Summary
Drosophila embryo dorsoventral (DV) polarity is defined by serine protease activity in the perivitelline space (PVS) between the embryonic membrane and the inner layer of the eggshell [1, 2, 3, 4, 5]. Gastrulation Defective (GD) cleaves and activates Snake (Snk). Activated Snk cleaves and activates Easter (Ea), exclusively on the ventral side of the embryo [6, 7, 8]. Activated Ea then processes Spätzle (Spz) into the activating ligand for Toll, a transmembrane receptor that is distributed throughout the embryonic plasma membrane [9]. Ventral activation of Toll depends upon the activity of the Pipe sulfotransferase in the ventral region of the follicular epithelium that surrounds the developing oocyte [10]. Pipe transfers sulfate residues to several protein components of the inner vitelline membrane layer of the eggshell [11]. Here we show that GD protein becomes localized in the ventral PVS in a Pipe-dependent process. Moreover, ventrally concentrated GD acts to promote the cleavage of Ea by Snk through an extracatalytic mechanism that is distinct from GD's proteolytic activation of Snk. Together, these observations illuminate the mechanism through which spatially restricted sulfotransferase activity in the developing egg chamber leads to localization of serine protease activity and ultimately to spatially specific activation of the Toll receptor in the Drosophila embryo.
Results and Discussion
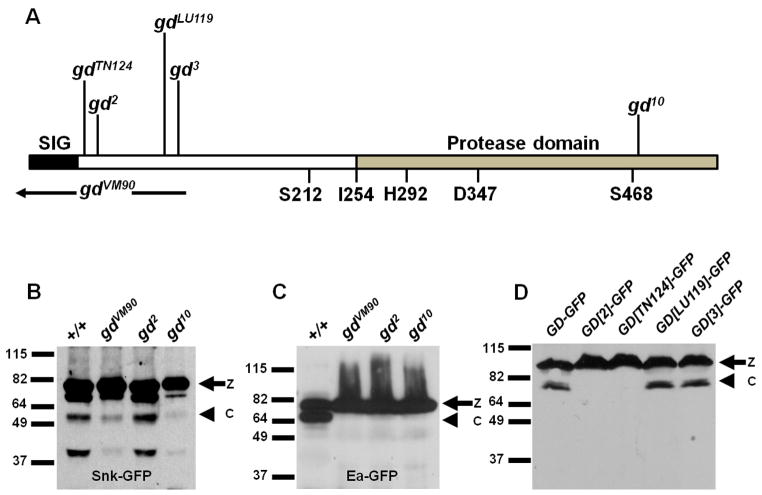
Mutations in the serine protease GD have been characterized according to their molecular lesions [12, 13, 14] (Figure 1A). Ponomareff et al. 2001 [14] additionally carried out a careful complementation analysis of available gd mutant alleles, which demonstrated the existence of two distinct classes of alleles capable of complementing one another. Missense mutations within the protease domain constitute the gd10 class while the gd2 class is comprised of missense mutations located near the amino terminal end of the protein [14]. Although females homozygous for strong gd2 or gd10 class alleles produce completely dorsalized embryos, embryos from gd2/gd10 class transheterozygotes exhibit normally polarized lateral and ventral pattern elements. The ability of gd2 class mutations to complement gd10 class mutations suggests that the gd2 class-encoded proteins retain the ability to process Snk. To test this, we examined the processing of a GFP-tagged version of Snk (Snk-GFP) [7] in the progeny of either wild-type females or females mutant for gd2, gd10 or gdVM90 [protein null, non-complementing allele] (Figure 1B). Consistent with previous observations [7], embryos maternally mutant for gdVM90 exhibited a low level of processed Snk-GFP whose formation does not depend upon GD protease activity. As expected, a similarly low level of processed Snk-GFP was observed in embryos from females carrying the protease-deficient gd10 allele. In contrast, embryos maternally mutant for gd2 exhibited a higher level of Snk-GFP processing that was comparable to the level observed in wild-type embryos. Thus, the gd2-encoded protein is capable of processing Snk, raising the question of which cleavage step in the protease cascade is affected in this mutant background. We therefore examined the processing of a GFP-tagged version of Ea (Ea-GFP) [7], which failed to undergo cleavage in all three mutant backgrounds (Figure 1C). The finding that Ea is not cleaved in gd2 maternal mutants, even though Snk is processed, suggests that in addition to processing Snk, wild-type GD functions to facilitate the processing of Ea by activated Snk. It is this second, extracatalytic function of GD that is disrupted by gd2 class mutations.
