Abstract
L-bifunctional enzyme (Ehhadh) is part of the classical peroxisomal fatty acid β-oxidation pathway. This pathway is highly inducible via peroxisome proliferator-activated receptor α (PPARα) activation. However, no specific substrates or functions for Ehhadh are known, and Ehhadh knockout (KO) mice display no appreciable changes in lipid metabolism. To investigate Ehhadh functions, we used a bioinformatics approach and found that Ehhadh expression covaries with genes involved in the tricarboxylic acid cycle and in mitochondrial and peroxisomal fatty acid oxidation. Based on these findings and the regulation of Ehhadh's expression by PPARα, we hypothesized that the phenotype of Ehhadh KO mice would become apparent after fasting. Ehhadh mice tolerated fasting well but displayed a marked deficiency in the fasting-induced production of the medium-chain dicarboxylic acids adipic and suberic acid and of the carnitine esters thereof. The decreased levels of adipic and suberic acid were not due to a deficient induction of ω-oxidation upon fasting, as Cyp4a10 protein levels increased in wild-type and Ehhadh KO mice.We conclude that Ehhadh is indispensable for the production of medium-chain dicarboxylic acids, providing an explanation for the coordinated induction of mitochondrial and peroxisomal oxidative pathways during fasting.
Keywords: peroxisome, fatty acid oxidation, mouse models, omega oxidation
Mitochondria and peroxisomes both perform fatty acid β-oxidation, but they have different roles in metabolism (1–3). Mitochondria oxidize the bulk of long-chain fatty acids present in our diet and fat stores, whereas peroxisomes oxidize specific carboxylic acids such as very long-chain fatty acids, branched-chain fatty acids, bile acids, and fatty dicarboxylic acids (DCAs) (1–3). Both organelles harbor multiple sets of different enzymes that function in β-oxidation. As such, peroxisomal enzymes differ in their ability to handle the specific peroxisomal carboxylic acid substrates. This knowledge has been derived from experiments with purified enzymes and from analysis of material from patients with single peroxisomal enzyme deficiencies and peroxisomal enzyme knockout (KO) mouse models (1–3).
There are four enzymatic steps in each cycle of peroxisomal β-oxidation: oxidation, hydration, dehydrogenation, and thiolytic cleavage (1–3). Rat and mouse peroxisomes have three acyl-CoA oxidases that perform the first step in β-oxidation. Acox1 handles the straight-chain substrates such as the very long-chain fatty acids and long-chain DCAs, Acox2 is active with the bile acid intermediates and the branched-chain fatty acids, and Acox3 accepts the branched-chain fatty acids (4). For the fourth step, rodents also have three different thiolases, including the 3-ketoacyl-CoA thiolases A (Acaa1a) and B (Acaa1b), and the sterol carrier protein x (Scpx or Scp2). Scp2 is active toward the 3-ketoacyl-CoA species of all peroxisomal substrates. In contrast, no specific substrates are known for Acaa1a and Acaa1b, although they are active toward the 3-ketoacyl-CoA esters of straight-chain fatty acids (5, 6).
For the second and third step of β-oxidation, the hydratase and 3-hydroxyacyl-CoA dehydrogenase activity, peroxisomes harbor D-bifunctional enzyme (Hsd17b4, also known as D-PBE, MFP2, or DBP) and L-bifunctional enzyme (Ehhadh, also known as L-PBE, MFP1, or LBP). Previous studies indicated that Ehhadh KO mice display no appreciable changes in lipid metabolism, and thus Hsd17b4 seems to be able to handle all peroxisomal substrates (7), which is in line with earlier biochemical studies using purified enzymes (8–12). Indeed, HSD17B4-deficient patients and Hsd17b4 KO mice accumulate very long-chain as well as branched-chain fatty acids and bile acid intermediates (13, 14). Thus, no specific substrates for Ehhadh have been identified, although the enzyme is active toward the enoyl-CoA esters of straight-chain fatty acids (15, 16).
