Abstract
Lipid droplets are cellular organelles that consists of a neutral lipid core covered by a monolayer of phospholipids and many proteins. They are thought to function in the storage, transport, and metabolism of lipids, in signaling, and as a specialized microenvironment for metabolism in most types of cells from prokaryotic to eukaryotic organisms. Lipid droplets have received a lot of attention in the last 10 years as they are linked to the progression of many metabolic diseases and hold great potential for the development of neutral lipid-derived products, such as biofuels, food supplements, hormones, and medicines. Proteomic analysis of lipid droplets has yielded a comprehensive catalog of lipid droplet proteins, shedding light on the function of this organelle and providing evidence that its function is conserved from bacteria to man. This review summarizes many of the proteomic studies on lipid droplets from a wide range of organisms, providing an evolutionary perspective on this organelle.
Keywords: apolipoprotein, mass spectrometry, triglycerides
The first observation of lipid droplets can be credited to van Leeuwenhoek when he viewed fat globules in milk using his self-made microscope in 1674 (1). Much more recently, lipid droplets have been found to exist as an intracellular organelle (2–5). About two decades ago, the identification of marker proteins provided a tool for scientists to study this cellular structure biochemically (6–8). In the last 10 years, established isolation methods and advanced proteomic technology have allowed scientists to study the protein composition of lipid droplets from many cell types and tissues, such as fibroblasts (9, 10), epithelia (10–13), adipocytes (14, 15), hepatocytes (16, 17), macrophages (18), pancreatic β-cells (19), mammary glands (20), livers (20, 21), white adipose tissues (22), and skeletal muscles (23), as well as from many other popular model organisms, including plants (24–28), insects (29–33), yeast (34–37), green algae (38–40), bacteria (41–43), and other sources (44–47) (Table 1). Several hundred proteins have been identified by these proteomic analyses, and some have been further verified using biochemical, imaging, and functional methods. Although there is diversity in the results from different studies, intriguingly, several functional groups of proteins are consistently and reproducibly represented. One such group includes lipid synthetic enzymes, suggesting that lipid droplets may function as a cellular site for some steps of lipid synthesis. Another exciting group that has been identified is proteins involved in membrane trafficking, such as the Rabs, SNAREs, and Arfs and their coatomers. The presence of these proteins on lipid droplets suggests that the organelle is able to change size, probably using fusion and fission mechanisms similar to cellular vesicles. Furthermore, this group may mediate the ability of the organelle to move within cells and to interact with other cellular organelles. A third group includes signaling proteins, such as RalA, 14-3-3, and Rap1B, and several protein kinases. The discovery of enzymes responsible for the synthesis of lipid signaling molecules provides further evidence of a signaling role for lipid droplets (48). A fourth, highly surprising group includes proteins involved in protein degradation, such as AUP1 (14, 17, 18, 49) and UBXDs (10, 50). A hypothesis that lipid droplets may mediate intracellular protein degradation was proposed (51), and interestingly, several studies support this possibility (52, 53). Collectively, these findings represent an important development in cell biology, as our understanding of lipid droplets has evolved from that of an inert storage structure to a functional cellular organelle.
TABLE 1.
Proteomic studies of lipid droplets
| Organisms | References |
| Mammals | |
| Mammalian cells | |
| Fibroblasts | (9, 10) |
| Epithelia | (10–13) |
| Adipocytes | (14, 15) |
| Hepatocytes | (16, 17) |
| Macrophage | (18) |
| Pancreatic β-cell | (19) |
| Skeletal muscle cells | (23) |
| Mammalian tissues | |
| Mammary gland | (20) |
| Liver | (20, 21) |
| White adipose tissue | (22) |
| Skeletal muscle | (23) |
| Plants | |
| Arabidopsis thaliana | (24) |
| Sesamum indicum | (25) |
| Brassica napus | (26–28) |
| Insects | |
| Drosophila melanogaster | (29, 30) |
| Bombyx mori | (31, 32) |
| Manduca sexta | (33) |
| Yeast | |
| Saccharomyces cerevisiae | (34–36) |
| Yarrowia lipolytica | (37) |
| Green algae | |
| Chlamydomonas reinhardtii | (38–40) |
| Bacteria | |
| Rhodococcus sp | (41) |
| Rhodococcus opacus | (42) |
| Rhodococcus ruber | (42) |
| Mycobacterium bovis | (43) |
| Other sources | |
| Coral–dinoflagellate endosymbiosis | (44) |
| Chloroplasts | (45, 46) |
| Chromoplasts | (46) |
| ER lumen | (47) |
Despite the consistent representation of these functional groups of proteins in lipid droplets from many sources, the proteomes that have been obtained so far have significant differences as well. These differences likely point to distinct properties and functions of different lipid droplets. Although the proteomic studies may shed light on the functions and diversity of the organelle, these complex proteomes await further analysis and validation using complimentary methods.
Here, we summarize proteomic studies of lipid droplets from different organisms and examine similarities in the putative structural proteins of this organelle from bacteria to man.
LIPID DROPLET PROTEOMICS
Mammalian cells and tissues
Lipid droplets have been observed in many mammalian cells and tissues, and it is currently thought that all mammalian cells are able to accumulate neutral lipids to form lipid droplets. The discovery of perilipin in 1991 (6) stimulated research on lipid droplets in adipocytes, and the cloning of the gene encoding the protein adipocyte differentiation-related protein (ADRP) in 1992 (7) and subsequent localization of this protein to lipid droplets (8) provided an essential marker protein that facilitated the study of lipid droplets in nonadipocytes. These proteins and three additional related proteins, including tail-interacting protein of 47 kDa (Tip47) (54), S3-12 (55), and OXPAT (56), previously termed PAT proteins (57), were recently renamed as perilipins (PLIN) 1–5 (58). Among them, PLIN1 has been well characterized. Its phosphorylation regulation and interaction with hormone-sensitive lipase (HSL) provide a molecular mechanism to govern lipid droplet triacylglycerol (TAG) hydrolysis (59).
