Summary
Ehrlichia chaffeensis infects monocytes/macrophages and causes human monocytic ehrlichiosis. To determine the role of type IV secretion (T4S) system in infection, candidates for T4S effectors were identified by bacterial two-hybrid screening of E. chaffeensis hypothetical proteins with positively charged C-terminus using E. chaffeensis VirD4 as bait. Of three potential T4S effectors, ECH0825 was highly upregulated early during exponential growth in a human monocytic cell line. ECH0825 was translocated from the bacterium into the host-cell cytoplasm and localized to mitochondria. Delivery of anti-ECH0825 into infected host cells significantly reduced bacterial infection. Ectopically expressed ECH0825 also localized to mitochondria and inhibited apoptosis of transfected cells in response to etoposide treatment. In double transformed yeast, ECH0825 localized to mitochondria and inhibited human Bax-induced apoptosis. Mitochondrial manganese superoxide dismutase (MnSOD) was increased over 9-fold in E. chaffeensis-infected cells, and the amount of reactive oxygen species (ROS) in infected cells was significantly lower than that in uninfected cells. Similarly, MnSOD was upregulated and the ROS level was reduced in ECH0825-transfected cells. These data suggest that, by upregulating MnSOD, ECH0825 prevents ROS-induced cellular damage and apoptosis to allow intracellular infection. This is the first example of host ROS levels linked to a bacterial T4S effector.
Keywords: Ehrlichia chaffeensis, type IV secretion effector, mitochondria, apoptosis, BAX, ROS, MnSOD
Introduction
Human monocytic ehrlichiosis is a tick-borne infectious disease characterized by fever, headache, myalgia, anorexia, and chills, and it is frequently accompanied by leukopenia, thrombocytopenia, anemia, and elevated levels of serum hepatic aminotransferases. Disease severity varies from asymptomatic to death, and severe morbidity is frequently documented (Maeda et al., 1987, Paddock et al., 2003). The etiologic agent of human monocytic ehrlichiosis, Ehrlichia chaffeensis, is a Gram-negative obligatory intracellular bacterium of the order Rickettsiales (Dawson et al., 1991). E. chaffeensis has unique tropism to infect human monocytes/macrophages, and it proliferates in membrane-bound inclusions in the host-cell cytoplasm, forming characteristic mulberry-like bacterial aggregates called morulae (Rikihisa, 2010a).
Human monocytes/macrophages are the first-line defense cells of innate immunity, and they are equipped with powerful antimicrobial mechanisms, including the phagocytosis and lysosomal destruction of invading pathogens and production of reactive oxygen species (ROS) (Cohen, 1994, Slauch, 2011). To evade these defenses, E. chaffeensis proliferates in an early endosome-like compartment containing early endosome antigen 1, transferrin receptor, transferrin, Rab5, and vacuolar-type H+ ATPase but not lysosomal markers or NADPH oxidase (Barnewall et al., 1997, Mott et al., 1999, Lin et al., 2007b). Having lost genes encoding lipopolysaccharide and peptidoglycan, E. chaffeensis prevents the activation of innate immunity by phagocytes and facilitates its adaptation to leukocytes and cells of the tick vector (Lin et al., 2003, Dunning Hotopp et al., 2006). In addition, E. chaffeensis does not induce superoxide generation in human monocytes and can block the production of superoxide by membrane NADPH oxidase in human monocytes in response to exogenous stimuli (Lin et al., 2007b).
Although E. chaffeensis subverts or overcomes several host antimicrobial defense mechanisms (Rikihisa, 2010b, Rikihisa, 2010a), the bacterial virulence factors responsible for the defense have not been documented. One of the important virulence factors for intracellular infection is the bacterial protein secretion system that directly delivers bacterial effector proteins into host eukaryotic cells (Rego et al., 2010). E. chaffeensis has genes encoding the type IV secretion (T4S) apparatus, which are expressed in the human acute leukemia monocytic cell line, THP-1, as well as in tick cells (Cheng et al., 2008, Bao et al., 2009, Rikihisa et al., 2010). However, T4S effectors of E. chaffeensis and their functions have not been examined except for a putative T4S effector, AnkA (Zhu et al., 2009, Rikihisa et al., 2010).
In this study, we identified three E. chaffeensis T4S effector candidates and characterized the biological activity of one of them, ECH0825, which was translocated from the bacterium to host cells, targeting host mitochondria. The study revealed a novel mechanism of inhibition of mitochondria-mediated apoptosis by ECH0825 through upregulation of the mitochondrial matrix protein manganese superoxide dismutase (MnSOD) and relieving ROS stress.
Results
Identification and characteristics of three putative E. chaffeensis T4S effectors
E. chaffeensis has genes encoding components of the T4S apparatus that are homologous to those of Anaplasma phagocytophilum and other members of α-Proteobacteria (Ohashi et al., 2002, Dunning Hotopp et al., 2006). VirD4 of Agrobacterium tumefaciens, which is the model organism for studying the T4S system, localizes to the bacterial inner membrane and is regarded as a coupling protein because it can bind T4S effectors to be delivered by the T4S apparatus (Cascales et al., 2004). In A. tumefaciens, the C-terminal transport signal for recruitment and translocation of T4S effector proteins is hydrophilic and has a net positive charge with a consensus motif of R-X(7)-R-X-X-R-X-X(n) (Vergunst et al., 2005). Two T4S effectors of A. phagocytophilum contain these characteristic C-terminal signals (Lin et al., 2007a, Niu et al., 2010); one effector, Ats-1, has been identified using A. phagocytophilum VirD4 as bait to screen an Anaplasma genomic prey library with the bacterial two-hybrid system (Niu et al., 2010). To identify T4S effectors of E. chaffeensis by the bacterial two-hybrid system, full-length E. chaffeensis virD4 (2,145 bp, GenBank YP_506872) was cloned into the bait plasmid. Ten selected genes encoding hypothetical proteins or conserved hypothetical proteins with a positively charged C-terminus from the E. chaffeensis genome were cloned into the prey plasmids (Table S1). The bait and each of the prey plasmids was co-transformed into the BacterioMatch II reporter strain and screened by M9 His-selective medium. This screen identified three T4S effector candidates: ECH0261 (GenBank YP_507082), ECH0767 (GenBank YP_507483), and ECH0825 (GenBank YP_507621) (Table S1). When the sequence encoding the C-terminal 250 amino acid residues of ECH0825 was cloned into the prey plasmid, the product interacted with VirD4; this was not the case, however, for the C-terminal 100 residues (Table S1). These results suggested that the central region of ECH0825 is required for interaction with VirD4. The three genes encoding T4S candidates were expressed by E. chaffeensis in THP-1 cells (Fig. 1A).
Fig. 1.
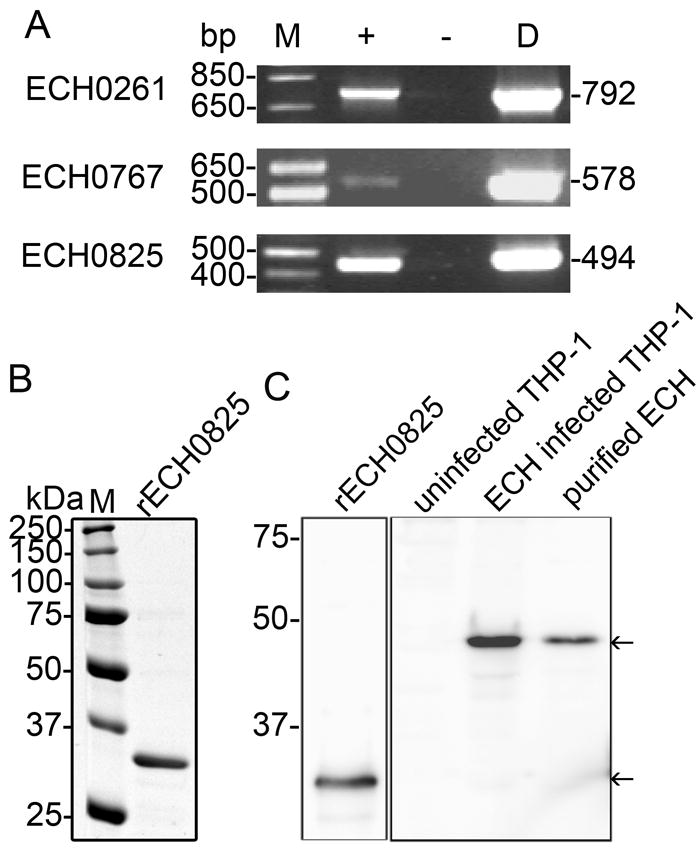
Three putative T4S effectors, including ECH0825, are expressed by E. chaffeensis in THP-1 cells.
A. ECH0261, ECH0767, and ECH0825 are transcribed by E. chaffeensis in THP-1 cells. M, molecular size marker; + and − indicate the presence or absence of reverse transcriptase, respectively; D, positive control (chromosomal DNA used as template) for PCR. Corresponding genes and sizes (in base pairs) of amplified products are indicated. No amplicon was detected without reverse transcriptase, indicating that the RNA preparation was not contaminated with genomic DNA.
B. rECH0825 (C-terminal 250 residues of ECH0825) expressed in E. coli was purified by immobilized Ni2+ affinity chromatography and subjected to SDS-PAGE followed by GelCode blue staining. M, protein molecular mass marker.
C. Western blot analysis of rECH0825, uninfected THP-1 cells, E. chaffeensis (ECH)-infected THP-1 cells, and purified ECH organisms using rabbit anti-ECH0825 antiserum. Molecular mass markers are indicated at the left. Arrows indicate rECH0825 and native ECH0825.