DCAs destined for peroxisomal β-oxidation are products of fatty acid ω-oxidation. Long-chain fatty acid ω-oxidation is initiated by microsomal cytochrome P450 enzymes of the Cyp4a family, producing an ω-hydroxyl group that is further metabolized into an ω-carboxyl group by the successive action of an alcohol and aldehyde dehydrogenase. DCA β-oxidation ends with the production of medium-chain DCAs such as adipic (C6-DCA) and suberic acid (C8-DCA). Available evidence suggests that Ehhadh might play a role in the β-oxidation of DCAs (17–19), based on observations that DCA β-oxidation is not deficient in HSD17B4-deficient fibroblasts and that in vitro Hsd17b4 and Ehhadh handle these substrates with comparable efficiency (18). Moreover, in vitro the β-oxidation of the DCAs of long-chain and polyunsaturated fatty acids is impaired in Ehhadh KO hepatocytes but not in Hsd17b4 KO hepatocytes (17, 19).
It has been known for decades that the peroxisomal enzymes Acox1, Ehhadh, and Acaa1b, often referred to as the classical β-oxidation pathway, are highly inducible by phthalate esters and drugs such as fibrates and Wy-14,643 (20). In the liver, this effect is mediated by peroxisome proliferator-activated receptor α (PPARα), a nuclear hormone receptor activated by these compounds. In normal physiology, PPARα is most likely activated by lipolysis-induced elevated levels of free fatty acids. PPARα mediates the adaptive response to fasting by increasing mitochondrial β-oxidation, ketogenesis, and microsomal ω-oxidation (21, 22). The function of the parallel induction of peroxisomal fatty acid β-oxidation is less well understood.
We used a bioinformatics approach to shed light on the physiological role of the classical peroxisomal β-oxidation pathway. This analysis positions Ehhadh in processes that generate energy from fatty acids. Combined with the fact that Ehhadh is PPAR inducible, we hypothesized that the phenotype of Ehhadh KO mice would become apparent after an overnight fast. Consequently, we measured metabolites in Ehhadh KO mice after fasting and show that Ehhadh is crucial for the formation of medium-chain DCAs such as C6-DCA and thus β-oxidation of the major long-chain DCAs produced during fasting.
MATERIALS AND METHODS
Generation of liver Ehhadh gene coexpression subnetworks
GeneNetwork (www.genenetwork.org) is a depository of datasets and tools for use in complex systems biology approaches in order to generate or predict higher order gene function (23, 24). We selected liver microarray data from three independently generated panels of genetically heterogeneous mice, an F2 intercross between C57BL/6 and BTBR mouse strains in which all cases are homozygous for the obese mutation in the leptin gene (the B6BTBRF2 cross; n = 41) (25), an intercross between C57BL/6 and DBA/2J (the BxD genetic reference population; n = 60 male mice) (26, 27), and an F2 intercross with C57BL/6J x C3H/HeJ on an ApoE null background (BHF2) (28). We used the (B6 x BTBR)F2-ob/ob Liver mRNA M430 RMA, UNC Agilent G4121A Liver Orig LOWESS Stanford (male mice), and UCLA BHF2 Liver Male mlratio datasets, respectively. The sole Ehhadh probe on each of the liver databases was selected and the top ∼500 Ehhadh covariates from each experiment were compared. The strongest Ehhadh liver covariates from each experiment (∼45 genes per cross) were used to construct coexpression subnetworks, which were subsequently merged using cytoscape (see Fig. 1 for the cutoff Pearson correlation coefficients used for the different crosses) (29). To determine which Gene Ontology (GO) terms were significantly overrepresented in the sets of Ehhadh correlates, the cytoscape plug-in Bingo (Biological Networks Gene Ontology tool) was used (30). Correlations between Ehhadh liver expression and various clinical phenotypes were determined in the B6BTBRF2 cross database using GeneNetwork.