Lipid droplets have been isolated successfully from several mammalian cells and tissues based on their low density and the presence of marker proteins (PLINs), and their proteins have been identified by mass spectrometry. An early proteomic study was performed using the mammary gland and liver (20). The proteins found in isolated lipid droplets from mammary gland were very similar to those of the milk fat globule membrane (MFGM) (20). In the same study, the first proteome of liver lipid droplets identified ADRP and fatty acid binding protein (FABP) (20). Three more lipid droplet proteomes were carried out using isolated lipid droplets from Chinese hamster ovary cells (CHO K2) (9), human hepatoma cell line Huh7 (16), and human squamous epithelial carcinoma cells A431 (11). A comparative proteomic study was conducted using 3T3-L1 adipocytes under basal and lipolytic conditions (14). In addition to several main lipid droplet proteins, many other proteins were identified in these studies, including lipid metabolism enzymes, such as adipose tri-glyceride lipase (ATGL); lipid synthases, such as acyl-CoA synthases; membrane-trafficking proteins, such as Rabs; and signaling proteins, such as 14-3-3 (9, 14), Rap1B (16), and protein kinases (11, 21). Interestingly, the proteomic study using CHO cells also demonstrated that oleate treatment stimulates CGI-58 translocation to lipid droplets (9). CGI-58 was later found to coactivate ATGL (60–64).
The discovery that Rab proteins are present on lipid droplets provided critical evidence in support of lipid droplets as a cellular organelle. The study using 3T3-L1 adipocytes found that some Rab proteins are recruited to lipid droplets during lipolysis, further suggesting that lipid droplet association of Rab proteins is physiological (14). More evidence that Rab proteins are lipid droplet proteins has since been provided by several functional studies. Rab18 was found to mediate the interaction between lipid droplets and endoplasmic reticulum (ER) (65, 66). The recruitment of early endosomes to lipid droplets has also been found to be Rab protein-dependent (67). In fact, the presence of Rab proteins in lipid droplet proteomes is consistent across all studies of mammalian lipid droplets. This observation supports the hypothesis that lipid droplets may play a role in neutral lipid transport between cellular organelles (68–70).
The ability of some proteins to move onto and off of the surface of lipid droplets suggests that, in addition to their possible role in neutral lipid trafficking, lipid droplets are probably dynamic and may be involved in other cellular activities. In addition to the translocation of Rabs to lipid droplets in lipolytic conditions (14), in vitro experiments have demonstrated that GTP can stimulate the translocation of Arf1 and its coatomers to lipid droplets (10). An RNA interference screening study conducted in Drosophila S2 cells revealed the importance of these proteins in lipid droplet morphology (71, 72). In addition, proteomic studies of phosphorylated proteins on lipid droplets isolated from Hela cells identified 45 phosphorylated proteins on the organelle, including ADRP, Rab, and ATGL (10, 12). Recently, phosphorylation of S406 on mouse ATGL has been found to be essential for the activation of the enzyme (73). Together, proteomic studies not only demonstrate the dynamic activities of lipid droplets but also provide important information for further mechanistic investigation of the organelle.
Lipid droplets may also play an active role in lipid synthesis. Lipid synthetic enzymes have been found on lipid droplets isolated from organisms ranging from bacteria to mammalian cells (34, 41, 74, 75). Some of these enzymes have then been localized to lipid droplets in living cells using GFP fusion proteins and immunofluorescence (16, 76, 77). Several in vitro studies have verified both the presence and activity of these enzymes on lipid droplets (78–80), suggesting that the lipid droplet is a cellular site for some steps of lipid synthesis.
Collectively, recent proteomic studies of mammalian cell lipid droplets have dramatically extended our understanding of lipid droplet properties and functions. Based on these and other studies, a picture is emerging of lipid droplets as an active organelle involved in lipid synthesis, catabolism, and trafficking.
Plants
Lipid droplets in plants are also referred to as lipid bodies, oil bodies, lipid particles, oleosomes, and spherosomes. Morphological and biochemical studies of the organelle became possible after the main proteins of plant lipid droplets were separated electrophoretically by Charles Slack in 1980 (81). Advances in mass spectroscopy protein identification enabled a series of investigations of plant lipid droplets. The first proteomic study was conducted with Arabidopsis thaliana seeds and identified eight proteins by LC-MS/MS (24), including four types of oleosins, ATS1 (a homologous gene to caleosin), 11-β-hydroxysteroid dehydrogenase-like protein (a homologous gene to steroleosin), TIP3.2 (probable aquaporin), and a predicted GPI-anchored protein. Up to 79% of the lipid droplet proteins present consisted of oleosins (24).
In 2005, MALDI-MS and MS/MS were used to analyze sesame seed lipid droplet proteins and found that oleosin and caleosin, both 17 kDa, are acetylated at the N-terminus, whereas steroleosin possesses a free methionine at its N-terminus. The authors speculated that these modifications enhance the stability of lipid droplets (25). In 2006, Katavic et al. analyzed the protein and lipid composition of lipid droplets from two Brassica napus cultivars and identified several new proteins, including a dehydrogenase and a myrosinase-associated protein which seemed to be involved in lipid droplet degradation (26). A proteomic investigation was conducted with rapeseed lipid droplets in 2009, which identified 25 proteins, including 15 oleosins, 3 steroleosins, and 1 caleosin. Two of the oleosins were found to be acetylated at their N-termini (27). A 2011 study examined the accumulation of lipids and proteins during seed formation by analyzing both whole seed and purified lipid droplets (28). The authors found that lipid and protein accumulation followed a sigmoidal pattern. The isolated lipid droplets of early-stage seeds and mature seeds had different oil and oleosin compositions. At the earlier stage, C18:2-containing lipids were the main lipids in the seeds, whereas C18:1-containing lipids predominated in mature seeds. Low molecular weight oleosins (BnS3 and BnS5) appeared earlier than high molecular weight oleosins (BnS1, BnS2, and BnS4) (28).