Presence of eukaryotic protein domains, and low total and C-terminal hydropathy values are important features of putative T4S substrates of A. marginale, Legionella pneumophila, and Bartonella henselae (Lockwood et al., 2011). Our bioinformatic analysis did not reveal any eukaryotic domains or conserved repeats in the three E. chaffeensis proteins (Table S2). Only ECH0825 had a total hydropathy value of less than -200, which is a typical characteristic for T4S substrates of A. marginale, L. pneumophila, and B. henselae (Lockwood et al., 2011). Genes encoding the three proteins were found to be conserved among the genus Ehrlichia. In addition ECH0825 had orthologous proteins in the genus Anaplasma, suggesting it has an important role in infection of broader members of the family Anaplasmataceae (Table S2). Taken together with our quantitative proteomic analysis data that show high expression of ECH0825 in mammalian cells (Lin et al., 2011), our subsequent experiments focused on determination of the putative T4S effector ECH0825 biological activity in mammalian cells.
ECH0825 is upregulated early during exponential growth
To confirm the expression of ECH0825 protein by E. chaffeensis in infected THP-1 cells, recombinant ECH0825 (rECH0825, C-terminal 250 residues) was affinity-purified (Fig. 1B) and used to immunize rabbits to produce anti-ECH0825 serum. Affinity-purified rabbit anti-ECH0825 IgG recognized rECH0825 at 30 kDa (Fig. 1C) and full-length ECH0825 in E. chaffeensis-infected THP-1 cells or in E. chaffeensis cells at 43 kDa (Fig. 1C). No immunoreactive proteins were detected in uninfected THP-1 cells, confirming the specificity of the ECH0825 antibody. In infected cells, in addition to the major 43-kDa band, weak degraded or cleaved smaller-sized bands were detected (Fig. 1C).
While infecting human cells, E. chaffeensis undergoes a biphasic developmental cycle (Rikihisa, 2010a). Synchronized cultures of E. chaffeensis in THP-1 cells showed that after E. chaffeensis entered host cells, the morula size increased until 40 h post-infection (p.i.), when larger morulae appeared to break up into several smaller ones (Fig. 2A). These small morulae subsequently increased in size until the infected cells ruptured at 76 h p.i. (Fig. 2A). Real-time PCR results revealed four stages of E. chaffeensis growth: lag stage, 0~27 h p.i.; pre-exponential growth stage, 27~52 h p.i.; exponential growths stage, 52~70 h p.i.; and stationary growth stage, 70~76 h p.i. Under our culture conditions, the number of bacteria increased over 600-fold at 76 h p.i. compared to 0 h p.i. (Fig. 2B).
Fig. 2.
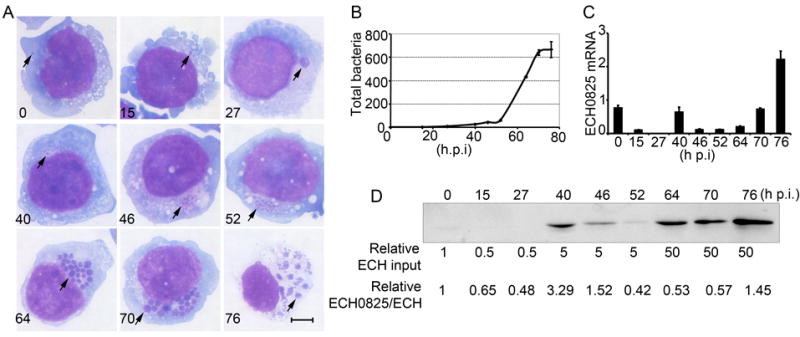
E. chaffeensis growth and temporal expression of ECH0825.
A. E. chaffeensis developmental cycle in synchronously infected THP-1 cells was examined by Diff-Quik staining. Arrows indicate bacteria or morulae. Scale bar: 5 μm.
B. Synchronous growth of E. chaffeensis determined by quantitative PCR. Genomic DNA extracted from infected THP-1 cells at different times p.i. was subjected to real-time PCR analysis. The data indicate the numbers of bacteria relative to the number at 0 h p.i. Data are expressed as the mean ± standard deviation (n = 3) and are representative of two independent experiments with similar results.
C. Temporal expression of ECH0825 in E. chaffeensis-infected THP-1 cells as determined by quantitative RT-PCR. Transcript amounts were normalized to the E. chaffeensis 16S rRNA gene. Data are expressed as the mean ± standard deviation (n = 3) and are representative of two independent experiments with similar results.
D. Temporal expression of ECH0825 in E. chaffeensis-infected THP-1 cells. Protein samples were subjected to western blot analysis based on bacteria number as determined by quantitative PCR. ECH input: relative ratios of E. chaffeensis loaded in SDS-PAGE wells.
The amount of ECH0825 transcript, normalized by bacterial number based on 16S rDNA level, was highest at 76 h p.i. with a small peak at 40 h p.i. (Fig. 2C). To determine ECH0825 protein levels in synchronously cultured E. chaffeensis, samples were loaded proportional to bacterial numbers as determined by quantitative PCR, and subjected to Western blotting. Protein band density for each sample was measured by densitometry and normalized by bacterial numbers. ECH0825 protein expression relative to bacterial DNA peaked at 40 h p.i. (Fig. 2D), corresponding to the pre-exponential growth stage.
ECH0825 is secreted into host cells and localizes to mitochondria
Double immunofluorescence labeling of E. chaffeensis and ECH0825 in E. chaffeensis-infected THP-1 cells showed that most of ECH0825 localized to bacterial inclusions, but a portion of ECH0825 was detected outside of inclusions, which was evident at 40 – 46 h p.i., corresponding to the pre-exponential growth stage (Fig. 3). Bioinformatic analysis using Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html) suggested that ECH0825 likely targets mitochondria with a probability score of 0.84 (range: 0 – 1.0). To determine the mitochondrial localization of secreted ECH0825 in THP-1 cells, immunofluorescence labeling was performed and examined using confocal microscopy. Using MnSOD as a mitochondrial marker, secreted ECH0825 was shown to localize to mitochondria at the beginning of exponential growth of E. chaffeensis in infected THP-1 cells (Figs. 4A and 4B). ECH0825 was also targeted to mitochondria in E. chaffeensis-infected human peripheral blood monocytes (Fig. 4C).
Fig. 3.
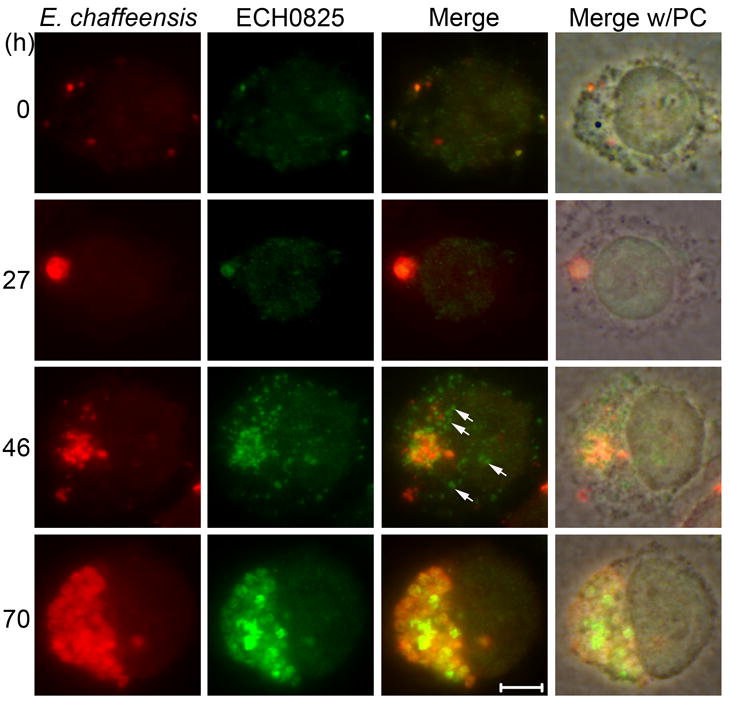
Translocation of ECH0825 from E. chaffeensis into the cytoplasm of THP-1 cells.
E. chaffeensis infected cells were subjected to double immunofluorescence labeling using dog anti-E. chaffeensis (Texas Red, red) and rabbit anti-ECH0825 (ECH0825; AF488, green) at different times p.i. Merge, merged images; Merge w/PC, image merged with phase contrast. Arrows indicate ECH0825 translocated to the cytoplasm of host cells. Scale bar: 5 μm.
Fig. 4.
