Abstract
Friend virus infection of adult immunocompetent mice is a well established model for studying genetic resistance to infection by an immunosuppressive retrovirus. This paper reviews both the genetics of immune resistance and the types of immune responses required for recovery from infection. Specific major histocompatibility complex (MHC) class I and II alleles are necessary for recovery, as is a non-MHC gene, Rfv-3, which controls virus-specific antibody responses. In concordance with these genetic requirements are immunological requirements for cytotoxic T lymphocyte, T helper, and antibody responses, each of which provides essential nonoverlapping functions. The complexity of responses necessary for recovery from Friend virus infection has implications for both immunotherapies and vaccines. For example, it is shown that successful passive antibody therapy is dependent on MHC type because of the requirement for T cell responses. For vaccines, successful immunization requires priming of both T cell and B cell responses. In vivo depletion experiments demonstrate different requirements for CD8+ T cells depending on the vaccine used. The implications of these studies for human retroviral diseases are discussed.
Scientific knowledge of retroviral infections in humans is relatively new and little is known about the types of immune responses required to successfully defend against these infections. Such knowledge would be extremely valuable for designing vaccines and immunomodulatory therapeutics. Studies of long term survivors of HIV infection are beginning to provide some insights (1–6), but such individuals are rare, and data are difficult to obtain. In general, cell-mediated responses rather than antibodies are considered the critical elements responsible for resolving most human viral infections. This is because humans with genetic deficiencies in T lymphocytes are very susceptible to many viral infections whereas those with antibody deficiencies are not (7, 8). However, antibody responses also appear essential for resistance against certain viruses such as enterovirus (9) and rabies virus (10), and there are numerous examples of antibodies curing or preventing viral infections (11–17). Thus, there remains controversy regarding which arms of the specific immune system are most important for resolving viral infections. Most likely this resolution depends on the specific virus and host involved, and often more than one aspect of the immune response is important, if not essential.
This review summarizes studies from the polycythemia-inducing strain of Friend virus (FV) complex, an immunosuppressive retrovirus model that induces leukemia in mice. The results indicate that resolution of retroviral infections may require more complex immunological responses than have been found for most other viruses. Numerous experiments using both genetic and immunological approaches demonstrate that immune resistance to FV requires multiple arms of the immune system, including CD4+ T cells, CD8+ T cells, and B cells, each providing essential nonoverlapping functions.
When adult mice of susceptible strains are infected with FV, their spleens rapidly enlarge because of virus-induced polyclonal proliferation of erythroid precursor cells (19–21). Subsequent proviral integration at the Spi-1 (ets) oncogene locus (22–27) combined with inactivation or mutation of the p53 tumor suppressor gene (28–30) produces fully malignant erythroleukemias. This process results in gross splenomegaly at 8–9 days postinfection and transplantable erythroleukemia cells as early as 15–20 days postinfection (31). Thus, a successful immune response must develop quickly enough to keep ahead of this transformation process.
Genes Involved in Recovery from FV Leukemia.
Mice have evolved a formidable array of genes involved in conferring immunological resistance to FV-induced disease, including at least four major histocompatibility complex (MHC) (H-2) genes (32–35) and one non-MHC gene, Rfv-3 (36). In addition, there are six genes (Fv-1–Fv-6) that confer resistance to infection through nonimmunological mechanisms (37–39). Adult mice with appropriate susceptibility alleles at the nonimmunological loci are infectable by FV and develop severe splenomegaly. Their subsequent survival is dependent on MHC and Rfv-3 genes that control immunological responsiveness. Mice having high recovery MHC and Rfv-3 genotypes, such as H-2b/b and Rfv-3r/s, spontaneously recover to near normal spleen size within several weeks and generally live out a normal life-span. Occasionally mice may eventually relapse, indicating the presence of persistent infection (40), but this aspect will not be further discussed. Experiments with MHC recombinant mice show that MHC regions H-2A, E, D, and T are important for recovery from acute FV infection.
The H-2D region of the mouse MHC has a very potent influence on recovery from FV infection because it encodes the class I molecules that present viral antigens to CTL (39). Of interest, the H-2D region also influences the kinetics of virus-specific CD4+ helper T cell responsiveness (41) and controls host susceptibility to FV-induced immunosuppression (ref. 42; Table 1). The H-2D region exhibits an unusual gene–dose effect whereby H-2Db/b mice show the highest recovery incidence, H-2Db/d mice are intermediate, and H-2Dd/d mice are lowest. Each of these genotypes differs in various FV-specific immune parameters (Table 1). One obvious way such a gene–dose effect might occur is through altering expression levels of the Db class I molecules used to present viral peptides to cytotoxic T lymphocyte (CTL). However, experiments to test this hypothesis in the FV system indicate that Db-associated high recovery did not require homozygous levels of Db expression (43). An alternative that also has been investigated is whether expression of low recovery alleles, such as Dd, might produce a negative influence on recovery. For instance, Dd gene products could delete potential Friend-specific T cells by negative selection during development in the thymus. Experiments with Dd transgenic mice showed that expression of Dd in an H-2b mouse did not adversely impact recovery (44). It is also possible that some of the effects associated with H-2D are mediated by other genes that are very closely linked to H-2D and have not been separated from H-2D in the MHC recombinants used for mapping experiments. Possibilities include the tumor necrosis factor complex and the H-2L gene.