The mechanism by which oleosin is targeted to the lipid droplet has been studied. The currently accepted model is that three separate domains are responsible for targeting the protein to lipid droplets: an N-terminal amphipathic domain, a C-terminal amphipathic α-helical domain, and a central hydrophobic lipid droplet-anchoring domain. The central hydrophobic domain probably embeds into the hydrophobic core, and the two terminal domains attach to the surface of the lipid droplets (82). These important secondary structures enable oleosins to attach tightly to lipid droplets, thus enhancing their stability.
Although these studies conducted in plants have identified dozens of lipid droplet proteins, their number falls far short of the hundreds of proteins identified in mammalian cells. The main protein of plant lipid droplets, oleosin, often represents up to 79% of the total protein of the organelle. Compared with its mammalian counterpart, the organelle in plants appears to lack other functional proteins, such as membrane-trafficking and lipid metabolic enzymes, suggesting that energy storage is the primary function of plant lipid droplets.
Insects
The popular genetically tractable animal model, Drosophila melanogaster, is a good system for lipid droplet research (83). Drosophila expresses two PLIN proteins, Lsd1 and Lsd2 (84), that are good marker proteins not only for localization of lipid droplets but also for determination of purity of isolated lipid droplets. Similar to their presence in mammals, lipid droplets in Drosophila are found in both fat and nonfat body cells. Although no one has successfully isolated lipid droplets from white adipose tissue in mammals, lipid droplets have been isolated from Drosophila fat body. In a proteomic analysis of fat body lipid droplets, 248 proteins were identified, including lipid synthetic enzymes and membrane-trafficking proteins, some of which were verified by intracellular localization studies (29). Another proteomic study was carried out using lipid droplets from Drosophila embryos. A particularly interesting finding from this work was that several histone proteins are present on embryonic lipid droplets (30). This finding distinguishes the lipid droplets in Drosophila embryos from those of other organisms. Among the proteins discovered were trafficking and transport proteins, such as Rab proteins, Sar1, and tubulin, which is consistent with the earlier discovery that lipid droplets move in a microtubule-dependent manner (85). This finding was recently confirmed by localizing the motor regulator Klar to lipid droplets (86). Further investigation of the differences in protein composition between the lipid droplets from the larval fat body and embryonic lipid droplets may provide a better understanding of the role of lipid droplets in Drosophila embryonic development.
Proteomic studies were conducted recently using isolated lipid droplets from silkmoth Bombyx mori (B. mori), and several proteins were identified (31, 32). During analysis of isolated lipid droplet protein phosphorylation, B. mori Lsd1 (Bmlsd1) was identified (32). More detailed proteomics was carried out using another insect species Manduca sexta (M. sexta) (33). The study involved three subtypes of lipid droplets that were isolated from the fat bodies of Manduca larvae and adult insects, as well as from Manduca ovary. Very similar to lipid droplet-associated proteins in other organisms, M. sexta lipid droplet-associated proteins include lipid synthetic and metabolic enzymes, membrane-trafficking proteins, and cell signaling proteins (33). Several apolipoproteins were identified as components of M. sexta lipid droplets, raising an interesting question about whether apolipoprotein particles in mammals originally had roles in lipid droplets. The identification of several ribosomal proteins in M. sexta lipid droplets needs to be further verified, but it raises provocative questions about whether lipid droplets are involved in protein translation.
Yeast
Although evolutionarily distant from mammals, yeast Saccharomyces cerevisiae (S. cerevisiae) is an excellent genetic model organism used in many studies related to human diseases. The recent prevalence of lipid metabolic diseases has motivated research on lipid metabolism in yeast, especially the study of lipid droplets (also referred to as lipid particles and lipid bodies). During the last decade, proteomic studies of yeast lipid droplets have shed new light on the role of lipid droplets in lipid synthesis (34) and membrane trafficking (35, 36, 87), and they have identified some proteins relevant to lipid metabolic disease in humans, such as the homolog of seipin (35). Mutants of seipin are related to lipodystrophy in humans (88) and to supersized lipid droplets in yeast (89). By the same token, lipin 1, the mutation of which causes lipodystrophy in mice (90, 91), has also been detected on lipid droplets (92, 93) and found to control lipid droplet formation in yeast (94).
Most of the lipid droplet proteins identified in proteomic studies in S. cerevisiae were verified using chromosomally integrated GFP-tagged proteins (76). Although one perilipin-like protein, MPL1 (Metarhizium anisopliae perilipin-like protein), was identified in the fungus Metarhizium anisopliae (95), no homologs for mammalian PLIN proteins have been identified in S. cerevisiae. One of the four major lipid droplet proteins identified in S. cerevisiae was absent in the S. cerevisiae erg6 strain, identifying that the protein is ERG6 (96). ERG6 was shown to be a sterol Δ24-methy-ltransferas (97) and to mediate the interaction between lipid droplets and mitochondria (98). Proteomic studies of lipid droplets from another yeast strain, Yarrowia lipolytica, also identified lipid synthetic enzymes and membrane-trafficking proteins (37). The major lipid droplet protein in this strain is ERG27, which is different from ERG6 identified in lipid droplets in S. cerevisiae (37).
Thus, a common feature of lipid droplet proteins identified by proteomic studies in yeast is that most of the proteins identified are involved in lipid synthesis, lipid metabolism, and membrane trafficking, indicating that lipid droplets in yeast are very active in lipid metabolism and transport.