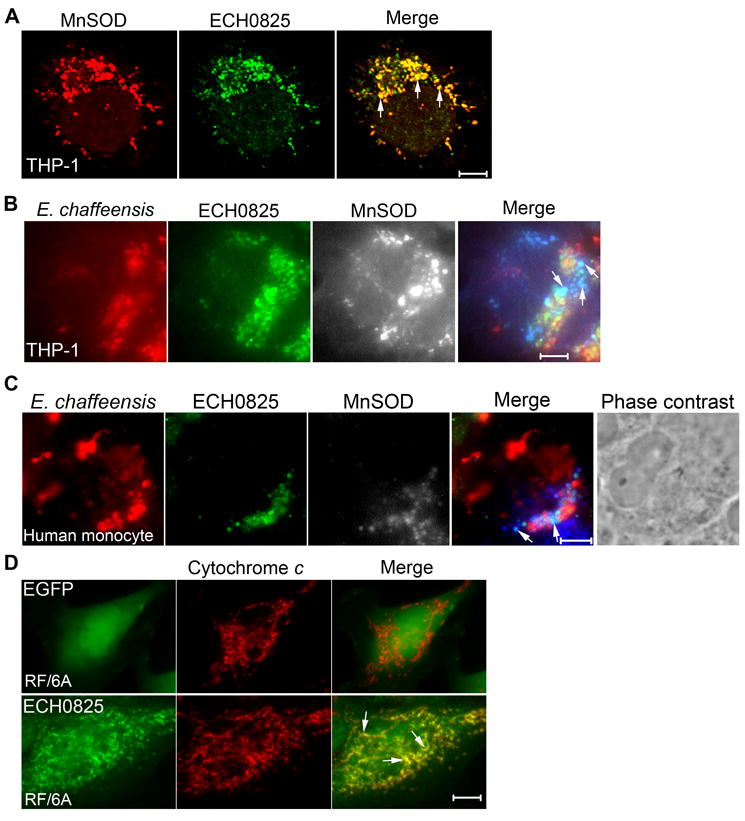
ECH0825 localizes to mitochondria in infected host cells.
A. Double immunofluorescence labeling of E. chaffeensis-infected THP-1 cells at 46 h p.i. using rabbit anti-ECH0825 (ECH0825; AF488, green) and monoclonal anti-MnSOD (MnSOD; AF555, red) was examined by confocal microscopy.
B, C. Triple immunofluorescence labeling of E. chaffeensis-infected THP-1 cells (B) and E. chaffeensis-infected human peripheral blood monocytes (C) using dog anti-E. chaffeensis (E. chaffeensis; Texas red, red), rabbit anti-ECH0825 (ECH0825; Alexa Fluor 488, green), and monoclonal anti-MnSOD (MnSOD; AF350, gray). Arrows indicate the localization of ECH0825 to host-cell mitochondria but not to intracellular E. chaffeensis.
D. Immunofluorescence labeling of RF/6A cells transfected with pECH0825 or pEGFP-N1; cells were labeled with anti-ECH0825 (ECH0825; AF488, green) and mouse monoclonal anti-cytochrome c (cytochrome c; AF555, red). Scale bar: 5 μm.
To examine whether the translocation of ECH0825 to host-cell mitochondria could occur independent from other E. chaffeensis proteins or bacterial infection, monkey endothelial cell line RF/6A, which can be readily infected with E. chaffeensis (Supplementary Fig. S1, and Fig. 10), was transfected with a plasmid encoding ECH0825 (pECH0825). The plasmid pECH0825 was constructed by replacing the GFP-coding gene in pEGFP-N1 with codon-optimized ECH0825 gene for mammalian expression because the AT-rich E. chaffeensis genomic ECH0825 gene yielded low protein levels when expressed in mammalian cells (data not shown). Control pEGFP-N1- or pECH0825-transfected RF/6A cells were labeled with cytochrome c, a mitochondrial inner membrane protein, and the results showed that ectopically expressed ECH0825, but not GFP, was translocated to mitochondria (Fig. 4D), indicating that ECH0825 is intrinsically targeted to host-cell mitochondria.
Fig. 10.
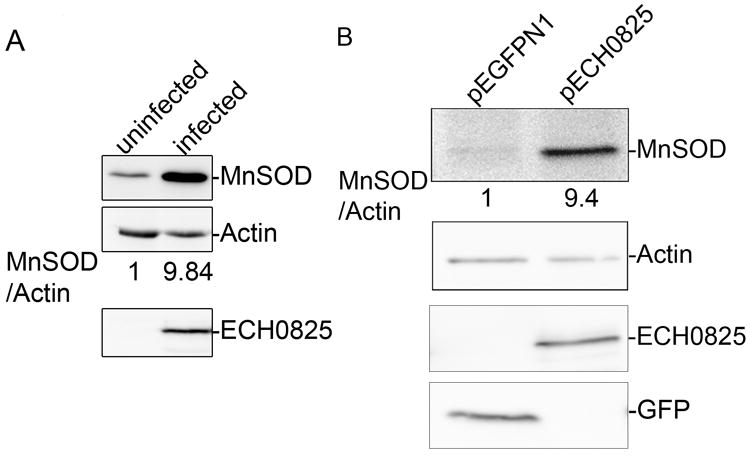
E. chaffeensis infection or ectopic expression of ECH0825 upregulate MnSOD expression.
Western blot analysis for MnSOD expression in (A) uninfected RF/6A cells or cells that were infected with E. chaffeensis (analysis done at 2 days p.i.) and (B) RF/6A cells transfected with pECH0825 or pEGFP-N1. Samples were probed with anti-MnSOD, anti-actin, anti-ECH0825 or anti-GFP by western blot analysis. The amount of protein was measured by densitometry.
ECH0825 has a direct effect on the efficiency of E. chaffeensis infection of THP-1 cells
To determine whether ECH0825 is required for E. chaffeensis infection, affinity-purified anti-ECH0825 or normal rabbit IgG was delivered into THP-1 cells by the Chariot protein delivery reagent to block ECH0825 function. Immunofluorescence labeling results showed that antibodies were delivered into host cells (Fig. 5A). At 1 day post-antibody delivery, the number of bacteria in cells treated with anti-ECH0825 was reduced more than 50% compared to cells treated with control IgG (Fig. 5B). This result suggests that host cytoplasmically delivered ECH0825 has a critical role in E. chaffeensis infection.
Fig. 5.
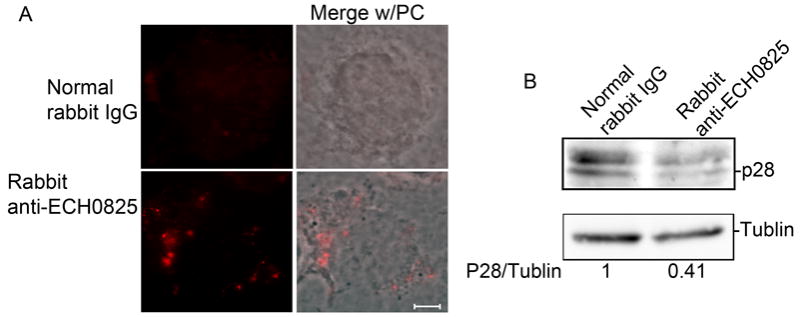
ECH0825 is required for E. chaffeensis infection.
Antibody against rECH0825, or normal rabbit IgG, was delivered to E. chaffeensis–infected THP-1 cells at 1 day p.i. by Chariot transfection reagent. A. Goat anti-rabbit conjugated with AF555 (red) was used to detect the delivered antibodies. Merge w/PC, image merged with phase contrast. B. Bacterial infection level was determined at 1 day post-delivery by western blot analysis with rabbit anti-P28 (E. chaffeensis major surface antigen).
ECH0825 inhibits etoposide-induced apoptosis by preventing Bax clumping, cytochrome c release, and loss of mitochondrial membrane potential
Mitochondria play a key role in regulating cellular apoptosis (Parone et al., 2003, Wang et al., 2009). Because ECH0825 localizes to mitochondria, we examined whether ECH0825 inhibits mitochondria-mediated apoptosis. pEGFP-N1- or pECH0825-transfected RF/6A cells were treated with etoposide for 1 day at 20 h post-transfection. Etoposide, a topoisomerase II inhibitor, can induce DNA double-strand breaks leading to the activation of caspase 2 and subsequent induction of Bax translocation to mitochondria and cytochrome c release, which results in apoptosis (Karpinich et al., 2002). In cells transfected with pEGFP-N1 control vector, nearly 50% of cells showed condensed or fragmented nuclei following etoposide treatment; however, only ~20% of pECH0825-transfected cells showed these nuclear effects (Figs 6A and 6B), indicating that ectopically expressed ECH0825 inhibits etoposide-induced apoptosis.
Fig. 6.
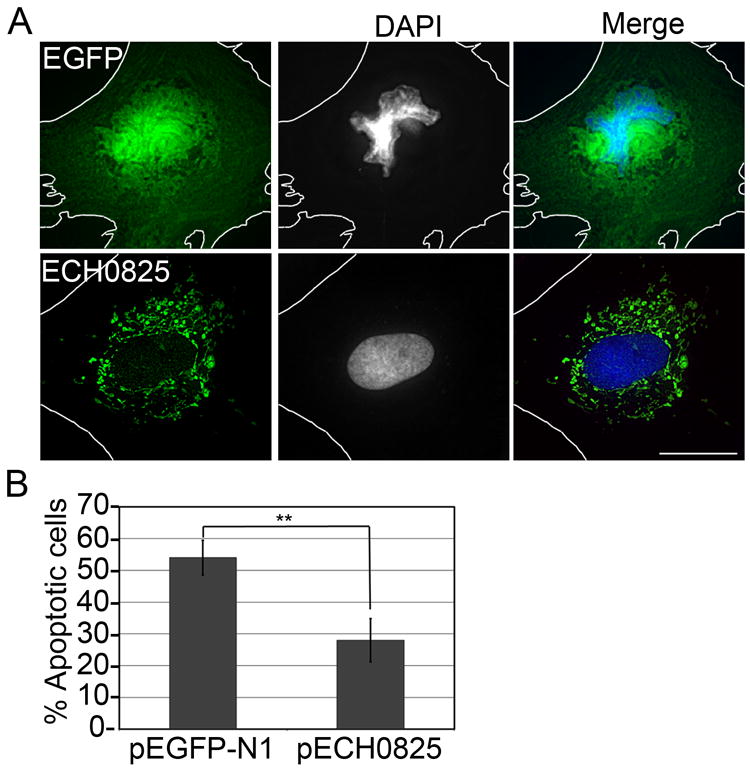
Ectopically expressed ECH0825 inhibits etoposide-induced apoptosis.