Table 1.
Gene dosage effects of H-2D genotype
| H-2D genotype | FV-specific T cell
responses
|
Recovery from FV*
|
FV-induced immunosuppression§ | ||
|---|---|---|---|---|---|
| CD4+ T cell proliferation† | CD8+ CTL‡ | Low FV dose | High FV dose | ||
| b/b | rapid | +++ | yes | yes | no |
| b/d | slow | ++ | yes | no | no |
| d/d | negative | + | no | no | yes |
Low dose, 100 spleen focus forming units; high dose, 1000 spleen focus forming units.
Kinetics of FV-specific CD4+ T cell proliferative responses after challenge with high dose of FV (rapid, 6 days; slow, 16 days).
The magnitude of FV-specific CTL responses is influenced by both the H-2D type and by the FV dose used for infection (41).
Significant decrease in antibody response to sheep red blood cell challenge.
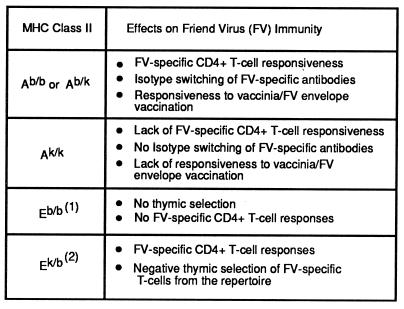
Two class II MHC genes, H-2A and H-2E, also play important roles in immunity to FV. For H-2A, high recovery is associated with the H-2b haplotype, and the effect is dominant (Table 2). The H-2Ab allele acts like a typical immune response gene influencing antigen presentation to CD4+ T cells (39). Mice with homozygous mutations in H-2A, such as H-2Abm12, or mice with low recovery alleles, such as H-2Ak, fail to mount T cell proliferative responses and have a low incidence of recovery (45).
Table 2.
MHC class II effects on FV immunity
(1) H-2E molecules are not expressed on the cell surface in H-2b mice because of lack of a functional E α gene (46, 47). (2) H-2E α chains form the k haplotype associate with β chains from the b haplotype to form functional cell surface heterodimers that present F-MuLV envelope peptides to CD4+ T cells (48). H-2Ek/k molecules may affect the FV-specific immune response also, but their role is unknown.
The situation with H-2E is more complex than H-2A because it has both positive and negative effects on FV immunity (35). Mice with an H-2b haplotype do not express H-2E heterodimers because of a defect in the gene encoding the α chain (46, 47). However, the H-2Eb β gene comes into play when a functional α chain gene is introduced by breeding with mice carrying another haplotype such as H-2a. H-2a/b heterozygous mice use a hybrid molecule comprised of an Ek α chain and an Eb β chain to present a Friend murine leukemia virus (F-MuLV) envelope peptide to CD4+ T cells (48). Blocking this presentation with specific antibodies reduces recovery, indicating an important positive role in FV immunity (35). However, despite this role, studies in transgenic and MHC recombinant mice have shown that the overall effect of expressing H-2E molecules is a decrease in recovery from FV infection. This reduction in recovery appears to occur through negative thymic selection of T cells that recognize H-2E. Thus, the positive and negative effects are temporally separated with positive effects occurring during the immune response and negative effects occurring during development of the T cell repertoire (Table 2). The H-2Qa-Tla region has a weak, but detectable, effect on recovery from FV leukemia (33). This is a rather large genetic region and the influence on recovery is not very strong, so the exact gene involved has not yet been determined.
In addition to the four MHC genes described above, the immune response to FV is also strongly influenced by a non-MHC gene, Rfv-3 (36). Mice require at least one resistance allele at this locus to make antiviral neutralizing antibodies to clear plasma viremia after FV infection. This effect is necessary, but not sufficient, for recovery from leukemia, as will be discussed further below.
Studies of FV-Specific Immunity.
Several studies have shown significant correlations between recovery from FV leukemia and various parameters of the FV-specific immune response. These include: (i) CTL responses, (ii) T cell proliferative responses, and (iii) production of virus-neutralizing antibodies. Subsequent investigations have established that each of these responses not only correlates with recovery but is also required.
CTL.
FV-specific CTL have been shown to recognize antigens in the context of both H-2Db and H-2Dd molecules (39). The primary CTL response from FV-infected recovering mice is directed against determinant(s) in the F-MuLV envelope protein (49, 50). A peptide from this protein has been described as an epitope for in vitro restimulated CTL (51). However, most primary CTL from infected mice do not recognize this epitope, and the major epitope recognized by CTL in recovering mice has not yet been identified. CTL responses also are developed during the rejection of a transplantable Friend tumor cell line, but in contrast to infection with live virus, the predominant CTL response is against an epitope encoded by the viral gag gene (52–56).