Figure 1. gd2 class mutations illuminate a role for GD in facilitating the cleavage of Ea by activated Snk.
(A) Structure of the GD protein showing the positions of the N-terminal gd2 class mutations gdTN124, gd2, gdLU119 and gd3, and the gd10 class mutation gd10 in the protease domain14. Genomic coding sequences missing from the non-complementing null mutation gdVM90 are indicated by the black bar. The gdVM90 mutation results from a 717 base pair deletion that overlaps the 5' end of the gene [14], and is likely to represent an RNA null allele (see Experimental Procedures). Also indicated are the positions of the GD signal peptide (SIG), the histidine (H292), aspartic acid (D347) and serine (S468) residues that comprise the active site of the catalytic region, and the amino acid residues to which the Ea signal peptide has been fused (S212 and I254) in the GDΔN211 and GDΔN253 constructs.
(B–D) Western blot analyses of embryonic extracts. Processing of Snk-GFP (B) and Ea- GFP (C) in embryos from females bearing the gd allele denoted above each lane carried in trans to gdVM90.
(D) Processing of GD-GFP and of mutant variants bearing the gd2 class mutations shown above each lane. z = zymogen, c = processed protease-bearing fragment.
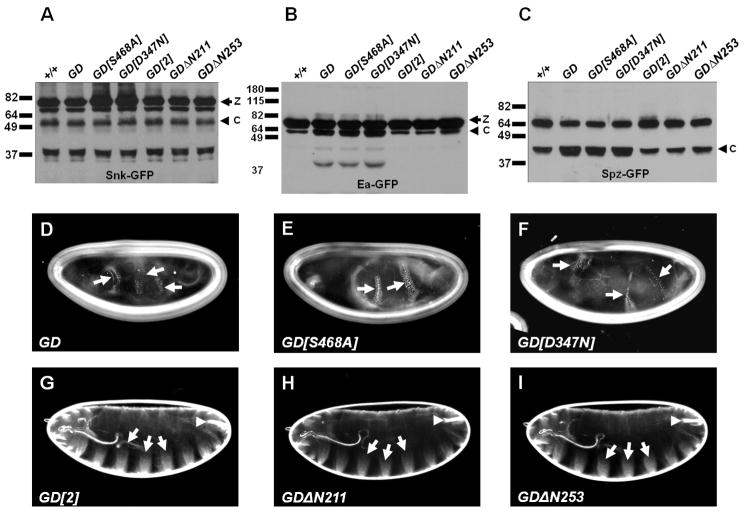
We next asked whether increasing the concentration of GD molecules that retain the extracatalytic function would increase Ea processing. We moderately overexpressed in wild-type females transgenes encoding wild-type GD, or two catalytically inactive versions of GD, GD[D347N] and GD[S468A], which each carry a mutation in one of the essential amino acids in the catalytic triad that makes up the active site of the protein [5, 15]. All three led to increases in processed Ea-GFP (Figure 2B) and Spz-GFP [7] (Figure 2C), indicating that GD proteins that lack protease activity can nevertheless promote Ea processing. All three transgenes were also capable of ventralizing the progeny of wild-type females (Figures 2D through F). Most of the cuticles produced by these embryos were completely encircled by ventral denticle belts. Skeletal elements of the larval head, and the dorsolaterally-derived Filzkörper material were absent from these embryos. These results indicate that the extracatalytic function does not require the presence of an active protease in the same GD molecule, provided there is another source of GD protease present in embryos to cleave and activate Snk. In contrast, overexpression of the mutant GD[2] protein, or of two pre-processed versions of GD, GDΔN211 and GDΔN253, in which a signal peptide is fused directly to amino acid 212 or 254 of the protein [5], failed to increase the levels of Ea-GFP or Spz-GFP processing (Figures 2B and 2C) or to ventralize the embryonic phenotype (Figures 2G, 2H and 2I). Thus, the ability to increase Ea processing in a concentration-dependent manner requires the presence of sequences within the N-terminal domain. Finally, in no case did overexpression of the tested GD transgenes lead to an increase in the levels of processed Snk-GFP above that present in wild-type embryos (Figure 2A), indicating that the effects on Ea processing were not achieved through increased levels of active Snk protease.
Figure 2. Proteolytically inactive versions of GD illuminate a role for the full-length protein in facilitating processing of Ea by Snk.