A. Double fluorescence labeling of pEGFP-N1- or pECH0825-transfected RF/6A cells treated with etoposide (100 μM) for 1 day; cells were labeled for DNA with DAPI (blue) and rabbit anti-ECH0825 (ECH0825; AF488, green). The white dashed lines denote contours of host cells. Scale bar: 15 μm.
B. Percentage of apoptotic cells with fragmented nuclei among pECH0825- or pEGFPN1-transfected cells. Data are presented as the mean ± standard deviation of three independent experiments. **Significantly different by Student’s t-test (p < 0.01).
Bax belongs to the pro-apoptotic Bcl-2 family and is diffusely distributed in the cytosol in healthy cells (Willis et al., 2005). Upon apoptotic stimuli, however, Bax is targeted to the outer mitochondrial membrane and oligomerizes to form pores, and cytochrome c is released to the cytosol (Lartigue et al., 2008). Because Bax was diffusely distributed in the cytosol in non-apoptotic RF/6A cells, it was nearly undetectable by fluorescence microscopy; clumped Bax, however, could be readily detected in severely apoptotic cells after etoposide treatment (Fig. 7A). More cells showed Bax clumping in pEGFP-N1-transfected negative-control cells than in pECH0825-transfected cells after etoposide treatment (Fig. 7B).
Fig. 7.
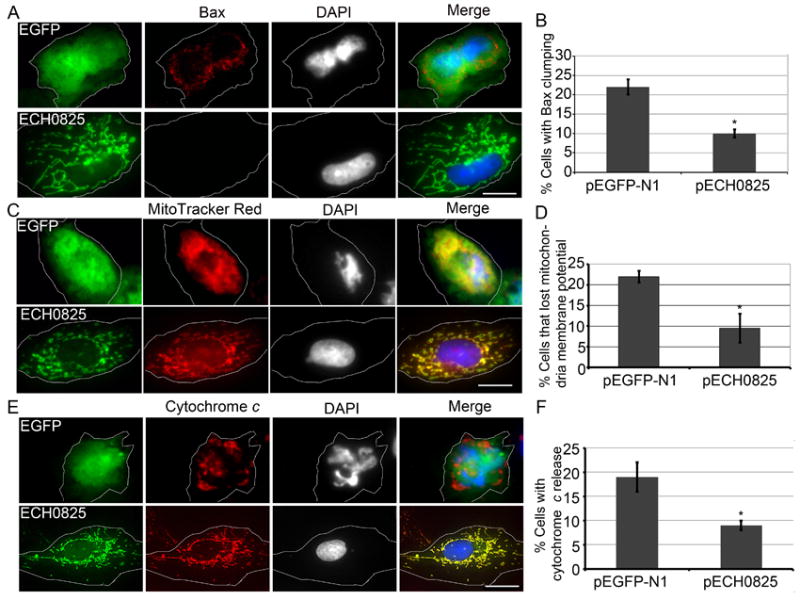
Ectopically expressed ECH0825 inhibits etoposide-induced Bax clumping, loss of mitochondrial membrane potential, and cytochrome c release.
Bax clumping (A, B), loss of mitochondrial membrane potential (C, D), and cytochrome c release (E, F) were inhibited in pECH0825-transfected cells compared to pEGFP-N1-transfected cells after 1 day of treatment with 100 μM etoposide. DAPI was used to label DNA (blue and gray); Bax was labeled with mouse anti-Bax and AF488-conjugated goat anti-mouse IgG; MitoTracker Red was used to stain mitochondria that had a high membrane potential. cytochrome c was labeled with mouse anti-cytochrome c and AF488-conjugated goat anti-mouse IgG; The white dashed lines denote contours of host cells. Scale bar, 15 μm. Percentage of cells with Bax clumping, loss of mitochondrial membrane potential, and cytochrome c release are shown in B, D, and F, representative data for which are presented as the mean ± standard deviation (n = 3) of two (D, F) or three (B) independent experiments. *Significantly different by Student’s t-test (p < 0.05).
Once Bax forms clumps and pores on the mitochondrial outer membrane, mitochondrial membrane potential is lost and apoptosis ensues (Antonsson, 2001). To monitor mitochondrial membrane potential, live cells were labeled with MitoTracker Red, which only labels mitochondria that have a high membrane potential (Poot et al., 1996, Keij et al., 2000). More cells showed loss of mitochondrial membrane potential in pEGFP-N1-transfected negative-control cells than in pECH0825-transfected cells after etoposide treatment (Fig. 7C and 7D).
Cytochrome c is a key component in the mitochondria-dependent apoptotic pathway (Jiang et al., 2004). Cytochrome c released into cytosol binds to Apaf-1 (apoptotic protease activating factor-1), and this bimolecular complex promotes the formation of the apoptosome, which activates initiator and executioner procaspases and results in apoptosis (Kumar, 2007). Cytochrome c became more diffuse after etoposide treatment in pEGFP-N1-transfected cells than in pECH0825-transfected cells (Figs. 7E and 7F). Taken together, these data suggested that ectopically expressed ECH0825 could prevent etoposide-induced apoptosis by preventing Bax clumping, cytochrome c release, and loss of mitochondrial membrane potential.
ECH0825 inhibits Bax-induced apoptosis in yeast
Although yeasts lack Apaf-1, p53, and Bcl-2 family proteins, the cell death–regulating activity of mitochondria in response to Bcl-2 family proteins is conserved in yeast; moreover, expression of human Bax induces growth arrest and cell death in yeast (Ligr et al., 1998). Yeast co-transformed with pYECH0825 and pBax (Fig. 8A) were used to study the effect of ECH0825 on human Bax-induced apoptosis. In transformed yeast cells, ECH0825 was also targeted to mitochondria as in RF/6A and THP-1 cells (Figs. 5 and 8B). After Bax was induced with galactose for 5 days, the number of yeast colonies co-transformed with the plasmids pYECH0825 and pBax was approximately 4-fold greater than that of yeast transformed with pGADT7 (control plasmid) and pBax (Fig. 8C), indicating that ECH0825 can inhibit human Bax-induced apoptosis in yeast.
Fig. 8.
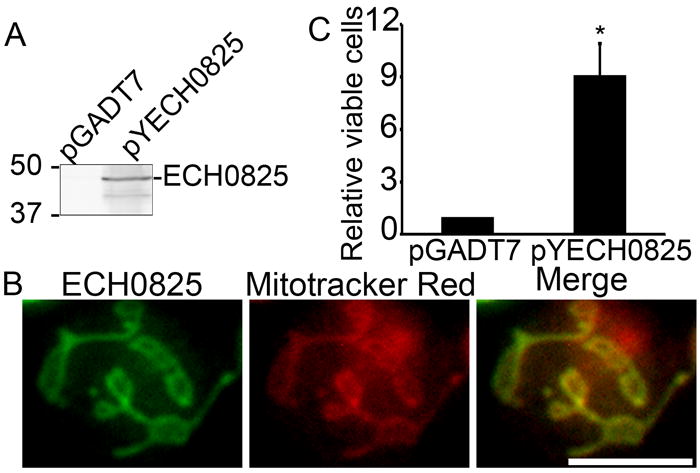
Ectopically expressed ECH0825 inhibits Bax-induced apoptosis in yeast.
A. Expression of ECH0825 in S. cerevisiae. Lysates of YPH499 cells transformed with pYECH0825 or pGADT7 AD (pGADT7) was subjected to western blot analysis with anti-ECH0825.
B. Localization of ECH0825 to mitochondria in S. cerevisiae. pYECH0825- or pGADT7 AD (pGADT7)-transformed YPH499 cells were loaded with MitoTracker Red and subjected to immunostaining with rabbit anti-ECH0825 and AF488–conjugated anti-rabbit IgG. Scale bar: 5 μm. C. ECH0825 partially rescues S. cerevisiae from Bax-induced growth arrest. YPH499 cells were co-transformed with pBax and pGADT7 AD (pGADT7) or pBax and pYECH0825. Recombinant yeast cells were cultured in the medium containing galactose to induce Bax expression. The number of viable cells was determined by a plate count technique. The numbers of viable cells at day 5 after Bax induction were compared to the number of viable cells on day 0. Data are expressed as the mean ± standard deviation (n = 3) and are representative of two independent experiments with similar results. *Significantly different by Student’s t-test (p < 0.05).
ECH0825 inhibits ROS production in mitochondria
Superoxide generated by aerobic respiration in mitochondria can cause DNA damage and is a critical factor for inducing host cell apoptosis (Cai et al., 1998, Thannickal et al., 2000, Wei, 2002 #695, Parone et al., 2003). Because etoposide increases the generation of ROS (Kurosu et al., 2003) and ECH0825 inhibits mitochondria-mediated apoptosis, we examined ROS levels in E. chaffeensis-infected cells using 2’, 7’-dichlorodihydrofluorescein diacetate (H2DCFDA), which can permeate the host-cell membrane and become trapped inside cells once hydrolyzed by cellular esterases to yield membrane-impermeable H2DCF. H2DCF can be oxidized by ROS to yield DCF, which can be detected by fluorescence spectrophotometry (Armstrong et al., 2007). The results showed that ROS production in E. chaffeensis-infected THP-1 cells was significantly lower than in uninfected cells (Fig. 9A), suggesting that E. chaffeensis can inhibit ROS production in its own cells as well as in host cells. Furthermore, ROS levels in ECH0825-transfected human embryonic kidney 293 (HEK293) cells, which can also be readily infected with E. chaffeensis (Miura et al., 2011), were significantly lower than in pEGFP-N1-transfected HEK293 cells (Fig. 9B), suggesting that ECH0825 in mitochondria reduces the ROS level in host cells.