CD8+ CTL responses correlate with reduction of splenomegaly in FV-infected animals (49) and probably act by direct killing of infected cells. In the FV model, CTL are detectable by direct assays without in vitro stimulation. The in vivo importance of the CD8+ T cell response has been demonstrated in resistant H-2b/b mice that were depleted of CD8+ cells before infection with FV. CD8 depletion increases mortality by greater than 70% (49).
T cell proliferation.
The rapid development of CD4+ T cell proliferative responses correlates with recovery from a high dose inoculation of FV (39, 41, 45) (Table 1). The CD4+ T cell response is specific for determinants in the F-MuLV envelope protein (45), and two T helper epitopes from the gp70 portion of envelope have been described at the peptide level (48, 57). One peptide binds to H-2Ab molecules and the other to H-2E molecules, thus providing ligands for recognition by CD4+ T cells. FV-specific CD4+ T cells play a central role in FV immunity, providing immunological help for CTL (50, 58) and B cells (35) and maybe also providing direct antiviral activity. Abrogation of these functions by in vivo depletion of CD4+ cells significantly compromises recovery from FV infection (49).
Cytokines.
For some murine leukemia viruses, type 1 T helper responses associated with specific cytokine profiles appear protective whereas type 2 responses do not (59). This issue has not been thoroughly addressed in the FV system, but studies on specific cytokines have been done. One study demonstrates depressed IL-2 and tumor necrosis factor-α levels in FV-infected BALB/c mice (60). Furthermore, in vivo therapy with tumor necrosis factor-α has been shown to produce temporary regression of FV-induced splenomegaly. However, the mechanism may have been through inhibition of hematopoiesis rather than immunomodulation of FV-specific responses (61). IL-6 and IFNγ levels are depressed in FV-infected DBA/2 mice, and therapy with a combination of IFNγ and lactoferrin increases natural killer (NK) cell activity and enhances survival (62). In other experiments, treatment of FV-infected mice with recombinant human IL-7 was shown to increase NK activity and produced long term survival in 20% of the mice (63). Thus, a major role for cytokines in recovery from FV is likely, but the specific mediators have not yet been completely determined. However, requirements for both CTL and IgG class antibodies in recovery from FV infection suggest that cytokines associated with TH-1 or TH-0 type responses might correlate with recovery.
Antibody and B cells.
Virus-neutralizing antibodies are required for recovery from FV infection, and their production is influenced by a non-MHC gene, Rfv-3 (Table 3). Rfv-3s/s mice have a suppressed FV-specific antibody response, even in the presence of the proper MHC type (H-2b/b) for virus-specific T cell responsiveness. Of interest, Rfv-3 appears to affect only the FV-specific antibody response and not responsiveness to other antigens (64). Failure to mount a virus-neutralizing antibody response to FV infection increases mortality by 90% or greater (58). The Rfv-3 gene has been mapped to chromosome 15 of the mouse, unlinked to the MHC, Ig, or T cell receptor loci (65). However, genetic linkage to several cytokine receptor genes (IL-2Rb, IL-3Rb1, and IL-3Rb2) suggests possible candidates for Rfv-3. It is of obvious interest to elucidate the mechanism by which a retrovirus can specifically suppress the antibody responses directed against it. In addition to the production of virus-neutralizing antibodies, B cells also appear to have important roles in antigen presentation and/or cytokine production. Both CD4+ and CD8+ T cell responses to FV-induced tumors are significantly reduced in B cell-depleted mice (66).
Table 3.
Recovery from FV induced leukemia is influenced by MHC genes (H-2) and Rfv-3
| Mouse strain | H-2 | Rfv-3† | Day 30
postinfection*
|
Recovery from FV leukemia | |
|---|---|---|---|---|---|
| FV viremia | FV neutralizing antibody | ||||
| A.BY | b/b | s/s | + | − | no |
| (C57BL/10 × A.BY)F1 | b/b | s/r | − | + | yes |
| A/WySn | a/a | s/s | + | − | no |
| (B10.A × A/WySn)F1 | a/a | s/r | − | + | no |
All of these mouse strains have similar levels of viremia at 10-14 days postinfection with FV.
s/r mice are similar to r/r mice in recovery from viremia and antibody production.
FV-Induced Immunosuppression.