(A–C) Western blot analyses to examine the processing of Snk-GFP, Ea-GFP or Spz-GFP in embryos also containing the modified versions of GD denoted above each lane. All constructs were expressed under the control of the germline-specific driver pCOG-Gal4:VP16. (D–N) Cuticles of larval progeny of females expressing the indicated GD transgenes under the control of pCOG-Gal4:VP16. Arrows show ventral denticles or ventral denticle material; arrowheads indicate Filzkörper, a dorsolateral structure.
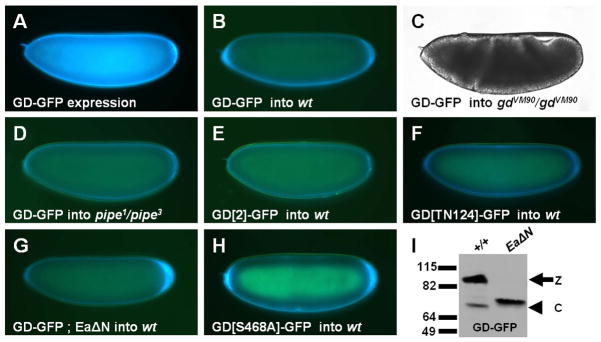
Cleavage of Ea is dependent upon the function of the Pipe sulfotransferase and is the first step in the protease cascade to be localized to the ventral side of the egg [6, 7, 8]. Previous work suggested that Pipe-sulfated, ventrally localized eggshell proteins contribute to the ventral processing of Ea by Snk [10, 11]. The observation that GD plays a role in Snk-mediated Ea processing raised the possibility that an interaction between GD and the sulfated cue in the ventral eggshell cue enables GD to bring about a productive encounter between Snk and Ea. Previous attempts to detect ventral enrichment of transgenically expressed GD-GFP in the PVS were hampered by high levels of fluorescence in the embryonic secretory pathway and throughout the PVS (Figure 3A) [16]. To eliminate this problem, we instead transplanted perivitelline fluid (PVF) from GD-GFP-expressing embryos into the PVS of wild-type, non-expressing cleavage stage embryos. Regardless of the site of injection, after 1–2 hours of incubation, recipient embryos exhibited a conspicuous accumulation of fluorescence in the ventral PVS (Figure 3B). Moreover, injection of PVF containing GD-GFP into the progeny of gd mutant females led to phenotypic rescue of polarized gastrulation movements (Figure 3C) and the differentiation of ventral and lateral cuticular pattern elements (data not shown). Strikingly, no ventral enrichment was observed following transplantation of PVF containing GD-GFP into embryos derived from pipe mutant mothers. Instead, the injected protein became uniformly distributed throughout the PVS (Figure 3D). Thus, ventral enrichment of GD-GFP requires the Pipe-sulfated ventral cue. Importantly, no ventral enrichment was observed following injection into wild-type embryos of PVF containing GFP-tagged proteins bearing any of the four gd2 class mutations (gd2, gdTN124, gd3 or gdLU119) (Figures 3E, 3F and data not shown), which lack GD's extracatalytic function. These data strongly support the hypothesis that interaction of GD with the ventral cue is essential for its ability to facilitate processing of Ea by Snk.
Figure 3. GD-GFP undergoes ventral enrichment in the egg PVS through a Pipe-dependent mechanism.
(A) Embryo from GD-GFP-expressing female. GFP signal appears blue in this and all subsequent panels displaying GFP distributions. (B–H) Recipient embryos of the denoted maternal genotypes injected with PVF containing the indicated versions of GD-GFP. (I) Western blot of GD-GFP processing in the embryos from wild-type (+/+) and EaΔN-expressing females.