Fig. 9.
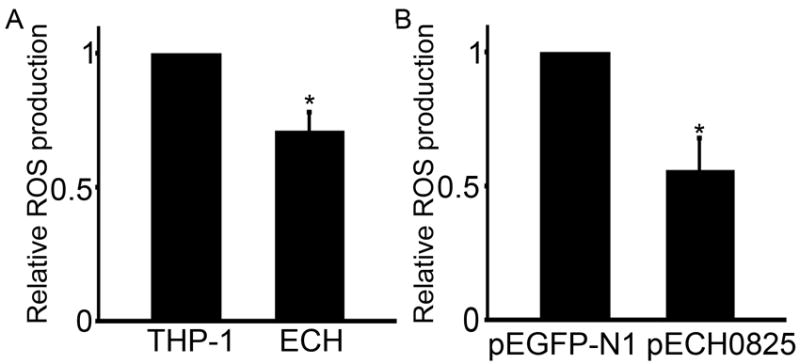
ROS production is reduced by E. chaffeensis infection or ectopically expressed ECH0825.
ROS production after 2 days of culture of E. chaffeensis-infected relative to uninfected THP-1 cells (A), and of pECH0825-transfected relative to pEGFP-N1-transfected HEK293 cells (B). ECH, E. chaffeensis-infected THP-1 cells. Data are expressed as the mean ± standard deviation (n = 3) and are representative of two independent experiments with similar results. *Significantly different by Student’s t-test (p < 0.05).
E. chaffeensis infection and ectopically expressed ECH0825 increase host MnSOD level
MnSOD in the mitochondrial matrix has an important role in converting membrane-impermeable superoxide anion generated through aerobic respiration to membrane-diffusible hydrogen peroxide, which is further detoxified by peroxidase and catalase (Oberley et al., 1988, Knirsch et al., 2001, Zelko et al., 2002). Mitochondrial MnSOD protects cells from apoptosis induced by many stimuli (Wong, 1995, Pani et al., 2000, Kops et al., 2002, Li et al., 2002, Tanaka et al., 2002). Western blotting results showed that, in E. chaffeensis-infected RF/6A cells, the protein amount of MnSOD was increased significantly compared to that in uninfected cells (Fig. 10A). Similar results were obtained with THP-1 cells (data not shown). Importantly, the MnSOD protein amount was also significantly upregulated in ECH0825-transfected RF6/A cells (Fig. 10B), suggesting that ECH0825 in mitochondria promotes an increase in the MnSOD level in the mitochondrial matrix to detoxify ROS. Quantitative RT-PCR results showed that mRNA amount of MnSOD (sod2) in ECH0825-transfected cells remained unchanged compared to GFP-transfected cells (data not shown), suggesting MnSOD upregulation by ECH0825 is not at the mRNA level but likely at protein level.
Discussion
In this study, we demonstrated that an E. chaffeensis T4S effector, ECH0825, can be translocated from the bacterium to the cytoplasm of host cells. Although the level of ECH0825 mRNA per bacterium was highest at the stationary stage, the level of ECH0825 protein per bacterium was highest at the pre-exponential growth stage, concurrent with the upregulation of mRNAs encoding components of the E. chaffeensis T4S apparatus (Cheng et al., 2008), suggesting that once the T4S components were expressed and the apparatus assembled, ECH0825 protein could be readily translocated to the cytoplasm of the host cell.
Host translocated ECH0825 had direct effects on efficiency of E. chaffeensis infection. At what stage of the intracellular infection, ECH0825 is involved, however, remains to be determined. Since the antibody added after bacterial uptake blocked the infection, ECH0825 is likely promoting more efficient replication or allowing additional cycles of replication as it blocks host cell apoptosis. The reduction of infection by intracellular antibody delivery is approximately 50%. More efficient delivery of antibody into the cell and perhaps into mitochondria (or alternative blocking approaches) would allow complete abrogation. However, there are other possibilities such as there are alternative proteins or pathways that allow continued infection/replication, but with lower efficiency. In fact L. pneumophila encodes multiple T4S effectors of overlapping functions to ensure their intracellular infection (Isberg et al., 2009).
Prevention of host-cell apoptosis provides a survival advantage for obligatory intracellular pathogens because it gives sufficient time for the bacteria to replicate. Bacterial pathogens have evolved several ways to prevent host-cell apoptosis by ensuring the integrity of mitochondria and preventing cytochrome c release, by activating cell survival pathways, or by preventing caspase activation (Faherty et al., 2008). A previous study showed that E. chaffeensis can upregulate the expression of nuclear factor-κB and other apoptosis inhibitors and can differentially regulate the expression of cyclins and cyclin-dependent kinase in THP-1 cells (Zhang et al., 2004). Several studies have shown that mitochondria associate closely with E. chaffeensis inclusions (Popov et al., 1995, Rikihisa, 1996, Liu et al., 2011). One of important functions of mitochondria is the regulation of apoptosis. A recent study showed that mitochondrial membrane potential is maintained in E. chaffeensis-infected DH82 cells treated with aphidicolin, an inhibitor of DNA polymerase-α; by contrast, the membrane potential is reduced in mock-infected DH82 cells, which show condensed nuclei, suggesting that E. chaffeensis can inhibit apoptosis of host cells (Liu et al., 2011). Our current study showed that ECH0825 is translocated to mitochondria and inhibits mitochondria-mediated apoptosis, which is accompanied by the inhibition of both cytoplasmic Bax translocation to (and clumping on) mitochondria and release of cytochrome c. This is similar to the mechanism by which the T4S effector Ats-1 inhibits apoptosis in A. phagocytophilum-infected human neutrophils and RF/6A cells (Niu et al., 2010). Indeed, ECH0825 and Ats-1 are orthologs, having 21% amino acid identity.
ROS can damage DNA and induce mitochondria-mediated apoptosis (Cai et al., 1998, Thannickal et al., 2000, Wei et al., 2002, Parone et al., 2003). The present study revealed that ROS production in E. chaffeensis-infected cells was lower than that in uninfected cells. E. chaffeensis can prevent ROS production by membrane NADPH oxidase during infection of human monocytes (Lin et al., 2007b). Indeed other studies have shown that intracellular bacteria, such as Chlamydia and A. phagocytophilum, can reduce ROS production to protect host cells by upregulating ferritin heavy chain (Carlyon et al., 2005, Vardhan et al., 2010). Ferritin can sequester cellular iron, which generates highly toxic hydroxyl radicals from H2O2 via the Harber-Weiss reaction (Arosio et al., 2002). In contrast, several bacterial pathogens, including Burkholderia cepacia, Borrelia hermsii, Listeria monocytogenes, Staphylococcus aureus, and Streptococcus pyogenes increase the ROS production, consequently kill the invading microorganisms and result in host apoptosis (Kobayashi et al., 2003, DeLeo, 2004).
Most of the toxic ROS found in eukaryotic cells are produced during ATP generation from oxygen in mitochondria, as 1-3% of electrons leak from the electron transport chain and interact with oxygen directly and yield superoxide (Halliwell et al., 1999). Other systems such as NADPH oxidase also produce ROS (Babior, 2004, Nauseef, 2008). To prevent oxidative damage to mitochondria and the cell, cells synthesize antioxidants. MnSOD in the mitochondrial matrix, catalyzes the conversion of superoxide anion to hydrogen peroxide (Knirsch et al., 2001, Zelko et al., 2002). In mitochondria, the action of MnSOD and the glutathione/glutathione peroxidase-1 system act cooperatively to constrain the levels of ROS (mainly O2− and H2O2) generated during normal aerobic metabolism (Oberley et al., 1988, Knirsch et al., 2001, Zelko et al., 2002). When cellular antioxidant defense systems cannot remove excess ROS, oxidative stress ensues, resulting in cell damage and apoptosis (Cai et al., 1998, Parone et al., 2003, Cui et al., 2011). Loss of MnSOD (Sod2−/−) is lethal in mice, whereas partial deficiency (Sod2+/−) increases the rate of apoptosis (Kokoszka et al., 2001, Van Remmen et al., 2001). MnSOD overexpression stabilizes the mitochondrial membrane, inhibits the permeability transition and apoptosis (Manna et al., 1998, Bruce-Keller et al., 1999, Epperly et al., 2002, Epperly et al., 2003), and blocks Fas-mediated apoptosis (Sato et al., 2004). MnSOD can regulate the levels of mitochondrial Bcl-2 family proteins, i.e., it can increase the Bcl-2 level and decrease the Bax level to suppress ROS-induced apoptosis (Xu et al., 2008, Sharma et al., 2009, Li et al., 2010). We observed a significant increase in mitochondrial MnSOD level in E. chaffeensis-infected cells, which may be responsible for reducing ROS levels in infected cells. Furthermore, our study showed that MnSOD was upregulated and ROS production was significantly lower in ECH0825-transfected cells compared with GFP-transfected cells, indicating a role for ECH0825 in MnSOD upregulation and ROS reduction. MnSOD, which is encoded by chromosomal DNA, is primarily transported to the mitochondrial matrix (Glick B, 1991). Our results showed that ectopically expressed ECH0825 mostly localized to mitochondria and mitochondria-translocated ECH0825 increased MnSOD protein amount, suggesting that ECH0825 can protect mitochondria and host cells from oxidative stress and apoptosis. This hypothesis is in agreement with previous reports that induction of mitochondrial MnSOD confers resistance to apoptosis in acute myeloblastic leukemia cells or HEK293 cells exposed to etoposide (Mantymaa et al., 2000, Chen et al., 2007) and that ROS induces apoptosis via selective activation of c-Jun N-terminal kinase (i.e., JNK), Bak, and Bax (Cai et al., 1998, Chen et al., 2007). Ats-1 of A. phagocytophilum localizes to the matrix of mitochondria and blocks Bax translocation to mitochondria (Niu et al., 2010), however, effects of Ats-1 on MnSOD or ROS have not been studied. It is possible that mitochondria-translocated Ats-1 also blocks host-cell apoptosis (Niu et al., 2010) by stabilizing host MnSOD. MnSOD is degraded in a caspase-dependent manner during Fas-mediated apoptosis (Pardo et al., 2006); However, the mechanism of MnSOD regulation by E. chaffeensis ECH0825 remains to be studied. Furthermore, we cannot exclude the alternate possibility that, in E. chaffeensis-infected DH82 cells, the reduced levels of ROS are a consequence of reduced energy generation in mitochondria. This possibility is supported by studies showing that mitochondrial DNA synthesis is inhibited and transcription of the mitochondrial genes nadph2 and cytB (encoding cytochrome b) is decreased in E. chaffeensis–infected cells compared to mock-infected cells (Liu et al., 2011).
