes | Yes | Yes | |
| SP 2022 | LRRFSTMPFMF S NINNV S NF | Yes | Yes | Yes | Yes | |
| SP 2023 | LRRFSTMPFMF A NINNV A NF | Yes | Yes | Yes | Yes | |
| SP 2024 | LRRFSTMPFMF I NINNV I NF | Yes | Yes | Yes | Yes | |
| SP 2025 | LRRFSTMPFMF T NINNVT NF | Yes | Yes | Yes | Yes | |
| SP 2026 | LRRFSTMPFMF(AllyGly)NINNV(AllyGly)NF | Yes | Yes | Yes | Yes | |
| SP 2027 | LRRFSTMPFMF V NINNV V NF | Yes | Yes | Yes | Yes | |
| SP 2034 | LRRFSTMPFdA FI NINNVINF | Yes | Yes | Yes | Yes | |
| SP 2035 | LRRFSTMPFA FI NINNVINF | Yes | Yes | Yes | Yes | |
| SP 2036 | LRRFSTAPFA FI NINNVINF | Yes | Yes | Yes | Yes | |
Acknowledgments
The authors thank all members of the laboratory for insightful discussions and review of the manuscript. We are grateful to ACEA Biosciences for the use of the Personal RT-CIS system which allowed us to perform migration and adhesion experiments and in particular to Dr. Yama Abassi for detailed technical discussions. "Dr. Popel serves as the Chief Scientific Officer of AsclepiX Therapeutics, LLC. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. This work was supported by the National Institutes of Health grants R21 CA131931, R21 CA152473, R01 CA138264, the Safeway Foundation, and the Edward N. and Della L. Thome Memorial Foundation.
References
- 1.Saladin PM, Zhang BD, Reichert JM. Current trends in the clinical development of peptide therapeutics. IDrugs. 2009;12:779–784. [PubMed] [Google Scholar]
- 2.Nabors LB, Fiveash JB, Markert JM, Kekan MS, Gillespie GY, Huang Z, et al. A phase 1 trial of ABT-510 concurrent with standard chemoradiation for patients with newly diagnosed glioblastoma. Arch Neurol. 2010;67:313–319. doi: 10.1001/archneurol.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley DA, Daignault S, Ryan CJ, Dipaola RS, Smith DC, Small E, et al. Cilengitide (EMD 121974, NSC 707544) in asymptomatic metastatic castration resistant prostate cancer patients: a randomized phase II trial by the prostate cancer clinical trials consortium. Invest New Drugs. 2010 doi: 10.1007/s10637-010-9420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reardon DA, Fink KL, Mikkelsen T, Cloughesy TF, O'Neill A, Plotkin S, et al. Randomized phase II study of cilengitide, an integrin-targeting arginine-glycine-aspartic acid peptide, in recurrent glioblastoma multiforme. J Clin Oncol. 2008;26:5610–5617. doi: 10.1200/JCO.2008.16.7510. [DOI] [PubMed] [Google Scholar]
- 5.Kenny LM, Coombes RC, Oulie I, Contractor KB, Miller M, Spinks TJ, et al. Phase I trial of the positron-emitting Arg-Gly-Asp (RGD) peptide radioligand 18F-AH111585 in breast cancer patients. J Nucl Med. 2008;49:879–886. doi: 10.2967/jnumed.107.049452. [DOI] [PubMed] [Google Scholar]
- 6.Rosca EV, Koskimaki JE, Pandey NB, Rivera CG, Tamiz AP, Popel AS. Anti-angiogenic peptides for cancer therapeutics. Current Pharmaceutical Biotechnology. 2011;12:1101–1116. doi: 10.2174/138920111796117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhutia SK, Maiti TK. Targeting tumors with peptides from natural sources. Trends Biotechnol. 2008;26:210–217. doi: 10.1016/j.tibtech.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Pernot M, Vanderesse R, Frochot C, Guillemin F, Barberi-Heyob M. Stability of peptides and therapeutic success in cancer. Expert Opin Drug Metab Toxicol. 2011;7:793–802. doi: 10.1517/17425255.2011.574126. [DOI] [PubMed] [Google Scholar]
- 9.Gentilucci L, De Marco R, Cerisoli L. Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr Pharm Des. 2010;16:3185–3203. doi: 10.2174/138161210793292555. [DOI] [PubMed] [Google Scholar]