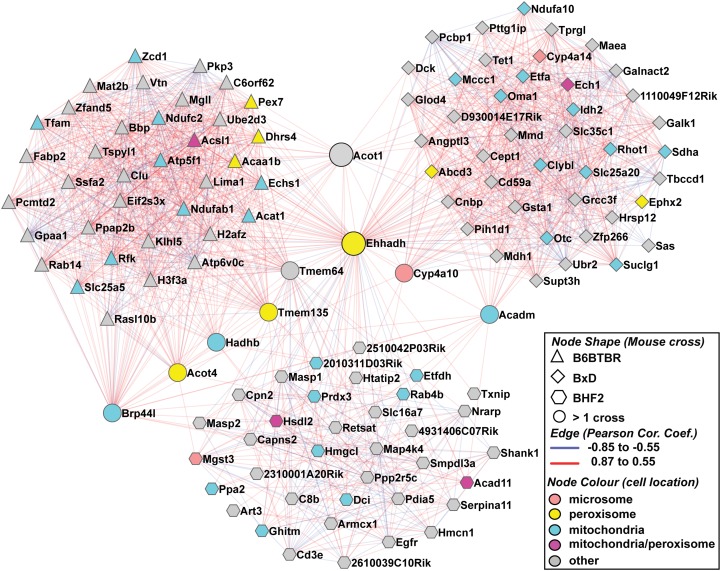
Fig.1.
The hepatic Ehhadh gene coexpression subnetwork. The merged Ehhadh gene subnetwork for an F2 intercross between C57BL/6 and BTBR mouse strains in which all cases are homozygous for the obese mutation in the leptin gene (the B6BTBRF2 cross; triangles), an intercross between C57BL/6 and DBA/2J (the BxD genetic reference population; diamonds), and an F2 intercross with C57BL/6J x C3H/HeJ on an ApoE null background (BHF2; hexagons). Nodes shared by more than one cross are shown as circles. The gene:gene interactions (edges) are indicated as color-coded correlation coefficients. Gene products known to localize in mitochondria are blue nodes, peroxisomal gene products are yellow nodes, and the microsomal Cyp4a gene products are pink nodes. Gene products with a dual peroxisomal and mitochondrial localization are purple nodes.
Animals
The generation of Ehhadh−/− (Ehhadh KO) mice has been described previously (7). Male WT (129S2/Hsd) and Ehhadh KO (129 background) were kept on regular chow and analyzed at 2 months of age. All mice were euthanized at 3:00 PM after 0, 6, or 20 h of fasting (n = 3 animals per time point). At this time, mice were anesthetized using pentobarbital, followed by blood, urine, and organ collection. All experiments were approved by the institutional review boards for animal experiments at the Academic Medical Center, Amsterdam, and the Northwestern University Feinberg School of Medicine, Chicago. All animals received humane care in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Metabolite measurements
Glucose and β-hydroxybutyrate were measured in perchloric acid deproteinized blood using established methods. Insulin (Ultra sensitive mouse insulin ELISA; Crystal Chem Inc., Downers Grove, IL) and acylcarnitines were measured in EDTA-plasma. Plasma, urine, and tissue acylcarnitines were measured on a tandem mass spectrometer (Waters Quattro Premier XE) using methods as described previously (31, 32). Urinary organic acids were methoximated using methoxyamine. After acidification, the organic acids were extracted twice using ethyl acetate/diethyl ether (1:1) and derivatized with N,O-Bis(trimethylsilyl)trifluoroacetamide with 1% Trimethylchlorosilane (BSTFA + TMCS; Thermo scientific). The methoxime trimethylsilyl esters were analyzed by GC/MS (Agilent HP 5973 mass spectrometer) and GC-flame ionization detection (Agilent 6890 SeriesGC) using CP-Sil 5 (Chrompack 7441) and CP-Sil 19 (Chrompack 7712) capillary columns. Creatinine was measured in urine using ultraperformance liquid chromatography-tandem MS with multiple reaction monitoring in positive electrospray ionization mode (Waters Quattro Premier XE). Heptafluorobutyric acid was used as an ion pair agent for the reverse-phase ultraperformance liquid chromatography. Stable isotope internal standards were used for quantification.