Green algae
Algae are a possible source of biodiesel, prompting interest in their lipid droplets and the TAG they contain. Two proteomic studies, which have recently been conducted on lipid droplets isolated from Chlamydomonas reinhardtii, identified about 250 proteins (38, 39). Similar to lipid droplet proteomes of other organisms, the proteins found in algae also include lipid synthetic enzymes, suggesting that lipid droplets in algae are also a dynamically active cellular organelle. An interesting discovery in these proteomic studies is a protein named major lipid droplet protein (MLDP), which is conserved across several types of microalgae. Suppressing MLDP expression with RNA interference induces an increase in lipid droplet size without altering cellular TAG content (38).
Bacteria
A broad evolutionary conservation of neutral lipid storage structures has been suspected for many years. However, it was not until recent proteomic, biochemical, and molecular studies in bacteria that this conservation across species was demonstrated to extend into the prokaryotic kingdom. The earliest of the studies on bacterial lipid droplets was conducted with Rhodococcus ruber (42). The isolated fraction was defined as “lipid inclusions.” Lipid droplets were next isolated from the hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guérin (43). Ten major proteins were identified, including 5 novel ones, a number far less than that of lipid droplets from other organisms. However, a proteomic study of the total proteins of lipid droplets purified from Rhodococcus sp. RHA1 identified 228 proteins (41). Two putative structural proteins, microorganism lipid droplet small (MLDS) and phage shock protein A (PspA) were identified, constituting about 15% of the total lipid droplet proteins. Deletion of MLDS resulted in the formation of enlarged lipid droplets, suggesting that this protein plays a critical role in lipid droplet dynamics (e.g., growth/metabolism). The lipid droplet-targeting domain of MLDS was determined to a region containing three putative α helixes. On the basis of both the primary amino acid sequence and predicted structure similarity, MLDS resembles mammalian apolipoprotein (APO)A-I/A-IV/E (41).
In addition, the proteomic study of RHA1 identified many ribosomal proteins and transcriptional regulators in isolated lipid droplets (41), similar to lipid droplet proteomes of other organisms (15, 18, 19, 30, 33, 36, 38). These data suggest that lipid droplets may provide intracellular membrane system for compartmentalization of cellular activities of certain bacteria.
Other sources
Proteomic studies have also been conducted on lipid droplets isolated from nonmodel organisms. In one, lipid droplets from a coral-dinoflagellate endosymbiont were analyzed (44). The protein composition was similar to that of other lipid droplets, including chaperones and proteins that are involved in lipid metabolism and trafficking.
Some cellular structures equivalent to lipid droplets have also been isolated and analyzed by mass spectrometry. These include plastoglobules in chloroplasts and chromoplasts in plants (45, 46) and luminal lipid droplets (LLD) in the endoplasmic reticulum of mammals (47). These observations suggest that lipid droplet-like structures can be generated inside of some other cellular organelles.
PUTATIVE STRUCTURAL PROTEINS OF LIPID DROPLETS
Proteomic and cell biology studies suggest that almost all organisms are able to accumulate neutral lipids in lipid droplets. However, the structural proteins of lipid droplets from different organisms are quite distinct. In mammals and insects, the predominant proteins are members of the perilipin family (58); however, other organisms do not express PLINs. In plants, the primary proteins are oleosin, caleosin, and steroleosin (24). In an insect fungal pathogen Metarhizium anisopliae, the Metarhizium perilipin-like protein (MPL1) was identified (95). In addition, proteomic studies have identified MLDP in green algae (38), Erg6 in yeast (96), and MLDS in bacteria (41) as possible structural proteins of the organelle. Although there are substantial differences in the amino acid sequences of mammalian perilipins compared with the main lipid droplet proteins from various species, when these proteins are expressed in different organisms, they all localize to lipid droplets. For example, mammalian PLIN 1, 2, and 3 can be targeted to bacterial lipid droplets (99), and Drosophila Lsd1 and Lsd2 can be targeted to the lipid droplets of CHO cells (100). Furthermore, Dictyostelium Lsd1 can be targeted to CHO cell lipid droplets (100), and five mycobacterial proteins can be targeted to yeast lipid droplets (43). Therefore, it appears that these lipid droplet structural proteins from diverse species have certain properties in common that allow them to be properly targeted to lipid droplets.
In mammals, APOs are the structural proteins of blood lipoprotein particles that are similar to lipid droplets in their general structure. Several lines of evidence suggest that lipid droplet structural proteins have properties in common with APOs. First, some APOs have been found on cellular lipid droplets. For example, APOA-V has been found on lipid droplets from hepatoma cells (101), APOA-I has been found on lipid droplets from white adipose tissue (22), and APOA-I and APOE have been found on lipid droplets of skeletal muscle cells (23). Second, both plant oleosins and bacterial MLDS have similar amphipathic helixes to the APOs (41, 82). Third, C-terminal sequence of PLIN 3, Tip47, forms a four-helix bundle that resembles the LDL receptor (LDLR) binding domain of APOE (102). Furthermore, C terminus of PLIN 2 (ADRP) shares a similar amino acid sequence with the APOE-like domain of Tip47 (103) and, hence, is predicted to form a similar structure. Together, it seems that these APO-like proteins are evolutionarily conserved proteins involved in lipid storage and trafficking and have evolved the ability to target intracellular or extracellular lipid-filled structures covered with a phospholipid monolayer.