FV suppresses both cellular and humoral immune responses in certain strains of mice (64, 67–70), and an important host gene has been mapped to H-2D (42). For example, H-2Dd/d mice are susceptible to FV-induced immunosuppression, but H-2Db/b mice are resistant (Table 1). After FV infection in H-2Dd/d mice, humoral immune responses to subsequent challenges with strong antigens such as sheep red blood cells are suppressed (70). Responses to T-independent antigens such as 2,4,6,-trinitrophenyl–Ficoll are affected as well, suggesting that immunosuppression need not act through decreased T cell help (64). The involvement of the H-2D region also suggests possible involvement of NK cells. Binding of the Ly-49A receptor on NK cells to H-2Dd molecules can induce global down-regulation of NK cell-mediated killing (71), and decreased NK activity has been associated with FV infection (63). FV-immunosuppressed mice also have been reported to have impaired antigen presentation by macrophages (72). Important to note, susceptibility to immunosuppression does not preclude successful treatment by immunotherapy (58) or protection by vaccination (42).
Immunotherapy.
Strain A mice lack virus-specific antibody responses because of their Rfv-3s/s type and fail to recover from FV infection. Immunotherapy using virus-neutralizing mAbs is effective at reducing mortality by 80–100% in A.BY mice, even when treatments are initiated as late as 10 days postinfection (58). Successful therapy requires both CD4+ and CD8+ T cells because depletion of either subset abrogates recovery. In contrast to the success of therapy in A.BY mice, immunotherapy is ineffective in the MHC congenic A strain A/Wy (H-2a/a, Rfv-3s/s), which is highly susceptible to FV-induced immunosuppression. The cause of the failure of antibody therapy in A/Wy mice appears to be weak T cell responses, which develop with slow kinetics relative to the A.BY strain. However, therapy becomes highly successful in A/Wy mice when the virus inoculum is reduced 5-fold. The resultant slowing of virus spread during antibody therapy allows immune responses to develop before becoming overwhelmed with the viral load. Furthermore, the treated animals are subsequently protected from a high dose challenge of virus. Thus, antibody therapy allows development of long term protective immunity.
Vaccination.
Experiments have shown that protection from FV infection can be elicited by several different types of vaccines including killed and attenuated viruses, viral proteins, peptides, and recombinant vaccinia vectors expressing FV genes (73–77). The study of vaccinated mice has allowed the identification of protective immunological epitopes and determination of the types of immunological responses necessary and/or sufficient for protection.
Protective epitopes have been localized to F-MuLV gag and env proteins by using recombinant vaccinia viruses expressing these genes (74, 76). F-MuLV envelope protects against infection better than gag, so most work has concentrated on envelope. The gp70 envelope protein contains at least one CTL epitope (49), three T helper epitopes (48, 57), and two neutralizing antibody epitopes (78–80). The potency of the T helper determinants has been demonstrated by successful vaccination with a small envelope peptide containing a T helper cell epitope (77).
Protection from FV-induced disease in vaccinated mice correlates with antibody responses, CD4+ T cell proliferative responses, and CD8+ CTL responses (74, 76, 81). Of interest, the requirement for CD8+ T cells in protection is dependent on the number of T helper epitopes in the vaccine (50). Mice immunized with a recombinant vaccinia vector expressing the full length F-MuLV envelope protein containing multiple immunological epitopes require CD4+ T cells for protection but not CD8+ T cells. However, if the number of immunological epitopes in the vaccine is reduced, CD8+ T cells as well as CD4+ T cells are critical for protection. Surprisingly, CD8+ T cell epitopes are not necessary in the vaccine even when CD8+ T cells are required for protection. This paradox appears to be due to the ability of vaccine-primed CD4+ T cells to provide immunological help for CD8+ T cells that are stimulated by the live virus challenge. Additional data also indicate that the expression of multiple CD4 epitopes in the vaccine is more important than expression of CD8 epitopes (50).
The method of immunization can dramatically alter the efficacy of vaccination, especially in terms of the ability to cross-protect different strains of mice. For example, immunization by tail scratch with recombinant vaccinia expressing the F-MuLV env protein protects H-2a/b mice but not MHC congenic H-2a/a mice (74) (Table 4). The nonresponsiveness of H-2a/a mice maps to the H-2A class II genes (42). On the other hand, when the same protein is biochemically purified and inoculated s.c. with complete Freund’s adjuvant or synthetic adjuvants, both strains of mice are protected (81, 82) (Table 4). Thus, there does not appear to be a complete lack of envelope responsive immune cells in H-2a/a mice, but their responsiveness is weak in the absence of adjuvant. Immunization with a live attenuated virus also protects mice of several MHC types, including H-2a/a mice (42, 74). The ability to protect regardless of MHC type correlates with induction of detectable, cell-mediated, and neutralizing antibody responses before challenge (74). Thus, the virus is faced with preexisting immunological effectors that can reduce the effective virus dose.
Table 4.
Protection of mice with different MHC types using various vaccines
Implications.
In conclusion, the FV model has yielded valuable information regarding genetic resistance to retroviral disease, but it is obvious that much remains to be discovered about the immunological mechanisms by which the genes impart their influence. Of particular interest are how the Rfv-3 gene causes susceptibility to suppression of the FV-specific antibody responses, how class I MHC genes influence FV-specific CD4+ T cell proliferative responses, and how the H-2D gene influences virus-induced general immunosuppression. The elucidation of these mechanisms may aid in the development of immunotherapies and vaccines that may be applicable to human diseases.