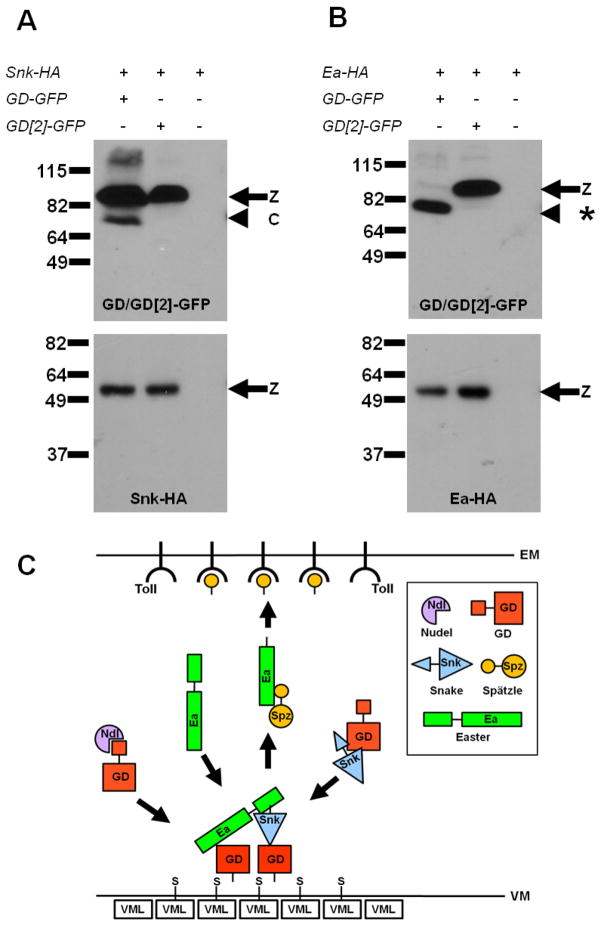
The proposed extracatalytic function of GD in promoting cleavage of Ea by Snk predicts that GD is present in a complex with Ea and Snk. To test this, we carried out co-immunoprecipitation studies and observed that GFP-tagged GD co-immunoprecipitated with both Snk-HA (Figure 4A) and Ea-HA (Figure 4B). Thus, GD is present in complexes with both downstream proteases. Interestingly, GD[2]-GFP also co-immunoprecipitated with both Snk-HA and Ea-HA (Figures 4A and 4B), suggesting that it is not the interaction of GD with Snk and Ea that requires the ventral cue but rather the productive catalytic interaction of Snk and Ea with each other.
Figure 4. GD complexes with both Ea and Snk.
(A, B) Extracts from embryos expressing Snk-HA or Ea-HA alone or together with GD-GFP or GD[2]-GFP were subjected to immune precipitation with GFP-Trap. Extracts were divided into two portions and Western blot analysis was carried out with anti-GFP (top panels) or anti-HA antibodies (bottom panels). When GD-GFP and Ea-HA are co-expressed, all of the GD-GFP zymogen is processed [*] due to a feedback mechanism [4, 5] in which high levels of activated Ea process GD-GFP. The same cleavage product is produced by EaΔN. Overexpressed GD[2]-GFP does not generate high enough levels of activated Ea to observe this effect. (C) Model for ventral processing of Ea by Snk. GD (red) is processed by Nudel (purple). GD cleaves Snk (blue). Processed GD binds to Pipe-sulfated (S) proteins in the ventral VM, including VML. Bound GD recruits and brings together Snk and Ea zymogen (green), resulting in Ea cleavage. Processed Ea cleaves Spz (yellow) to form the active Toll ligand, which binds and activates Toll in the ventral embryonic membrane (EM).
The results described above indicate that sequences in its N-terminal domain are required for ventral localization and the extracatalytic function of GD. Like the other serine proteases in this pathway, GD undergoes processing. GD zymogen is cleaved in a process that is dependent upon the Nudel serine protease [5, 7] to generate a 44–48 kD C-terminal fragment that has been considered to be the active form of GD [4, 5]. We observed processing of GD-GFP from 90 kD to 73 kD, but no cleavage of GD[2]-GFP or GD[TN124]-GFP was detected (Figure 1D). As our experiments previously demonstrated that Snk-GFP was cleaved normally in embryos from gd2 mothers (Figure 1B), this surprising result indicates that GD does not require processing to activate its proteolytic function. This finding raised the intriguing possibility that the cleavage event that GD normally undergoes is instead required for it to undergo ventral localization and exert its extracatalytic function. To test this possibility, we examined the processing and localization of GD-GFP co-expressed with EaΔN, a constitutively active version of Ea [17] that cleaves GD at an ectopic site when they are co-expressed in tissue culture cells [4, 5]. When we co-expressed GD-GFP with EaΔN in Drosophila embryos, almost all of the GD-GFP zymogen was processed to a fragment that was slightly larger than that seen when GD-GFP was expressed alone (Figure 3I). When PVF from these embryos was transplanted to the PVS of wild-type embryos, no ventral enrichment of fluorescence was observed in the injected embryos (Figure 3G). This observation is consistent with the hypothesis that processing of GD at a specific location is a prerequisite for its ventral localization, perhaps to expose determinants that interact with the ventral cue. In contrast, GFP-tagged versions of proteins bearing the two other gd2 class mutations, gd3 and gdLU119, did exhibit processed forms (Figure 1D), although they did not undergo ventral localization (data not shown). The exact site at which GD is cleaved is not known, but the size of the processed fragment is consistent with cleavage taking place between the amino acids affected by the gd2 and gdLU119 mutations (see Figure 1A). Taken together, these results suggest that the amino acids affected in gd2 and gdTN124 are required for normal processing of GD, and that cleavage of GD produces a molecule that interacts with the ventral cue through determinants that are disrupted by the gd3 and gdLU119 mutations. Notably, GD’s own proteolytic activity does not play a role in this process. GD[S468A]-GFP, a catalytically inactive version of GD-GFP in which the active site serine was converted to alanine, exhibited normal ventral enrichment when PVF containing this protein was introduced into the PVS of wild-type embryos (Figure 3H).