The fact that ROS levels are significantly reduced in E. chaffeensis-infected cells than uninfected cells suggests that E. chaffeensis is also equipped with an antioxidant mechanism to protect itself. It is possible that ECH0825 also acts as an antioxidant inside the bacteria as inside mitochondria that are evolved from a Rickettsial ancestor (Andersson et al., 1998). Interestingly, genes encoding FeSOD (sodB) and components of the T4S apparatus are co-transcribed in both E. chaffeensis and A. phagocytophilum (Ohashi et al., 2002), perhaps to coordinate bacterial and host antioxidant-based survival mechanisms.
Currently, identified T4S effector molecules of A. phagocytophilum and E. chaffeensis are Ats-1/ECH0825 and AnkA, which possess C-terminal positively charged amino acid residues and can be recognized by the T4S coupling protein VirD4 (Rikihisa et al., 2009, Rikihisa et al., 2010). They are abundantly produced and secreted into the mammalian host cytoplasm, are not toxic to host cells, and can manipulate host cell processes to aid the infection process. At the cellular level, the two effectors have distinct subcellular localization: Ats-1 and ECH0825 target host mitochondria and AnkA is in the cytoplasm and nucleus, and distinct signaling mechanisms in host cells (Rikihisa et al., 2009, Zhu et al., 2009, Rikihisa et al., 2010, Scharf et al., 2011). Thus in these obligatory intracellular pathogens, the T4S system has evolved as a critical host-subversive virulence factor.
T4S apparatus genes are highly expressed by E. chaffeensis in tick cells as well as in mammalian cells (Bao et al., 2009), thus T4S system is likely important in tick stage of E. chaffeensis. In addition to ECH0825, the present study identified two T4S candidates of E. chaffeensis (Table S1). Although ECH0261 and ECH0767 proteins are not highly expressed in mammalian cells (Lin et al., 2011), the recent microarray study reported that ECH0767 is highly transcribed in tick (AAE2 and ISE6) cells (Kuriakose et al., 2011). Further experiments are ongoing to address the potential role of ECH0767 in E. chaffeensis infection of tick cells. Future studies are expected to provide better understanding of roles of multiple T4S effectors during the entire life cycle of the vector-borne obligatory intracellular pathogens.
Experimental Procedures
E. chaffeensis and cell culture
E. chaffeensis Arkansas (Anderson et al., 1991) was propagated in THP-1 (ATCC, Manassas, VA), and RF/6A cell (ATCC) was cultured as described (Kumagai et al., 2010, Niu et al., 2010). HEK293 cells were cultured in DMEM (Invitrogen, Carlsbad, CA) supplemented with 2 mM l-glutamine and 10% fetal bovine serum (FBS, Atlanta biological, Lawrenceville, GA).
Synchronous culture of E. chaffeensis and reverse transcription polymerase chain reaction (RT-PCR)
The procedure for synchronous culture of E. chaffeensis was modified from Dr. Cheng (Cheng et al., 2008). An approximate multiplicity of infection of 50 was used in this study. To disrupt host cells and liberate E. chaffeensis, infected THP-1 cells on ice were sonicated twice for 10 s at setting 2 with a W-380 sonicator (Heat System-Ultrasonics, Farmington, NY). The supernatant was collected after centrifugation at 700 × g and passed through a 2.7-μm filter (Whatman, Florham Park, NJ) to remove cell debris. The bacterial suspension on ice was further sonicated for 30 s twice at setting 4.5 to disrupt reticulate cells of E. chaffeensis (Cheng et al., 2008). E. chaffeensis was centrifuged at 10,000 × g for 10 min and used to infect 1 × 108 THP-1 cells in 5 ml of culture medium in a T-25 flask. After incubating at 37°C for 1 h with gentle shaking every 10 min, the cells were washed three times with fresh medium to remove unbound or un-internalized bacteria and resuspended at a density of 3 to 4 × 105 cells/ml. After unbound or un-internalized bacteria were removed, this time point was designated as 0 h p.i. Infected THP-1 cells were harvested at designated times p.i. and divided into several aliquots. Cells for RNA extraction were suspended in RNAlater (Qiagen, Valencia, CA) and stored at −20°C. Cells for DNA extraction and western blotting were stored at −80°C. Expression levels of ECH0825, ECH261 and ECH0767 were determined by RT-PCR analysis, and quantitative PCR was performed as described (Cheng et al., 2006). All primers are listed in Table S3.
Bacterial two-hybrid screen
E. chaffeensis proteins that interact with VirD4 were determined using the BacterioMatch II two-hybrid system (Stratagene/Agilent, La Jolla, CA). E. chaffeensis chromosomal DNA was extracted using QIAamp DNA Mini kit (Qiagen). E. chaffeensis virD4 was cloned into bait vector pBT (Stratagene/Agilent) to construct the pBT-VirD4 bait plasmid. Ten selected E. chaffeensis genes (Table S1) encoding hypothetical proteins or conserved hypothetical proteins having a net positive charge in the C-terminal region (Vergunst et al., 2005) were cloned into prey vector pTRG (Stratagene/Agilent) to construct pTRG-prey plasmids: the full-length gene was cloned for genes of ≤750 bp, and the 3’ 750 bp was cloned for genes of ≥ 750 bp (Table S1). pBT-VirD4 and pTRG-prey were co-transformed into the reporter strain XL1-Blue MRF’ Kan. Dual transformants were plated on an M9 His-selective medium and non-selective medium.
Production of rabbit antisera against ECH0825 and western blot analysis
The 3’-terminal 750 bp of ECH0825 was cloned into pET-33b(+) (Novagen, San Diego, CA), and rECH0825 protein was purified by Ni-affinity chromatography (Sigma-Aldrich, St. Louis, MO) as described (Cheng et al., 2006). Purified rECH0825 was subjected to SDS-PAGE (12% acrylamide). rECH0825 (~200 μg) recovered from gel slices was used for the first immunization of rabbits after being mixed with TiterMax (TiterMax USA, Norcross, GA) in 1:1 ratio (v/v). Similarly, 100 μg rECH0825 was mixed with Freund’s incomplete adjuvant (Sigma-Aldrich) in 1:1 ratio (v/v) and used to immunize rabbits for the second to fourth times at 2-week intervals. The rabbit anti-ECH0825 serum was affinity-purified with rECH0825 bound to a HiTrap NHS-activated HP column (GE Healthcare, Piscataway, NJ).
Proteins in lysates from uninfected or E. chaffeensis-infected, or antibody-delivered THP-1 or RF/6A cells, or from pECH0825- or pEGFP-N1-transfected RF/6A cells were subjected to SDS-PAGE and Western blotting using primary antibodies including rabbit-anti-rP28 (diluted 1:2,000 (Ohashi et al., 1998)), rabbit anti-ECH0825 (1:2,000), mouse anti-MnSOD (1:1,000; Alexis Biochemicals, San Diego, CA), rabbit anti-actin (1:2,000; Sigma-Aldrich), mouse anti-α-tubulin (1:2,000; Santa Cruz Biotechnology, Santa Cruz, CA), or mouse anti-GFP (1:2,000; Clone B-2, Santa Cruz Biotechnology), and peroxidase-conjugated secondary antibodies (1:2,000; KPL, Gaithersburg, MD). The procedure for antibody detection, image capture, and densitometry was carried out as described (Lin et al., 2007a).