Although results from FV studies cannot be directly related to human infections such as HIV, consideration of human data in light of the FV results may lead to new interpretations and even better designs for human experiments. For example, it is now known that both non-MHC (83–85) and MHC genes (86) influence the rate of HIV infection and progression to AIDS in humans. Furthermore, there is no reason to suspect that the immune responses required to deal with HIV would be any less complex than those illustrated for FV in mice. Thus, by analogy with the results of FV immunotherapy, part of the reason for the failures of passive antibody therapies in AIDS patients may be related to the high virus loads and low T cell counts in the patients studied (87–95). The FV results suggest that HIV immunotherapy might be more successful if initiated early during the course of infection before virus-induced CD4+ T cell depletion.
The best hope for controlling the worldwide pandemic of AIDS lies in development of an effective vaccine. One message that might be gleaned from the FV experiments is that a successful HIV vaccine would most likely be one that stimulates multiple immune system components with a broad spectrum of antigens. Priming with multiple CD4+ T cell epitopes might be very important because of the central role these cells play in amplifying both CTL and antibody responses. One of the best FV vaccines is the live attenuated virus, and live attenuated viruses have been the most successful vaccines in the simian immunodeficiency virus model as well (18). However, there are several concerns about using such a vaccine for HIV in humans. These include reversion to virulence, insertional mutagenesis, recombination with endogenous retroviral sequences to produce new infectious viruses, and pathogenesis in immunocompromised hosts. Ideally, one might construct a live nonretroviral vector to deliver HIV antigens that would replicate for longer periods of time than recombinant vaccinia and still avoid the major drawbacks of retroviral vectors cited above. Continuous expression over a 2- to 3-week period would more closely mimic immunization by a live attenuated retrovirus and allow development of potent immune effectors. Optimal retroviral protection may require the presence of specific effectors rather than just immunological memory, so further studies will be required to determine how such effectors can be persistently maintained.
ABBREVIATIONS
- FV
Friend virus
- MHC
major histocompatibility complex
- CTL
cytotoxic T lymphocyte
- F-MuLV
Friend murine leukemia virus
- NK cell
natural killer cell
References
- 1.Michael N L, Chang G, d’Arcy L A, Ehrenberg P K, Mariani R, Busch M P, Birx D L, Schwartz D H. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz D, Sharma U, Busch M, Weinhold K, Matthews T, Lieberman J, Birx D, Farzedagen H, Margolick J, Quinn T, Davis B, Bagasra O, Pomerantz R, Viscidi R. AIDS Res Hum Retroviruses. 1994;10:1703–1711. doi: 10.1089/aid.1994.10.1703. [DOI] [PubMed] [Google Scholar]
- 4.Harrer T, Harrer E, Kalams S A, Elbeik T, Staprans S I, Feinberg M B, Cao Y, Ho D D, Yilma T, Caliendo A M, Johnson R P, Buchbinder S P, Walker B D. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 5.Toso J F, Chen C H, Mohr J R, Piglia L, Oei C, Ferrari G, Greenberg M L, Weinhold K J. J Infect Dis. 1995;172:964–973. doi: 10.1093/infdis/172.4.964. [DOI] [PubMed] [Google Scholar]
- 6.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, Takiguchi M, Schultz T, McMichael M, Whittler H. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 7.Oldstone M B A. Curr Topics Microbiol Immunol. 1994;189:1–8. doi: 10.1007/978-3-642-78530-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Good R A. Immunol Today. 1991;12:283–286. doi: 10.1016/0167-5699(91)90127-F. [DOI] [PubMed] [Google Scholar]
- 9.McKinney R E, Jr, Katz S L, Wilfert C M. Rev Infect Dis. 1987;9:334–356. doi: 10.1093/clinids/9.2.334. [DOI] [PubMed] [Google Scholar]
- 10.Bahmanyar M, Fayaz A, Nour-Salehi S, Mohammadi M, Koprowski H. J Am Med Assoc. 1976;236:2751–2754. [PubMed] [Google Scholar]