Our results demonstrate that GD provides two essential functions in the DV pathway: Activation of the Snk protease and ventrally localized Ea cleavage. These findings support a model in which processing of GD generates a fragment with a C-terminal protease domain and N-terminal sequences that interact with the sulfated ventral cue to localize GD to the ventral region of the PVS (Figure 4C). GD may bind directly to carbohydrates associated with vitelline membrane protein that have been sulfated as a result of Pipe enzymatic action in ventral cells of the follicle layer. Consistent with this possibility, GD protein has been shown to bind to heparin [4, 18] and to anionic components of a highly purified Drosophila eggshell matrix preparation [18].
We propose that ventrally-localized GD binds to both Ea and Snk and plays a direct role in promoting an interaction between them. It is possible that a single GD molecule binds either to Ea or to Snk and that ventral localization of GD acts to concentrate GD-bound Ea and Snk and bring them into proximity. Alternatively, GD bound only to Snk or Ea may undergo a conformational change when it interacts with the Pipe-sulfated ventral cue that results in an enhancement of Snk proteolytic activity or an increased susceptibility of Ea to cleavage by Snk. Lastly, GD bound simultaneously to both Ea and Snk may respond to the ventral cue by undergoing a conformational change that brings Snk and Ea into productive juxtaposition and results in Ea cleavage. A mechanism in which GD interacts with Pipe and the sulfated ventral cue to promote productive interaction between Ea and Snk can also explain the results of RNA injection studies carried out by Han et al. 2000 [15] and by DeLotto, 2010 [19]. Those investigators showed that injection of very high levels of in vitro synthesized RNA encoding the GD zymogen could lateralize, or reorient the polarity of embryos produced by pipe mutant females. They interpreted those results to indicate that Pipe is normally required for activation of GD on the ventral side of the embryo, but that at high concentrations, GD could become enzymatically active by an alternative Pipe-independent mechanism. Our current results suggest instead that high concentrations of GD can promote interactions between Easter and Snake, or conformational changes in those proteins that lead to Snake-mediated cleavage of Easter, even in the absence of Pipe and the ventral cue. In conclusion, by demonstrating that the GD serine protease is localized within the ventral PVS in a Pipe-dependent manner and that the interaction of GD with the Pipe-sulfated ventral cue enables it to bring about the ventrally restricted processing of Easter, the work reported here explains how ventrally localized sulfotransferase activity in the follicle cell layer leads to spatially localized activation of the Toll receptor and to the formation of the Drosophila embryonic DV axis.
Experimental Procedures
Drosophila strains and maintenance
All stocks were maintained employing standard conditions and procedures. The wild-type Drosophila melanogaster stock used was a w/w mutant derivative of Oregon R. Stocks bearing the following mutations are described in more detail on Flybase (http://flybase.org/): gd2, gd3, gdLU119, gdTN124, gdVM90, gdVO27 (bearing the same amino acid change as gd10, G469E. For simplicity, we refer to this mutant as gd10 throughout this manuscript), pip1 (formerly pip386), pip3. The strains carrying the Gal4 driver insertions nos-Gal4:VP16 and pCOG-Gal4:VP16 are described in Rorth, (1998) [20]. Stocks carrying the following transgenes have been described in Cho et al. (2010) [7]: pUASp-Ea-GFP, pUASp-GD-GFP, pUASp-Snk-GFP, pUASp-Spz-GFP, pUASp-Ea-HA, pUASp-Snk-HA.