Delivery of anti-ECH0825 IgG into host cells using Chariot reagent
Chariot protein delivery reagent (Active Motif, Carlsbad, CA) was used to deliver anti-ECH0825 IgG into E. chaffeensis-infected THP-1 cells at 1 d p.i. as described (Lin et al., 2007a). Briefly, 6 μl of Chariot in 100 μl H2O was mixed with 100 μl of phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) containing 5 μg of affinity-purified rabbit anti-ECH0825 IgG or control normal rabbit IgG. Following incubation for 30 min at room temperature, the complex was added to 5 × 105 of E. chaffeensis-infected THP-1 cells at 1 day p.i. in 0.4 ml of medium lacking serum. After culturing for 2.5 h, cells were supplied with another 1.4 ml RPMI 1640 medium containing 10% FBS and 2 mM l-glutamine. At 2.5 h post-transfection, an aliquot of cells (~50 μl) was cytocentrifuged onto slides for immunofluorescence labeling, and the remaining cells were cultured. The number of bacteria was determined at 1 day post-transfection by Western blotting.
ECH0825 transfection and apoptosis analysis
Full-length ECH0825 was codon-optimized for expression in human cells and synthesized by GenScript (Piscataway, NJ). Codon-optimized or wild-type ECH0825 was cloned into pEGFP-N1 (Clontech, Mountain View, CA) by replacing the gene encoding enhanced GFP (EGFP) to create plasmid pECH0825. The sequence of the cloned fragment was confirmed by DNA sequencing. After Escherichia coli strain DH5α (Invitrogen) was transformed, plasmids were extracted using the EndoFree Plasmid Maxi kit (Qiagen). RF/6A or HEK293 cells were transfected by electroporation with Gene Pulser Xcell System (Bio-Rad, Hercules, CA) as described (Niu et al., 2010).
pEGFP-N1- or pECH0825-transfected RF/6A cells were seeded at 1 × 105 cells/ml into each well of a 6-well plate, and at 20 h post-transfection cells were treated with 100 μM etoposide (Sigma-Aldrich) for 1 day and fixed with 2% paraformaldehyde in PBS. The integrity of nuclei was determined by staining cells with 300 nM of 4’, 6-diamidino-2-phenylindole, dilactate (DAPI, Invitrogen) for 5 min, and cells were observed by fluorescence microscopy to quantify cellular apoptosis.
Immunofluorescence analysis
Cells were fixed with 2% paraformaldehyde at room temperature for 30 min; for double immunofluorescence analysis, the cells were incubated with rabbit anti-ECH0825 and mouse anti-MnSOD, or dog anti-E. chaffeensis (Barnewall et al., 1997) and mouse anti-cytochrome c (Santa Cruz Biotechnology) or mouse anti-Bax (BD Transduction Laboratory, San Diego, CA) diluted 1:100 in PGS (PBS supplemented with 0.1% gelatin and 0.1% saponin) for 1 h at room temperature. For triple immunofluorescence analysis, cells were incubated with mouse anti-MnSOD, rabbit anti-ECH0825, and dog anti-E. chaffeensis. The secondary antibodies, Alexa Fluor 350 (AF350)-conjugated goat anti-mouse IgG (Invitrogen), AF555-conjugated goat anti-mouse IgG (Invitrogen), AF488-conjugated goat anti-rabbit IgG (Invitrogen), and Texas Red-conjugated goat anti-dog IgG (Rockland, Gilbertsville, PA), each diluted 1:100 in PGS, were incubated at room temperature for 1 h. To determine the mitochondrial membrane potential, live RF/6A cells were incubated with 400 nM MitoTracker Red CMXRos (Invitrogen) in Advanced-MEM medium (Invitrogen) for 30 min at 37°C. After washing with PBS, cells were fixed in 3% paraformaldehyde in Advanced-MEM medium without serum. Images were captured by a Nikon Eclipse E400 fluorescence microscope with a xenon-mercury light source (Nikon Instruments, Melville, NY), a DeltaVision Deconvolution microscope system (Applied Precision, Issaquah, WA), or an LSM 510 laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY).
Co-transformation with pYECH0825 and pBax and induction of Bax in Saccharomyces cerevisiae
The DNA fragments of codon-optimized ECH0825 were amplified and cloned into yeast constitutive expression vector pGADT7 AD (Clontech) by replacing the GAL4 AD sequence, resulting in the recombinant plasmid termed pYECH0825. Primers used are listed in Table S3. A pair of plasmids, pBax (Niu et al., 2010) and pGADT7 AD, or pBax and pYECH0825, was co-transformed into S. cerevisiae haploid strain YPH499 (ATCC) by using the YEASTMAKER Yeast Transformation System 2 (Clontech). The procedures for yeast culture, MitoTracker Red CMXRos labeling, yeast immunofluorescence analysis, and Bax induction were as described (Niu et al., 2010) except that anti-ECH0825 was used.
ROS assay with H2DCFDA
The level of ROS in whole cells was detected using H2DCFDA (Invitrogen). Briefly, after 2 days of culture, 3 × 105 uninfected and E. chaffeensis-infected THP-1 cells, or pEGFP-N1- and pECH0825-transfected HEK293 cells, were harvested and washed with PBS. The pellet was resuspended with 200 μl prewarmed PBS with 10 μM H2DCFDA or DMSO control and incubated for 30 min at 37°C under 5% CO2. After washing cells with PBS and resuspending them with 200 μl prewarmed PBS, the fluorescence intensity of DCF, corresponding to the ROS level, was measured with a Spectra Max GeminiXS Microplate Fluorometer (Molecular Devices, Sunnyvale, CA) at excitation and emission wavelengths of 492 nm and 520 nm, respectively, with a cutoff of 515 nm.
Supplementary Material
E. chaffeensis-infected RF/6A cells at 2 d p.i. were fixed and labeled with rabbit anti-P28 (red). Images were acquired by fluorescence microscopy. Scale bar: 10 μm.
Acknowledgments
This work was supported by National Institutes of Health grant R01 AI054476.
References
- Anderson BE, Dawson JE, Jones DC, Wilson KH. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Antonsson B. Bax and other pro-apoptotic Bcl-2 family “killer-proteins” and their victim the mitochondrion. Cell Tissue Res. 2001;306:347–361. doi: 10.1007/s00441-001-0472-0. [DOI] [PubMed] [Google Scholar]
- Armstrong JS, Whiteman M. Measurement of reactive oxygen species in cells and mitochondria. Methods Cell Biol. 2007;80:355–377. doi: 10.1016/S0091-679X(06)80018-X. [DOI] [PubMed] [Google Scholar]
- Arosio P, Levi S. Ferritin, iron homeostasis, and oxidative damage. Free Radic Biol Med. 2002;33:457–463. doi: 10.1016/s0891-5849(02)00842-0. [DOI] [PubMed] [Google Scholar]
- Babior BM. NADPH oxidase. Curr Opin Immunol. 2004;16:42–47. doi: 10.1016/j.coi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol. 2009;191:278–286. doi: 10.1128/JB.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun. 1997;65:1455–1461. doi: 10.1128/iai.65.4.1455-1461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Geddes JW, Knapp PE, McFall RW, Keller JN, Holtsberg FW, et al. Anti-death properties of TNF against metabolic poisoning: mitochondrial stabilization by MnSOD. J Neuroimmunol. 1999;93:53–71. doi: 10.1016/s0165-5728(98)00190-8. [DOI] [PubMed] [Google Scholar]
- Cai J, Jones DP. Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J Biol Chem. 1998;273:11401–11404. doi: 10.1074/jbc.273.19.11401. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Ryan D, Archer K, Fikrig E. Effects of Anaplasma phagocytophilum on host cell ferritin mRNA and protein levels. Infect Immun. 2005;73:7629–7636. doi: 10.1128/IAI.73.11.7629-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Kumagai Y, Lin M, Zhang C, Rikihisa Y. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell Microbiol. 2006;8:1241–1252. doi: 10.1111/j.1462-5822.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Wang X, Rikihisa Y. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J Bacteriol. 2008;190:2096–2105. doi: 10.1128/JB.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS. Molecular events in the activation of human neutrophils for microbial killing. Clin Infect Dis. 1994;18(Suppl 2):S170–179. doi: 10.1093/clinids/18.supplement_2.s170. [DOI] [PubMed] [Google Scholar]
- Cui M, Zhang Y, Liu S, Xie W, Ji M, Lou H, Li X. 1-oxoeudesm-11(13)-ene-12,8alpha-lactone-induced apoptosis via ROS generation and mitochondria activation in MCF-7 cells. Arch Pharm Res. 2011;34:1323–1329. doi: 10.1007/s12272-011-0812-x. [DOI] [PubMed] [Google Scholar]
- Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol. 1991;29:2741–2745. doi: 10.1128/jcm.29.12.2741-2745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR. Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis. 2004;9:399–413. doi: 10.1023/B:APPT.0000031448.64969.fa. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperly MW, Bernarding M, Gretton J, Jefferson M, Nie S, Greenberger JS. Overexpression of the transgene for manganese superoxide dismutase (MnSOD) in 32D cl 3 cells prevents apoptosis induction by TNF-alpha, IL-3 withdrawal, and ionizing radiation. Exp Hematol. 2003;31:465–474. doi: 10.1016/s0301-472x(03)00041-9. [DOI] [PubMed] [Google Scholar]
- Epperly MW, Sikora CA, DeFilippi SJ, Gretton JA, Zhan Q, Kufe DW, Greenberger JS. Manganese superoxide dismutase (SOD2) inhibits radiation-induced apoptosis by stabilization of the mitochondrial membrane. Radiat Res. 2002;157:568–577. doi: 10.1667/0033-7587(2002)157[0568:msdsir]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Faherty CS, Maurelli AT. Staying alive: bacterial inhibition of apoptosis during infection. Trends Microbiol. 2008;16:173–180. doi: 10.1016/j.tim.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B, S G. Import of proteins into mitochondria. Annu Rev Genet. 1991;25:21–24. doi: 10.1146/annurev.ge.25.120191.000321. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. New York, USA: Oxford University Press; 1999. [Google Scholar]
- Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- Karpinich NO, Tafani M, Rothman RJ, Russo MA, Farber JL. The course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochrome c. J Biol Chem. 2002;277:16547–16552. doi: 10.1074/jbc.M110629200. [DOI] [PubMed] [Google Scholar]
- Keij JF, Bell-Prince C, Steinkamp JA. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry. 2000;39:203–210. doi: 10.1002/(sici)1097-0320(20000301)39:3<203::aid-cyto5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Knirsch L, Clerch LB. Tyrosine phosphorylation regulates manganese superoxide dismutase (MnSOD) RNA-binding protein activity and MnSOD protein expression. Biochemistry. 2001;40:7890–7895. doi: 10.1021/bi010197n. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoszka JE, Coskun P, Esposito LA, Wallace DC. Increased mitochondrial oxidative stress in the Sod2 (+/-) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proc Natl Acad Sci U S A. 2001;98:2278–2283. doi: 10.1073/pnas.051627098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. Cyclic di-GMP Signaling Regulates Invasion of Ehrlichia chaffeensis into Human Monocytes. J Bacteriol. 2010 doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One. 2011;6:e24136. doi: 10.1371/journal.pone.0024136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu T, Fukuda T, Miki T, Miura O. BCL6 overexpression prevents increase in reactive oxygen species and inhibits apoptosis induced by chemotherapeutic reagents in B-cell lymphoma cells. Oncogene. 2003;22:4459–4468. doi: 10.1038/sj.onc.1206755. [DOI] [PubMed] [Google Scholar]
- Lartigue L, Medina C, Schembri L, Chabert P, Zanese M, Tomasello F, et al. An intracellular wave of cytochrome c propagates and precedes Bax redistribution during apoptosis. J Cell Sci. 2008;121:3515–3523. doi: 10.1242/jcs.029587. [DOI] [PubMed] [Google Scholar]
- Li C, Wright MM, Jackson RM. Reactive species mediated injury of human lung epithelial cells after hypoxia-reoxygenation. Exp Lung Res. 2002;28:373–389. doi: 10.1080/01902140290092001. [DOI] [PubMed] [Google Scholar]
- Li WJ, Shin MK, Oh SJ. Time dependent bladder apoptosis induced by acute bladder outlet obstruction and subsequent emptying is associated with decreased MnSOD expression and Bcl-2/Bax ratio. J Korean Med Sci. 2010;25:1652–1656. doi: 10.3346/jkms.2010.25.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65. doi: 10.1016/s0014-5793(98)01227-7. [DOI] [PubMed] [Google Scholar]
- Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007a;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Kikuchi T, Brewer HM, Norbeck AD, Rikihisa Y. Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front Microbiol. 2011;2:24. doi: 10.3389/fmicb.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y. Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell Microbiol. 2007b;9:861–874. doi: 10.1111/j.1462-5822.2006.00835.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang Z, Jiang Y, Zhang L, Popov VL, Zhang J, et al. Obligate intracellular bacterium Ehrlichia inhibiting mitochondrial activity. Microbes Infect. 2011;13:232–238. doi: 10.1016/j.micinf.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One. 2011;6:e27724. doi: 10.1371/journal.pone.0027724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med. 1987;316:853–856. doi: 10.1056/NEJM198704023161406. [DOI] [PubMed] [Google Scholar]
- Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappaB and activated protein-1. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- Mantymaa P, Siitonen T, Guttorm T, Saily M, Kinnula V, Savolainen ER, Koistinen P. Induction of mitochondrial manganese superoxide dismutase confers resistance to apoptosis in acute myeloblastic leukaemia cells exposed to etoposide. Br J Haematol. 2000;108:574–581. doi: 10.1046/j.1365-2141.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Miura K, Matsuo J, Rahman MA, Kumagai Y, Li X, Rikihisa Y. Ehrlichia chaffeensis Induces Monocyte Inflammatory Responses through MyD88, ERK, and NF-kappaB but Not through TRIF, Interleukin-1 Receptor 1 (IL-1R1)/IL-18R1, or Toll-Like Receptors. Infect Immun. 2011;79:4947–4956. doi: 10.1128/IAI.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun. 1999;67:1368–1378. doi: 10.1128/iai.67.3.1368-1378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 2010;6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberley LW, Oberley TD. Role of antioxidant enzymes in cell immortalization and transformation. Mol Cell Biochem. 1988;84:147–153. doi: 10.1007/BF00421049. [DOI] [PubMed] [Google Scholar]
- Ohashi N, Zhi N, Lin Q, Rikihisa Y. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect Immun. 2002;70:2128–2138. doi: 10.1128/IAI.70.4.2128-2138.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N, Zhi N, Zhang Y, Rikihisa Y. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect Immun. 1998;66:132–139. doi: 10.1128/iai.66.1.132-139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock CD, Childs JE. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev. 2003;16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani G, Bedogni B, Anzevino R, Colavitti R, Palazzotti B, Borrello S, Galeotti T. Deregulated manganese superoxide dismutase expression and resistance to oxidative injury in p53-deficient cells. Cancer Res. 2000;60:4654–4660. [PubMed] [Google Scholar]
- Pardo M, Melendez JA, Tirosh O. Manganese superoxide dismutase inactivation during Fas (CD95)-mediated apoptosis in Jurkat T cells. Free Radic Biol Med. 2006;41:1795–1806. doi: 10.1016/j.freeradbiomed.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Parone P, Priault M, James D, Nothwehr SF, Martinou JC. Apoptosis: bombarding the mitochondria. Essays Biochem. 2003;39:41–51. doi: 10.1042/bse0390041. [DOI] [PubMed] [Google Scholar]
- Poot M, Zhang YZ, Kramer JA, Wells KS, Jones LJ, Hanzel DK, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- Popov VL, Chen SM, Feng HM, Walker DH. Ultrastructural variation of cultured Ehrlichia chaffeensis. J Med Microbiol. 1995;43:411–421. doi: 10.1099/00222615-43-6-411. [DOI] [PubMed] [Google Scholar]
- Rego AT, Chandran V, Waksman G. Two-step and one-step secretion mechanisms in Gram-negative bacteria: contrasting the type IV secretion system and the chaperone-usher pathway of pilus biogenesis. Biochem J. 2010;425:475–488. doi: 10.1042/BJ20091518. [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. Ehrlichiae. Slovak Academy of Sciences; 1996. pp. 272–286. [Google Scholar]
- Rikihisa Y. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol. 2010a;8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet Parasitol. 2010b;167:155–166. doi: 10.1016/j.vetpar.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol. 2010;13:59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y, Lin M, Niu H, Cheng Z. Type IV secretion system of Anaplasma phagocytophilum and Ehrlichia chaffeensis. Ann N Y Acad Sci. 2009;1166:106–111. doi: 10.1111/j.1749-6632.2009.04527.x. [DOI] [PubMed] [Google Scholar]
- Sato T, Machida T, Takahashi S, Iyama S, Sato Y, Kuribayashi K, et al. Fas-mediated apoptosome formation is dependent on reactive oxygen species derived from mitochondrial permeability transition in Jurkat cells. J Immunol. 2004;173:285–296. doi: 10.4049/jimmunol.173.1.285. [DOI] [PubMed] [Google Scholar]
- Scharf W, Schauer S, Freyburger F, Petrovec M, Schaarschmidt-Kiener D, Liebisch G, et al. Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J Clin Microbiol. 2011;49:790–796. doi: 10.1128/JCM.02051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DR, Sunkaria A, Bal A, Bhutia YD, Vijayaraghavan R, Flora SJ, Gill KD. Neurobehavioral impairments, generation of oxidative stress and release of pro-apoptotic factors after chronic exposure to sulphur mustard in mouse brain. Toxicol Appl Pharmacol. 2009;240:208–218. doi: 10.1016/j.taap.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Slauch JM. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol. 2011;80:580–583. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Matsumura I, Ezoe S, Satoh Y, Sakamaki T, Albanese C, et al. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol Cell. 2002;9:1017–1029. doi: 10.1016/s1097-2765(02)00522-1. [DOI] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, et al. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol. 2001;281:H1422–1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- Vardhan H, Bhengraj AR, Jha R, Srivastava P, Jha HC, Mittal A. Higher expression of ferritin protects Chlamydia trachomatis infected HeLa 229 cells from reactive oxygen species mediated cell death. Biochem Cell Biol. 2010;88:835–842. doi: 10.1139/o10-027. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med (Maywood) 2002;227:671–682. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GH. Protective roles of cytokines against radiation: induction of mitochondrial MnSOD. Biochim Biophys Acta. 1995;1271:205–209. doi: 10.1016/0925-4439(95)00029-4. [DOI] [PubMed] [Google Scholar]
- Xu Z, Lin S, Wu W, Tan H, Wang Z, Cheng C, et al. Ghrelin prevents doxorubicin-induced cardiotoxicity through TNF-alpha/NF-kappaB pathways and mitochondrial protective mechanisms. Toxicology. 2008;247:133–138. doi: 10.1016/j.tox.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Sinha M, Luxon BA, Yu XJ. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect Immun. 2004;72:498–507. doi: 10.1128/IAI.72.1.498-507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. chaffeensis-infected RF/6A cells at 2 d p.i. were fixed and labeled with rabbit anti-P28 (red). Images were acquired by fluorescence microscopy. Scale bar: 10 μm.