- 11.Fujinami R S, Rosenthal A, Lampert P W, Zurbriggen A, Yamada M. J Virol. 1989;63:2081–2087. doi: 10.1128/jvi.63.5.2081-2087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine B, Hardwick J M, Trapp B D, Crawford T O, Bollinger R C, Griffin D E. Science. 1991;254:856–860. doi: 10.1126/science.1658936. [DOI] [PubMed] [Google Scholar]
- 13.Lodmell D L, Ewalt L C. J Virol. 1985;55:788–795. doi: 10.1128/jvi.55.3.788-795.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palladino G, Mozdzanowska K, Washko G, Gerhard W. J Virol. 1995;69:2075–2081. doi: 10.1128/jvi.69.4.2075-2081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Z Q, Knowles B B, McCarrick J W, Ertl H C. Virology. 1995;214:398–404. doi: 10.1006/viro.1995.0049. [DOI] [PubMed] [Google Scholar]
- 16.Gobet R, Cerny A, Ruedi E, Hengartner H, Zinkernagel R M. Exp Cell Biol. 1988;56:175–180. doi: 10.1159/000163477. [DOI] [PubMed] [Google Scholar]
- 17.Perillo R P, Campbell C R, Strang S, Bodicky C J, Costigan D J. Arch Int Med. 1984;144:81–85. doi: 10.1001/archinte.144.1.81. [DOI] [PubMed] [Google Scholar]
- 18.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 19.Li J-P, D’Andrea A D, Lodish H F, Baltimore D. Nature (London) 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 20.Hoatlin M E, Kozak S L, Lilly F, Chakraborti A, Kozak C A, Kabat D. Proc Natl Acad Sci USA. 1990;87:9985–9989. doi: 10.1073/pnas.87.24.9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferro F E, Jr, Kozak S L, Hoatlin M E, Kabat D. J Biol Chem. 1993;268:5741–5747. [PubMed] [Google Scholar]
- 22.Lavigueur A, Bernstein A. Oncogene. 1991;6:2197–2201. [PubMed] [Google Scholar]
- 23.Paul R, Schuetze S, Kozak S L, Kozak C A, Kabat D. J Virol. 1991;65:464–467. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuetze S, Paul R, Gliniak B C, Kabat D. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreau-Gachelin F, Ray D, de Both N J, van der Feltz M J, Tambourin P, Tavitian A. Leukemia. 1990;4:20–23. [PubMed] [Google Scholar]
- 26.Moreau-Gachelin F, Ray D, Mattei M G, Tambourin P, Tavitian A. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- 27.Moreau-Gachelin F, Tavitian A, Tambourin P. Nature (London) 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 28.Munroe D G, Peacock J W, Benchimol S. Mol Cell Biol. 1990;10:3307–3313. doi: 10.1128/mcb.10.7.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson P, Benchimol S. Cancer Surv. 1992;12:137–151. [PubMed] [Google Scholar]
- 30.Johnson P, Chung S, Benchimol S. Mol Cell Biol. 1993;13:1456–1463. doi: 10.1128/mcb.13.3.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tambourin P, Wendling F, Moreau-Gachelin F. Blood Cells. 1981;7:133–144. [PubMed] [Google Scholar]
- 32.Lilly F. J Exp Med. 1968;127:465–473. doi: 10.1084/jem.127.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazawa M, Nishio J, Wehrly K, David C S, Chesebro B. J Immunol. 1992;148:1964–1967. [PubMed] [Google Scholar]
- 34.Chesebro B, Wehrly K. J Immunol. 1978;120:1081–1085. [PubMed] [Google Scholar]
- 35.Perry L L, Miyazawa M, Hasenkrug K, Wehrly K, David C S, Chesebro B. J Virol. 1994;68:4921–4926. doi: 10.1128/jvi.68.8.4921-4926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chesebro B, Wehrly K. Proc Natl Acad Sci USA. 1979;76:425–429. doi: 10.1073/pnas.76.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-David Y, Bernstein A. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 38.Hoatlin M E, Kabat D. Trends Microbiol. 1995;3:51–57. doi: 10.1016/s0966-842x(00)88875-7. [DOI] [PubMed] [Google Scholar]
- 39.Chesebro B, Miyazawa M, Britt W J. Annu Rev Immunol. 1990;8:477–499. doi: 10.1146/annurev.iy.08.040190.002401. [DOI] [PubMed] [Google Scholar]
- 40.Chesebro B, Bloom M, Wehrly K, Nishio J. J Virol. 1979;32:832–837. doi: 10.1128/jvi.32.3.832-837.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Britt W J, Chesebro B. J Exp Med. 1983;157:1736–1745. doi: 10.1084/jem.157.6.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison R P, Earl P L, Nishio J, Lodmell D L, Moss B, Chesebro B. Nature (London) 1987;329:729–732. doi: 10.1038/329729a0. [DOI] [PubMed] [Google Scholar]