Reverse transcription polymerase chain reaction of RNA
40 pairs of ovaries each were dissected from gdVM90/gdVM90 females and from w/w, Oregon R females, respectively. Total RNA was isolated from the ovaries using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). 5 micrograms of total RNA were subjected to reverse transcription using the SupersScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen Life Technologies, Carlsbad, CA). One tenth of the cDNA produced was then subjected to PCR-mediated amplification using the following two oligonucleotides:
5'-TCGATCACCAGGGGATCGTGGCCTTGGC-3'
5'-TCAAATTACAAAGGCCGTGATCCAGTCCAG-3'
An 831 base pair amplification produce corresponding to the serine protease catalytic domain was produced using cDNA from w/w, Oregon R-derived females. No amplification product was observed using the cDNA from gdVM90/gdVM90 females.
Plasmid constructs
For the construction of pUASp-GD[2]-GFP, pUASp-GD[TN124]-GFP, pUASp-GD[LU119]-GFP and pUASp-GD[3]-GFP the two oligonucleotides:
5'-ATTCCCGCGGCCGCAAAATGAGGCTGCACCTGGCGGCGATCC-3' and 5'-CCGTGGATACGCACCGCCGGATCCG-3' were used for the PCR mediated amplification of genomic DNA isolated from adult female flies homozygous for the gd2 gdTN124, gdLU119 and gd3 mutations, respectively. This resulted in the amplification of 447 bp DNA fragments bearing each of the four mutations. The DNA fragments were digested with NotI and BamHI and the resultant fragments were ligated to similarly digested pUASp-GD-GFP [7].
For the construction of pUASp-EaΔN, the two oligonucleotides:
5'-CCGATTGCGGCCGCAAAATGCTAAAGCCATCGATTATCTG-3' and 5'-GGACATTCTAGATCAGGACTCAATAGTGTTTTG-3' were used for PCR-mediated amplification of a DNA fragment carrying the EaΔN cDNA construct5. The resultant PCR product was digested with NotI and XbaI and ligated to similarly digested pUASp [20].
For the construction of pUASp-GD, the two oligonucleotides:
5'-ATTCCCGCGGCCGCAAAATGAGGCTGCACCTGGCGGCGATCC-3' and 5'-ACACATTCTAGATGTGATTCAAATTACAAAGGCCG-3' were used for PCR mediated amplification of a DNA fragment carrying the gd cDNA. The resultant PCR product was digested with NotI and XbaI and ligated to similarly digested pUASp. For the construction of pUASp-GD[2], the NotI/BamHI DNA fragment bearing the gd2 mutation described above was ligated to NotI/BamHI digested PUASp-GD. For the construction of pUASp-GD[D347N], and pUASp-GD[S468A] the same two oligonucleotides were used for PCR mediated amplification of gd coding sequences from the plasmids pRMHA-3N-GDDN, and pRMHA-3N-GDSA, respectively (kind gifts of Dr. Ellen LeMosy) [5]. In both cases the resultant PCR fragment was digested with NotI and XbaI and ligated to similarly digested pUASp.
In the case of pUASp-GD[S468A]-GFP the two oligonucleotides:
5'-ATTCCCGCGGCCGCAAAATGAGGCTGCACCTGGCGGCGATCC-3' and 5'-GATGTGAGATCTATTACAAAGGCCGTGATCCAG-3' were used for PCR-mediated amplification of gd coding sequences from pRMHA-3N-GDSA [5]. The resultant PCR fragment was digested with NotI and BglII and ligated to NotI/BamHI digested pUASp-GFP [7].
For the construction of pUASp-GDΔN211 and pUASP-GDΔN253, the two oligonucleotides:
5'-CCGATTGCGGCCGCAAAATGCTAAAGCCATCGATTATCTG-3' (encoding the amino terminus and signal peptide of Ea) and
5'-ACACATTCTAGATGTGATTCAAATTACAAAGGCCG-3' were used for PCR mediated amplification of gd coding sequences from the plasmids GD-212 and GD-254 respectively (kind gifts of Dr. Ellen LeMosy) [5]. In both cases the resultant PCR fragment was digested with NotI and XbaI and ligated to similarly digested pUASp.