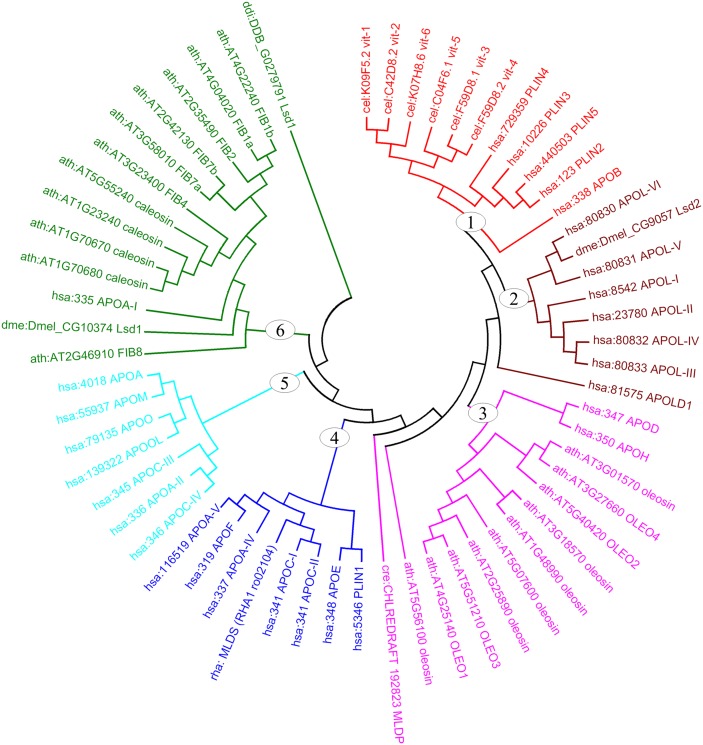
To better understand the evolutionary relationships among these APO-like proteins, we selected 61 proteins, identified as abundant lipid droplet proteins, from seven species and grouped them based on protein similarity using Molecular Evolutionary Genetics Analysis version 4.0 (MEGA4) software (Fig. 1) (104). These proteins were mainly clustered into six groups, each group including at least one human APO. Four human PLIN family proteins were similar to the Caenorhabditis elegans APOs, 6 vit family proteins (group 1). In group 2, Drosophila Lsd2 showed similarity to 7 mammalian APOL subfamily proteins. Ten plant oleosin proteins were close to mammalian APOD and APOH in group 3. An interesting point was that bacterial MLDS (RHA1 ro02104) was found to be close to mammalian APOC-I/APOC-II in group 4, indicating that APO-like proteins may have occurred evolutionarily earlier than PLIN family proteins. Plant fibrillarin (FIB) family proteins were found in the plastoglobules of A. thaliana and have no enzymatic activity (46, 105). These 7 FIB proteins were clustered with 4 plant caleosin proteins, Drosophila Lsd1, and mammalian APOA-I in group 6. Sequence alignment analysis showed that sequence similarity among different groups was very low. Therefore, their apparent similarity may be due more to the similarity of their predicted structures than to similarity of amino acid sequences. Taken together, this analysis speculates that these APO-like proteins may be evolutionarily conserved as the structural proteins of lipid droplets.
Fig. 1.
Phylogenetic analysis of APO-like proteins in seven species. These proteins are mainly clustered into six groups annotated clockwise in red (group 1), maroon (group 2), fuchsia (group 3), blue (group 4), aqua (group 5), and green (group 6). Cluster analysis was conducted using MEGA4. APO, apolipoprotein; ath, Arabidopsis thaliana; cel, Caenorhabditis elegans; cre, Chlamydomonas reinhardtii; ddi, Dictyostelium discoideum; dme, Drosophila melanogaster; hsa, Homo sapiens; OLEO, oleosin; rha, Rhodococcus sp. RHA1. The sequences and names of proteins are downloaded from KEGG database (106).
CONCLUDING REMARKS
Proteomic studies have contributed greatly to current lipid droplet research, giving rise to i) the discovery of many proteins that define this storage structure as a multifunctional cellular organelle; ii) the development of new tools/methods that not only facilitate lipid droplet studies but also extend our knowledge of the organelle from prokaryotic to eukaryotic organisms; and iii) the identification of novel major lipid droplet proteins that show similarities to APOs.
Subsequent proteomic studies of lipid droplets should focus on i) the study of important tissues and organs, such as white adipose tissue, heart, and bone marrow, and on popular model organisms, such as Caenorhabditis elegans and zebra fish; ii) the discovery of key regulators or proteins that govern lipid droplet biogenesis and aspects of morphology, including membrane curvature and budding, as well as dynamics of fusion and fission; and iii) the identification of major proteins that mediate interaction between lipid droplets and other cellular organelles.
The results from proteomic studies discussed herein have clearly portrayed lipid droplets as a multifunctional organelle that is conserved from bacteria to man, and imply that they may be a very ancient cellular organelle, if not the earliest, with a membrane monolayer for compartmentalization of cellular activities. It will be intriguing to determine functions of bacterial lipid droplets to confirm this hypothesis. Similarities between the putative structural proteins of lipid droplets and the APOs of lipoprotein particles suggest that all structures with a neutral lipid core covered by a monolayer of phospholipids may have APO-like proteins as their structural proteins. These similarities will likely enable the study of lipid droplets from a broad range of sources, facilitating further investigations of their cellular functions and roles in the pathology of metabolic diseases as well as in biofuel development.
Acknowledgments
The authors thank Drs. Joy Fleming and John K. Zehmer for their critical reading of this manuscript and useful suggestions.
Footnotes
Abbreviations:
- ADRP
- adipocyte differentiation-related protein
- ATGL
- adipose triglyceride lipase
- CHO
- Chinese hamster ovary
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- MLDS
- microorganism lipid droplet small
- PLIN
- perilipin
- TAG
- triacylglycerol
This work was supported by Ministry of Science and Technology of China Grant Nos. 2009CB919000 and 2011CBA00900 and by National Natural Science Foundation of China Grant Nos. 30971431, 31000365, and 31100068. The authors declare that there are no competing financial interests.