- 43.Hasenkrug K J, Sprangrude G J, Nishio J, Brooks D M, Chesebro B. J Virol. 1994;68:2059–2064. doi: 10.1128/jvi.68.4.2059-2064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazawa M, Nishio J, Wehrly K, Jay G, Melvold R W, Chesebro B. Eur J Immunogenet. 1992;19:159–164. doi: 10.1111/j.1744-313x.1992.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 45.Miyazawa M, Nishio J, Chesebro B. J Exp Med. 1988;168:1587–1605. doi: 10.1084/jem.168.5.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones P P, Murphy D B, McDevitt H O. Immunogenetics. 1981;12:321–337. doi: 10.1007/BF01561674. [DOI] [PubMed] [Google Scholar]
- 47.Mathis D J, Benoist C, Williams V E d, Kanter M, McDevitt H O. Proc Natl Acad Sci USA. 1983;80:273–277. doi: 10.1073/pnas.80.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwashiro M, Kondo T, Shimizu T, Yamagishi H, Takahashi K, Matsubayashi Y, Masuda T, Otaka A, Fujii N, Ishimoto A, Miyazawa M, Robertson M J, Chesebro B, Kuribayashi K. J Virol. 1993;67:4533–4542. doi: 10.1128/jvi.67.8.4533-4542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson M N, Spangrude G J, Hasenkrug K, Perry L, Nishio J, Wehrly K, Chesebro B. J Virol. 1992;66:3271–3277. doi: 10.1128/jvi.66.6.3271-3277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasenkrug K J, Brooks D M, Nishio J, Chesebro B. J Virol. 1996;70:368–372. doi: 10.1128/jvi.70.1.368-372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan K S, Lilly F. Virology. 1991;181:91–100. doi: 10.1016/0042-6822(91)90473-o. [DOI] [PubMed] [Google Scholar]
- 52.Greenberg P D, Cheever M A. Surv Immunol Res. 1985;4:283–296. [PubMed] [Google Scholar]
- 53.Klarnet J P, Kern D E, Okuno K, Holt C, Lilly F, Greenberg P D. J Exp Med. 1989;169:457–467. doi: 10.1084/jem.169.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klarnet J P, Kern D E, Okuno K, Holt C, Lilly F, Greenberg P D. J Exp Med. 1989;169:457–467. doi: 10.1084/jem.169.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kondo T, Uenishi H, Shimizu T, Hirama T, Iwashiro M, Kuribayashi K, Tamamura H, Fujii N, Fujisawa R, Miyazawa M, Yamagishi H. J Virol. 1995;69:6735–6741. doi: 10.1128/jvi.69.11.6735-6741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, Qin H, Chesebro B, Cheever M A. J Virol. 1996;70:7773–7782. doi: 10.1128/jvi.70.11.7773-7782.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu T, Uenishi H, Teramura Y, Iwashiro M, Kuribayashi K, Tamamura H, Fujii N, Yamagishi H. J Virol. 1994;68:7704–7708. doi: 10.1128/jvi.68.12.7704-7708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasenkrug K J, Brooks D M, Chesebro B. Proc Natl Acad Sci USA. 1995;92:10492–10495. doi: 10.1073/pnas.92.23.10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marshall D J, Gaulton G N. Leukemia. 1996;10:1860–1866. [PubMed] [Google Scholar]
- 60.Faxvaag A, Espevik T, Dalen A. Cell Immunol. 1993;150:247–256. doi: 10.1006/cimm.1993.1194. [DOI] [PubMed] [Google Scholar]
- 61.Johnson C S, Chang M J, Furmanski P. Blood. 1988;72:1875–1883. [PubMed] [Google Scholar]
- 62.Lu L, Shen R N, Zhou S Z, Srivastava C, Harrington M, Miyazawa K, Wu B, Lin Z H, Ruscetti S, Broxmeyer H E. Int J Hematol. 1991;54:117–124. [PubMed] [Google Scholar]
- 63.Lu L, Zhou Z, Wu B, Xiao M, Shen R N, Williams D E, Kim Y J, Kwon B S, Ruscetti S, Broxmeyer H E. Int J Cancer. 1992;52:261–265. doi: 10.1002/ijc.2910520218. [DOI] [PubMed] [Google Scholar]
- 64.Morrison R P, Nishio J, Chesebro B. J Exp Med. 1986;163:301–314. doi: 10.1084/jem.163.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hasenkrug K J, Valenzuela A, Letts V A, Nishio J, Chesebro B, Frankel W N. J Virol. 1995;69:2617–2620. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schultz K R, Klarnet J P, Gieni R S, HayGlass K T, Greenberg P D. Science. 1990;249:921–923. doi: 10.1126/science.2118273. [DOI] [PubMed] [Google Scholar]
- 67.Butler R C, Frier J M, Chapekar M S, Graham M O, Friedman H. Infect Immun. 1983;39:1260–1264. doi: 10.1128/iai.39.3.1260-1264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mortensen R F, Ceglowski W S, Friedman H. J Immunol. 1974;112:2077–2086. [PubMed] [Google Scholar]