- 10.Abraham S, Guo F, Li LS, Rader C, Liu C, Barbas CF, 3rd, et al. Synthesis of the next-generation therapeutic antibodies that combine cell targeting and antibody-catalyzed prodrug activation. Proc Natl Acad Sci U S A. 2007;104:5584–5589. doi: 10.1073/pnas.0700223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruby VJ, Sharma SD, Toth K, Jaw JY, al-Obeidi F, Sawyer TK, et al. Design, synthesis, and conformation of superpotent and prolonged acting melanotropins. Ann N Y Acad Sci. 1993;680:51–63. doi: 10.1111/j.1749-6632.1993.tb19674.x. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Oliver E, Kitchens KM, Vere J, Alkan SS, Tamiz AP. Structure-activity relationship studies of permeability modulating peptide AT-1002. Bioorg Med Chem Lett. 2008;18:4584–4586. doi: 10.1016/j.bmcl.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Gautier B, Goncalves V, Diana D, Di Stasi R, Teillet F, Lenoir C, et al. Biochemical and structural analysis of the binding determinants of a vascular endothelial growth factor receptor peptidic antagonist. J Med Chem. 2010;53:4428–4440. doi: 10.1021/jm1002167. [DOI] [PubMed] [Google Scholar]
- 14.Mirochnik Y, Aurora A, Schulze-Hoepfner FT, Deabes A, Shifrin V, Beckmann R, et al. Short pigment epithelial-derived factor-derived peptide inhibits angiogenesis and tumor growth. Clin Cancer Res. 2009;15:1655–1663. doi: 10.1158/1078-0432.CCR-08-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haviv F, Bradley MF, Kalvin DM, Schneider AJ, Davidson DJ, Majest SM, et al. Thrombospondin-1 mimetic peptide inhibitors of angiogenesis and tumor growth: design, synthesis, and optimization of pharmacokinetics and biological activities. J Med Chem. 2005;48:2838–2846. doi: 10.1021/jm0401560. [DOI] [PubMed] [Google Scholar]
- 16.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 17.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 18.Karagiannis ED, Popel AS. A systematic methodology for proteome-wide identification of peptides inhibiting the proliferation and migration of endothelial cells. Proc Natl Acad Sci USA. 2008;105:13775–13780. doi: 10.1073/pnas.0803241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karagiannis ED, Popel AS. Identification of novel short peptides derived from the alpha 4, alpha 5, and alpha 6 fibrils of type IV collagen with anti-angiogenic properties. Biochem Biophys Res Commun. 2007;354:434–439. doi: 10.1016/j.bbrc.2006.12.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koskimaki JE, Karagiannis ED, Rosca EV, Vesuna F, Winnard PT, Jr, Raman V, et al. Peptides derived from type IV collagen, CXC chemokines, and thrombospondin-1 domain-containing proteins inhibit neovascularization and suppress tumor growth in MDA-MB-231 breast cancer xenografts. Neoplasia. 2009;11:1285–1291. doi: 10.1593/neo.09620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koskimaki JE, Karagiannis ED, Tang BC, Hammers H, Watkins DN, Pili R, et al. Pentastatin-1, a collagen IV derived 20-mer peptide, suppresses tumor growth in a small cell lung cancer xenograft model. BMC Cancer. 2010;10:29. doi: 10.1186/1471-2407-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosca EV, Koskimaki JE, Pandey NB, Wolff AC, Popel AS. Development of a biomimetic peptide derived from collagen IV with anti-angiogenic activity in breast cancer. Cancer Biology & Therapy. 2011;12(9):808–817. doi: 10.4161/cbt.12.9.17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnaoutova I, George J, Kleinman HK, Benton G. The endothelial cell tube formation assay on basement membrane turns 20: state of the science and the art. Angiogenesis. 2009;12:267–274. doi: 10.1007/s10456-009-9146-4. [DOI] [PubMed] [Google Scholar]
- 24.Eikesdal HP, Sugimoto H, Birrane G, Maeshima Y, Cooke VG, Kieran M, et al. Identification of amino acids essential for the antiangiogenic activity of tumstatin and its use in combination antitumor activity. Proc Natl Acad Sci U S A. 2008;105:15040–15045. doi: 10.1073/pnas.0807055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera CG, Rosca EV, Pandey NB, Koskimaki JE, Bader JS, Popel AS. Novel peptide-specific QSAR analysis applied to collagen IV peptides with antiangiogenic activity. J Med Chem. 2011;54(19):6492–6500. doi: 10.1021/jm200114f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Rosca EV, Pandey NB, Popel AS. Small peptides derived from somatotropin conserved domain-containing proteins inhibit blood and lymphatic endothelial cell proliferation, migration, adhesion and tube formation. Int J Biochem Cell Biol. 2011;43(12):1812–1821. doi: 10.1016/j.biocel.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