REFERENCES
- 1.Kernohan E. A., Lepherd E. E. 1969. Size distribution of fat globules in cow's milk during milking, measured with a Coulter counter. J. Dairy Res. 36: 177–182 [Google Scholar]
- 2.Murphy D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40: 325–438 [DOI] [PubMed] [Google Scholar]
- 3.Martin S., Parton R. G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7: 373–378 [DOI] [PubMed] [Google Scholar]
- 4.Beckman M. 2006. Cell biology. Great balls of fat. Science. 311: 1232–1234 [DOI] [PubMed] [Google Scholar]
- 5.Farese R. V., Jr., Walther T. C. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266: 11341–11346 [PubMed] [Google Scholar]
- 7.Jiang H. P., Serrero G. 1992. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc. Natl. Acad. Sci. USA. 89: 7856–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasaemle D. L., Barber T., Wolins N. E., Serrero G., Blanchette-Mackie E. J., Londos C. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38: 2249–2263 [PubMed] [Google Scholar]
- 9.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279: 3787–3792 [DOI] [PubMed] [Google Scholar]
- 10.Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6: 3256–3265 [DOI] [PubMed] [Google Scholar]
- 11.Umlauf E., Csaszar E., Moertelmaier M., Schuetz G. J., Parton R. G., Prohaska R. 2004. Association of stomatin with lipid bodies. J. Biol. Chem. 279: 23699–23709 [DOI] [PubMed] [Google Scholar]
- 12.Kim S. C., Chen Y., Mirza S., Xu Y., Lee J., Liu P., Zhao Y. 2006. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5: 3446–3452 [DOI] [PubMed] [Google Scholar]
- 13.Orban T., Palczewska G., Palczewski K. 2011. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem. 286: 17248–17258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasaemle D. L., Dolios G., Shapiro L., Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279: 46835–46842 [DOI] [PubMed] [Google Scholar]
- 15.Hou J., Liu Y., Lu Z., Liu X., Liu J. 2012. Biochemical characterization of RNase HII from Aeropyrum pernix. Acta Biochim. Biophys. Sin. (Shanghai). 44: 339–346 [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644: 47–59 [DOI] [PubMed] [Google Scholar]
- 17.Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J. Biochem. 139: 921–930 [DOI] [PubMed] [Google Scholar]
- 18.Wan H. C., Melo R. C., Jin Z., Dvorak A. M., Weller P. F. 2007. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 21: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson S., Resjo S., Gomez M. F., James P., Holm C. 2012. Characterization of the lipid droplet proteome of a clonal insulin-producing beta-cell line (INS-1 832/13). J. Proteome Res. 11: 1264–1273 [DOI] [PubMed] [Google Scholar]
- 20.Wu C. C., Howell K. E., Neville M. C., Yates J. R., 3rd, McManaman J. L. 2000. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 21: 3470–3482 [DOI] [PubMed] [Google Scholar]
- 21.Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernandez M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269 [DOI] [PubMed] [Google Scholar]
- 22.Kanshin E., Wang S., Ashmarina L., Fedjaev M., Nifant'ev I., Mitchell G. A., Pshezhetsky A. V. 2009. The stoichiometry of protein phosphorylation in adipocyte lipid droplets: analysis by N-terminal isotope tagging and enzymatic dephosphorylation. Proteomics. 9: 5067–5077 [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Wang Y., Li J., Yu J., Pu J., Li L., Zhang S., Peng G., Yang F., Liu P. 2011. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J. Proteome Res. 10: 4757–4768 [DOI] [PubMed] [Google Scholar]
- 24.Jolivet P., Roux E., D'Andrea S., Davanture M., Negroni L., Zivy M., Chardot T. 2004. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42: 501–509 [DOI] [PubMed] [Google Scholar]
- 25.Lin L. J., Liao P. C., Yang H. H., Tzen J. T. 2005. Determination and analyses of the N-termini of oil-body proteins, steroleosin, caleosin and oleosin. Plant Physiol. Biochem. 43: 770–776 [DOI] [PubMed] [Google Scholar]
- 26.Katavic V., Agrawal G. K., Hajduch M., Harris S. L., Thelen J. J. 2006. Protein and lipid composition analysis of oil bodies from two Brassica napus cultivars. Proteomics. 6: 4586–4598 [DOI] [PubMed] [Google Scholar]
- 27.Jolivet P., Boulard C., Bellamy A., Larre C., Barre M., Rogniaux H., d'Andrea S., Chardot T., Nesi N. 2009. Protein composition of oil bodies from mature Brassica napus seeds. Proteomics. 9: 3268–3284 [DOI] [PubMed] [Google Scholar]
- 28.Jolivet P., Boulard C., Bellamy A., Valot B., d'Andrea S., Zivy M., Nesi N., Chardot T. 2011. Oil body proteins sequentially accumulate throughout seed development in Brassica napus. J. Plant Physiol. 168: 2015–2020 [DOI] [PubMed] [Google Scholar]
- 29.Beller M., Riedel D., Jansch L., Dieterich G., Wehland J., Jackle H., Kuhnlein R. P. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 5: 1082–1094 [DOI] [PubMed] [Google Scholar]
- 30.Cermelli S., Guo Y., Gross S. P., Welte M. A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795 [DOI] [PubMed] [Google Scholar]
- 31.Fónagy A., Ohnishi A., Esumi Y., Suzuki Y., Matsumoto S. 2005. Further studies of lipid droplets in the bombykol-producing pheromone gland of Bombyx mori. Ann. N. Y. Acad. Sci. 1040: 310–314 [DOI] [PubMed] [Google Scholar]
- 32.Ohnishi A., Hull J. J., Kaji M., Hashimoto K., Lee J. M., Tsuneizumi K., Suzuki T., Dohmae N., Matsumoto S. 2011. Hormone signaling linked to silkmoth sex pheromone biosynthesis involves Ca2+/calmodulin-dependent protein kinase II-mediated phosphorylation of the insect PAT family protein Bombyx mori lipid storage droplet protein-1 (BmLsd1). J. Biol. Chem. 286: 24101–24112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soulages J. L., Firdaus S. J., Hartson S., Chen X., Howard A. D., Arrese E. L. 2012. Developmental changes in the protein composition of Manduca sexta lipid droplets. Insect Biochem. Mol. Biol. 42: 305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181: 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grillitsch K., Connerth M., Kofeler H., Arrey T. N., Rietschel B., Wagner B., Karas M., Daum G. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta. 1811: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Athenstaedt K., Jolivet P., Boulard C., Zivy M., Negroni L., Nicaud J. M., Chardot T. 2006. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics. 6: 1450–1459 [DOI] [PubMed] [Google Scholar]