- 69.Mortensen R F, Ceglowski W S, Friedman H. J Immunol. 1973;111:1810–1819. [PubMed] [Google Scholar]
- 70.Ceglowski W S, Friedman H. J Immunol. 1968;101:594–604. [PubMed] [Google Scholar]
- 71.Yokoyama W M, Daniels B F, Seaman W E, Hunziker R, Margulies D H, Smith H R. Semin Immunol. 1995;7:89–101. doi: 10.1006/smim.1995.0013. [DOI] [PubMed] [Google Scholar]
- 72.Jones S M, Moors M A, Ryan Q, Klyczek K K, Blank K J. Viral Immunol. 1992;5:201–211. doi: 10.1089/vim.1992.5.201. [DOI] [PubMed] [Google Scholar]
- 73.Hunsmann G, Schneider J, Schulz A. Virology. 1981;113:603–612. doi: 10.1016/0042-6822(81)90188-4. [DOI] [PubMed] [Google Scholar]
- 74.Earl P L, Moss B, Morrison R P, Wehrly K, Nishio J, Chesebro B. Science. 1986;234:728–731. doi: 10.1126/science.3490689. [DOI] [PubMed] [Google Scholar]
- 75.Ruan K S, Lilly F. Proc Natl Acad Sci USA. 1992;89:12202–12206. doi: 10.1073/pnas.89.24.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyazawa M, Nishio J, Chesebro B. J Virol. 1992;66:4497–4507. doi: 10.1128/jvi.66.7.4497-4507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyazawa M, Fujisawa R, Ishihara C, Takei Y A, Shimizu T, Uenishi H, Yamagishi H, Kuribayashi K. J Immunol. 1995;155:748–758. [PubMed] [Google Scholar]
- 78.Chesebro B, Wehrly K, Cloyd M, Britt W, Portis J, Collins J, Nishio J. Virology. 1981;112:131–144. doi: 10.1016/0042-6822(81)90619-x. [DOI] [PubMed] [Google Scholar]
- 79.Britt W J, Chesebro B. J Immunol. 1983;130:2363–2367. [PubMed] [Google Scholar]
- 80.Robertson M N, Miyazawa M, Mori S, Caughey B, Evans L H, Hayes S F, Chesebro B. J Virol Methods. 1991;34:255–271. doi: 10.1016/0166-0934(91)90105-9. [DOI] [PubMed] [Google Scholar]
- 81.Ishihara C, Miyazawa M, Nishio J, Chesebro B. J Immunol. 1991;146:3958–3963. [PubMed] [Google Scholar]
- 82.Ishihara C, Miyazawa M, Nishio J, Azuma I, Chesebro B. Vaccine. 1992;10:353–356. doi: 10.1016/0264-410x(92)90378-w. [DOI] [PubMed] [Google Scholar]
- 83.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 84.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 85.Paxton W A, Dragic T, Koup R A, Moore J P. AIDS Res Hum Retroviruses. 1996;12:1203–1207. doi: 10.1089/aid.1996.12.1203. [DOI] [PubMed] [Google Scholar]
- 86.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O’Brien S J, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann D L. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 87.Zolla-Pazner S, Pinter A, Mizuma H. J Virol Methods. 1987;17:45–53. doi: 10.1016/0166-0934(87)90067-x. [DOI] [PubMed] [Google Scholar]
- 88.Jackson G G, Perkins J T, Rubenis M, Paul D A, Knigge M, Despotes J C, Spencer P. Lancet. 1988;ii:647–652. doi: 10.1016/s0140-6736(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 89.Levy J, Youvan T, Lee M L. Blood. 1994;84:2130–2135. [PubMed] [Google Scholar]
- 90.Hinkula J, Bratt G, Gilljam G, Nordlund S, Broliden P A, Holmberg V, Olausson-Hansson E, Albert J, Sandstrom E, Wahren B. J Acquired Immune Defic Syndr. 1994;7:940–951. [PubMed] [Google Scholar]
- 91.Jacobson J M, Colman N, Ostrow N A, Simson R W, Tomesch D, Marlin L, Rao M, Mills J L, Clemens J, Prince A M. J Infect Dis. 1993;168:298–305. doi: 10.1093/infdis/168.2.298. [DOI] [PubMed] [Google Scholar]
- 92.Vittecoq D, Mattlinger B, Barre-Sinoussi F, Courouce A M, Rouzioux C, Doinel C, Bary M, Viard J P, Bach J F, Rouger P, Lefrere J J. J Infect Dis. 1992;165:364–368. doi: 10.1093/infdis/165.2.364. [DOI] [PubMed] [Google Scholar]
- 93.Vittecoq D, Chevret S, Morand-Joubert L, Heshmati F, Audat F, Bary M, Dusautoir T, Bismuth A, Viard J P, Barre-Sinoussi F, Bach J F, Lefrere J J. Proc Natl Acad Sci USA. 1995;92:1195–1199. doi: 10.1073/pnas.92.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karpas A, Hewlett I K, Hill F, Gray J, Byron N, Gilgen D, Bally V, Oates J K, Gazzard B, Epstein J E. Proc Natl Acad Sci USA. 1990;87:7613–7617. doi: 10.1073/pnas.87.19.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karpas A, Gray J, Byron N, Gilgen D, Bally V, Oates J K, Gazzard B. Biotherapy. 1990;2:159–172. doi: 10.1007/BF02173455. [DOI] [PubMed] [Google Scholar]