For the construction of pUASp-GD-HA, the two oligonucleotides:
'-ATTCCCGCGGCCGCAAAATGAGGCTGCACCTGGCGGCGATCC-3' and 5'-GGATGTTCTAGAAATTACAAAGGCCGTGATCCAGTCC-3' were used for PCR mediated amplification of a DNA fragment carrying the gd cDNA. The resultant PCR product was digested with NotI and XbaI and ligated to similarly digested pUASp-HA [7], a pUASp derivative that carries three tandemly arranged copies of the HA epitope of Influenza Hemagglutinin inserted at the XbaI site of pUASp.
Transgenic fly lines carrying insertions of the constructs described above were generated by conventional P-element mediated transformation [21].
Perivitelline injection
The various GFP-tagged versions of GD examined in these studies were expressed in the germline of adult females under the control of the nos-Gal4:VP16 driver line. PVF was obtained from the progeny of these females at gastrula or germband extension stages and injected into the dorsal or ventral PVS of cleavage/early syncytial blastoderm stage embryos as described in Stein et al. (1991) [22] and Stein and Nüsslein-Volhard (1992) [23]. Embryos were allowed to incubate for approximately 1.5 hours at room temperature, then subjected to fluorescence or brightfield microscopy on a Zeiss Axioplan2 imaging microscope.
Embryonic phenotypes
Larval cuticles were prepared according to Van der Meer (1977) [24].
Western blotting and Immunoprecipitation
Collection of eggs, preparation of extracts, determination of protein concentrations in extracts, and Western Blot analysis of tagged versions of GD, Snk, Ea and Spz were carried out as described in the Supplemental Information accompanying Cho et al. 2010 [7]. Following electroblotting to nitrocellulose membranes, blots were incubated with monoclonal primary antibodies against either GFP (1/1000)(Monoclonal B-2, catalogue no. sc-9996, Santa Cruz Biotechnology, Santa Cruz, CA) or against the HA epitope (1/1000) (Monoclonal 16B12, catalogue no. MMS-101P, Covance Inc., Emeryville, CA). Blots were washed and incubated with Goat Anti-Mouse IgG (1/5000) (cat. no. 31430, Thermo Scientific, Rockford, IL), followed by detection using the Pierce Super Signal Detection System (Pierce, Rockford, IL).
Co-Immunoprecipitation and Immunoblotting
For co-immunoprecipitation studies, embryos were collected 2–4 hours after egg deposition and homogenized as for Western blot analysis. Following determination of protein concentrations, a volume of extract containing 100 μg protein was incubated with GFP-Trap beads (Chromotek, GMBH, Martinsried, Germany) at 4°C for 2 hours as described by the manufacturer. Precipitates were then divided into two aliquots and subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblotting with monoclonal anti-GFP and monoclonal anti-HA, respectively, as described above.
Highlights.
Gastrulation Defective becomes enriched ventrally in the egg perivitelline space
Localization of GD depends on Pipe and on determinants near the N terminus of GD
GD facilitates the processing of Easter independent of its role in processing Snake
GD cleavage is not needed to process Snake but may be required for GD localization
Acknowledgments
We are grateful to Robert DeLotto, Carl Hashimoto, Ellen LeMosy and Trudi Schüpbach for providing Drosophila stocks and/or DNA clones. This work was supported by a grant from the National Institutes of Health (GM077337).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morisato D, Anderson KV. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu Rev Genet. 2001;29:371–399. doi: 10.1146/annurev.ge.29.120195.002103. [DOI] [PubMed] [Google Scholar]
- 2.Roth S. The origin of dorsoventral polarity in Drosophila. Phil Trans R Soc Lond B. 2003;358:1317–1329. doi: 10.1098/rstb.2003.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moussian B, Roth S. Dorsoventral Axis Formation in the Drosophila Embryo- Shaping Transducing a Morphogen Gradient. Curr Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Dissing M, Giordano H, DeLotto R. Autoproteolysis and feedback in a protease cascade directing Drosophila dorsal-ventral cell fate. EMBO J. 2001;20:2387–2393. doi: 10.1093/emboj/20.10.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeMosy EK, Tan YQ, Hashimoto C. Activation of a protease cascade involved in patterning the Drosophila embryo. Proc Nat Acad Sci USA. 2001;98:5055–5060. doi: 10.1073/pnas.081026598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeMosy EK. Spatially dependent activation of the patterning protease, Easter. FEBS Lett. 2006;580:2269–2272. doi: 10.1016/j.febslet.