- 38.Moellering E. R., Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell. 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen H. M., Baudet M., Cuine S., Adriano J. M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M., et al. 2011. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics. 11: 4266–4273 [DOI] [PubMed] [Google Scholar]
- 40.Fan H. X., Miao L. L., Liu Y., Liu H. C., Liu Z. P. 2011. Gene cloning and characterization of a cold-adapted beta-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microb. Technol. 49: 94–99 [DOI] [PubMed] [Google Scholar]
- 41.Ding Y., Yang L., Zhang S., Wang Y., Du Y., Pu J., Peng G., Chen Y., Zhang H., Yu J., et al. 2012. Identification of the major functional proteins of prokaryotic lipid droplets. J. Lipid Res. 53: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalscheuer R., Waltermann M., Alvarez M., Steinbuchel A. 2001. Preparative isolation of lipid inclusions from Rhodococcus opacus and Rhodococcus ruber and identification of granule-associated proteins. Arch. Microbiol. 177: 20–28 [DOI] [PubMed] [Google Scholar]
- 43.Low K. L., Shui G., Natter K., Yeo W. K., Kohlwein S. D., Dick T., Rao S. P., Wenk M. R. 2010. Lipid droplet-associated proteins are involved in the biosynthesis and hydrolysis of triacylglycerol in Mycobacterium bovis bacillus Calmette-Guerin. J. Biol. Chem. 285: 21662–21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng S. E., Chen W. N., Chen H. K., Lu C. Y., Mayfield A. B., Fang L. S., Chen C. S. 2011. Lipid bodies in coral-dinoflagellate endosymbiosis: proteomic and ultrastructural studies. Proteomics. 11: 3540–3555 [DOI] [PubMed] [Google Scholar]
- 45.Vidi P. A., Kanwischer M., Baginsky S., Austin J. R., Csucs G., Dormann P., Kessler F., Brehelin C. 2006. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281: 11225–11234 [DOI] [PubMed] [Google Scholar]
- 46.Ytterberg A. J., Peltier J. B., van Wijk K. J. 2006. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Gilham D., Lehner R. 2007. Proteomic and lipid characterization of apolipoprotein B-free luminal lipid droplets from mouse liver microsomes: implications for very low density lipoprotein assembly. J. Biol. Chem. 282: 33218–33226 [DOI] [PubMed] [Google Scholar]
- 48.Bozza P. T., Magalhaes K. G., Weller P. F. 2009. Leukocyte lipid bodies - biogenesis and functions in inflammation. Biochim. Biophys. Acta. 1791: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spandl J., Lohmann D., Kuerschner L., Moessinger C., Thiele C. 2011. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J. Biol. Chem. 286: 5599–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zehmer J. K., Bartz R., Bisel B., Liu P., Seemann J., Anderson R. G. 2009. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 122: 3694–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ploegh H. L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 448: 435–438 [DOI] [PubMed] [Google Scholar]
- 52.Ohsaki Y., Cheng J., Suzuki M., Fujita A., Fujimoto T. 2008. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J. Cell Sci. 121: 2415–2422 [DOI] [PubMed] [Google Scholar]
- 53.Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. 2010. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285: 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolins N. E., Rubin B., Brasaemle D. L. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276: 5101–5108 [DOI] [PubMed] [Google Scholar]
- 55.Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., Bickel P. E. 2003. Adipocyte protein S3-12 coats nascent lipid droplets. J. Biol. Chem. 278: 37713–37721 [DOI] [PubMed] [Google Scholar]
- 56.Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., et al. 2006. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 55: 3418–3428 [DOI] [PubMed] [Google Scholar]
- 57.Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. 1999. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10: 51–58 [DOI] [PubMed] [Google Scholar]
- 58.Kimmel A. R., Brasaemle D. L., McAndrews-Hill M., Sztalryd C., Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51: 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. 2009. Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 284: 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319 [DOI] [PubMed] [Google Scholar]
- 62.Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. 2009. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J. Biol. Chem. 284: 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z., Zhou L. 2011. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286: 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., et al. 2011. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286: 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118: 2601–2611 [DOI] [PubMed] [Google Scholar]
- 66.Martin S., Driessen K., Nixon S. J., Zerial M., Parton R. G. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280: 42325–42335 [DOI] [PubMed] [Google Scholar]
- 67.Liu P., Bartz R., Zehmer J. K., Ying Y. S., Zhu M., Serrero G., Anderson R. G. 2007. Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta. 1773: 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zehmer J. K., Huang Y., Peng G., Pu J., Anderson R. G., Liu P. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 9: 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodman J. M. 2008. The gregarious lipid droplet. J. Biol. Chem. 283: 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy S., Martin S., Parton R. G. 2009. Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta. 1791: 441–447 [DOI] [PubMed] [Google Scholar]