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho YS, Stevens LM, Stein D. Pipe-dependent ventral processing of Easter by Snake is the defining step in Drosophila embryo DV axis formation. Curr Biol. 2010;20:1133–1137. doi: 10.1016/j.cub.2010.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen P, Tian S, Tully SE, Crvatt BF, LeMosy EK. Activation of Snake in a serine protease cascade that defines the dorsoventral axis is atypical and pipe-independent in Drosophila embryos. FEBS Lett. 2010;584:3557–3560. doi: 10.1016/j.febslet.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto C, Gerttula S, Anderson KV. Plasma membrane localization of the Toll protein in the syncytial Drosophila embryo: importance of transmembrane signaling for dorsal-ventral pattern formation. Development. 1991;111:1021–1028. doi: 10.1242/dev.111.4.1021. [DOI] [PubMed] [Google Scholar]
- 10.Sen J, Goltz JS, Stevens L, Stein D. Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal- ventral polarity. Cell. 1998;95:471–481. doi: 10.1016/s0092-8674(00)81615-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Stevens LM, Stein D. Sulfation of eggshell components by Pipe defines dorsal-ventral polarity in the Drosophila embryo. Curr Biol. 2009;19:1200–1205. doi: 10.1016/j.cub.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konrad K, Goralski T, Mahowald A. Developmental genetics of the gastrulation defective locus in Drosophila melanogaster. Dev Biol. 1988;127:133–142. doi: 10.1016/0012-1606(88)90195-9. [DOI] [PubMed] [Google Scholar]
- 13.Konrad KD, Goralski TJ, Mahowald AP, Marsh JL. The gastrulation defective gene of Drosophila melanogaster is a member of the serine protease superfamily. Proc Natl Acad Sci USA. 1998;95:6819–6824. doi: 10.1073/pnas.95.12.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponomareff G, Giordano H, DeLotto Y, DeLotto R. Interallelic complementation at the Drosophila melanogaster gastrulation defective locus defines discrete functional domains of the protein. Genetics. 2001;159:635–645. doi: 10.1093/genetics/159.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JH, Si HL, Tan YQ, LeMosy EK, Hashimoto C. Gastrulation Defective is a serine protease involved in activating the receptor Toll to polarize the Drosophila embryo. Proc Natl Acad Sci USA. 2000;97:9093–9097. doi: 10.1073/pnas.97.16.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein D, Charatsi I, Cho YS, Zhang Z, Nguyen J, DeLotto R, Luschnig S, Moussian B. Localization and activation of the Drosophila protease Easter require the ER-resident Saposin-like protein Seele. Curr Biol. 2010;20:1953–1958. doi: 10.1016/j.cub.2010.09.069. [DOI] [PubMed] [Google Scholar]
- 17.Chasan R, Jin Y, Anderson KV. Activation of the Easter zymogen is regulated by five other genes to define dorsal-ventral polarity in the Drosophila embryo. Development. 1992;115:607–616. doi: 10.1242/dev.115.2.607. [DOI] [PubMed] [Google Scholar]
- 18.Sukumari-Ramesh S, LeMosy EK. Gastrulation defective protease interacts with anionic components of the Drosophila ovary extracellular matrix. Protein Pept Lett. 2009;16:437–443. doi: 10.2174/092986609787848135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLotto R. Gastrulation defective, a complement factor C2/B-like protease, interprets a ventral pre-pattern in Drosophila. EMBO Rep. 2001;2:721–726. doi: 10.1093/embo-reports/kve153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rorth P. Gal4 in the Drosophila female germ line. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 21.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable elements. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 22.Stein D, Roth S, Vogelsang E, Nüsslein-Volhard C. The polarity of the dorsoventral axis in the Drosophila embryo is defined by an extracellular signal. Cell. 1991;65:725–735. doi: 10.1016/0092-8674(91)90381-8. [DOI] [PubMed] [Google Scholar]
- 23.Stein D, Nüsslein-Volhard C. Multiple extracellular activities in Drosophila egg perivitelline fluid are required for the establishment of embryonic dorsal-ventral polarity. Cell. 1992;68:429–440. doi: 10.1016/0092-8674(92)90181-b. [DOI] [PubMed] [Google Scholar]
- 24.van der Meer JM. Optical clean and permanent whole mount preparations for phase-contrast microscopy of cuticular structures of insect larvae. Dros Inf Serv. 1977;52:160. [Google Scholar]