- 71.Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 453: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beller M., Sztalryd C., Southall N., Bell M., Jackle H., Auld D. S., Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahmadian M., Abbott M. J., Tang T., Hudak C. S., Kim Y., Bruss M., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., et al. 2011. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 13: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bozza P. T., Bakker-Abreu I., Navarro-Xavier R. A., Bandeira-Melo C. 2011. Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot. Essent. Fatty Acids. 85: 205–213 [DOI] [PubMed] [Google Scholar]
- 75.Zhang S., Du Y., Wang Y., Liu P. 2010. Lipid droplet - a cellular organelle for lipid metabolism. Acta Biophys. Sin. 26: 97–105 [Google Scholar]
- 76.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. 2003. Global analysis of protein localization in budding yeast. Nature. 425: 686–691 [DOI] [PubMed] [Google Scholar]
- 77.Accioly M. T., Pacheco P., Maya-Monteiro C. M., Carrossini N., Robbs B. K., Oliveira S. S., Kaufmann C., Morgado-Diaz J. A., Bozza P. T., Viola J. P. 2008. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 68: 1732–1740 [DOI] [PubMed] [Google Scholar]
- 78.Sorger D., Daum G. 2002. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286: 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., et al. 2007. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48: 1280–1292 [DOI] [PubMed] [Google Scholar]
- 81.Slack C. R., Bertaud W. S., Shaw B. D., Holland R., Browse J., Wright H. 1980. Some studies on the composition and surface properties of oil bodies from the seed cotyledons of safflower (Carthamus tinctorius) and linseed (Linum ustatissimum). Biochem. J. 190: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murphy D. J., Keen J. N., O'Sullivan J. N., Au D. M., Edwards E. W., Jackson P. J., Cummins I., Gibbons T., Shaw C. H., Ryan A. J. 1991. A class of amphipathic proteins associated with lipid storage bodies in plants. Possible similarities with animal serum apolipoproteins. Biochim. Biophys. Acta. 1088: 86–94 [DOI] [PubMed] [Google Scholar]
- 83.Kühnlein R. P. 2011. The contribution of the Drosophila model to lipid droplet research. Prog. Lipid Res. 50: 348–356 [DOI] [PubMed] [Google Scholar]
- 84.Lu X., Gruia-Gray J., Copeland N. G., Gilbert D. J., Jenkins N. A., Londos C., Kimmel A. R. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 12: 741–749 [DOI] [PubMed] [Google Scholar]
- 85.Welte M. A., Gross S. P., Postner M., Block S. M., Wieschaus E. F. 1998. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 92: 547–557 [DOI] [PubMed] [Google Scholar]
- 86.Yu Y. V., Li Z., Rizzo N. P., Einstein J., Welte M. A. 2011. Targeting the motor regulator Klar to lipid droplets. BMC Cell Biol. 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wiederhold E., Veenhoff L. M., Poolman B., Slotboom D. J. 2010. Proteomics of Saccharomyces cerevisiae organelles. Mol. Cell. Proteomics. 9: 431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Agarwal A. K., Garg A. 2004. Seipin: a mysterious protein. Trends Mol. Med. 10: 440–444 [DOI] [PubMed] [Google Scholar]
- 89.Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., Yang H. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reue K., Xu P., Wang X. P., Slavin B. G. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41: 1067–1076 [PubMed] [Google Scholar]
- 91.Garg A. 2004. Acquired and inherited lipodystrophies. N. Engl. J. Med. 350: 1220–1234 [DOI] [PubMed] [Google Scholar]
- 92.Wang H., Zhang J., Qiu W., Han G. S., Carman G. M., Adeli K. 2011. Lipin-1gamma isoform is a novel lipid droplet-associated protein highly expressed in the brain. FEBS Lett. 585: 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valdearcos M., Esquinas E., Meana C., Gil-de-Gomez L., Guijas C., Balsinde J., Balboa M. A. 2011. Subcellular localization and role of lipin-1 in human macrophages. J. Immunol. 186: 6004–6013 [DOI] [PubMed] [Google Scholar]
- 94.Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., Goodman J. M. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang C., St Leger R. J. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 282: 21110–21115 [DOI] [PubMed] [Google Scholar]
- 96.Leber R., Zinser E., Zellnig G., Paltauf F., Daum G. 1994. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 10: 1421–1428 [DOI] [PubMed] [Google Scholar]
- 97.Zinser E., Paltauf F., Daum G. 1993. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 175: 2853–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pu J., Ha C. W., Zhang S., Jung J. P., Huh W. K., Liu P. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hänisch J., Wältermann M., Robenek H., Steinbüchel A. 2006. Eukaryotic lipid body proteins in oleogenous actinomycetes and their targeting to intracellular triacylglycerol inclusions: impact on models of lipid body biogenesis. Appl. Environ. Microbiol. 72: 6743–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., Kimmel A. R. 2002. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277: 32253–32257 [DOI] [PubMed] [Google Scholar]
- 101.Nelbach L., Shu X., Konrad R. J., Ryan R. O., Forte T. M. 2008. Effect of apolipoprotein A-V on plasma triglyceride, lipoprotein size, and composition in genetically engineered mice. J. Lipid Res. 49: 572–580 [DOI] [PubMed] [Google Scholar]
- 102.Hickenbottom S. J., Kimmel A. R., Londos C., Hurley J. H. 2004. Structure of a lipid droplet protein; the PAT family member TIP47. Structure. 12: 1199–1207 [DOI] [PubMed] [Google Scholar]
- 103.Chong B. M., Russell T. D., Schaack J., Orlicky D. J., Reigan P., Ladinsky M., McManaman J. L. 2011. The adipophilin C terminus is a self-folding membrane-binding domain that is important for milk lipid secretion. J. Biol. Chem. 286: 23254–23265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 105.Deruère J., Romer S., d'Harlingue A., Backhaus R. A., Kuntz M., Camara B. 1994. Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell. 6: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38: D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]