Immunoblot analysis
Liver homogenates were prepared in PBS using a dispersion tool followed by sonication. Samples were further prepared according to the instructions for electrophoresis of NuPAGE Bis-Tris mini gels (4–12%; Life Technologies). Cyp4a10 was detected using goat anti-rat Cyp4a1 (Daiichi Pure Chemicals, Tokyo, Japan). Acox1 was detected with an antibody specific for the 50 kDa rat protein (gift of P. van Veldhoven, Katholieke Universiteit Leuven, Belgium). Ehhadh was detected using an IgG fraction against the rat protein (gift of T. Hashimoto, Shinshu University School of Medicine, Matsumoto, Nagano, Japan). Hsd17b4 was detected using an IgG fraction against human Hsd17b4 (gift of T. Hashimoto). Rabbit anti-Abcd3 (or PMP70) was raised against a 21 amino acid synthetic rat peptide (Zymed, Carlsbad, CA). Antibodies were visualized using the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Statistical analysis
The six groups of mice were compared using the nonparametric Kruskal-Wallis (ANOVA) test followed by Dunn's multiple comparison test to determine statistical significance between the WT and Ehhadh KO samples after 20 h of fasting.
RESULTS
The Ehhadh gene coexpression subnetwork indicates a role in the generation of energy
Genes that participate in a similar biochemical process or physiological function often have coregulated expression. To search for clues on the role of Ehhadh, we generated gene coexpression subnetworks using published liver microarray data. We selected three independent mouse experiments: an F2 intercross between C57BL/6 and BTBR mouse strains in which all cases are homozygous for the obese mutation in the leptin gene (the B6BTBRF2 cross [25]), an intercross between C57BL/6 and DBA/2J (the BxD genetic reference population [26, 27]), and an F2 intercross with C57BL/6J x C3H/HeJ on an ApoE null background (BHF2 [28]). These experiments make use of genetic variation between four inbred strains (C57BL/6, BTBR, DBA, and C3H) and thereby enable the study of the effect of this genetic variation on mutant backgrounds known to greatly influence metabolism (ApoE null and ob/ob backgrounds). Due to these inherent genetic variations, Ehhadh expression varied 3.7-, 2.2-, and 1.5-fold within these respective crosses (Supplementary Fig. Ia-c). The top ∼500 Ehhadh liver covariates from each experiment were compared. Although generally distinct in their identity, the three sets of Ehhadh gene covariates shared several genes (Supplementary Fig. IIA) as well as GO enrichments (data not shown). To depict these observations graphically, the strongest Ehhadh liver covariates from each experiment (top ∼45 genes per cross) were used to construct three coexpression subnetworks, which were subsequently merged into one network and analyzed further by GO enrichment (Fig. 1). Overall, the combined network contained several established PPARα target genes (e.g., Acadm and Hadhb) and was significantly enriched for GO terms such as fatty acid oxidation, tricarboxylic acid cycle, mitochondria, and peroxisomes (Supplementary Fig. IIB-D). The merged gene expression subnetwork also contained Cyp4a10 and Cyp4a14. These genes encode members of the cytochrome P450 family that perform the first rate-limiting step in the ω-oxidation of long-chain fatty acids. Thus, these coexpression subnetworks are consistent with a function of Ehhadh in the generation of energy from fatty acids.
To demonstrate that genetic variation in Ehhadh could be related to variation in metabolic traits, we queried Genenetwork to determine if Ehhadh liver expression correlated with clinical endpoints, which had been archived in the B6BTBRF2 cross database (25). This analysis revealed a significant and negative correlation of hepatic Ehhadh expression with fasting plasma glucose in this population of mice, suggesting that variation in expression of Ehhadh and its partners in the coexpression network could affect glucose metabolism possibly via its effects on generation of energy from fatty acids (Supplementary Fig. III).
Decreased medium-chain dicarboxylic acid levels in Ehhadh KO mice upon fasting
Ehhadh is a well established PPARα target gene (33, 34). Its expression is induced during fasting. In combination with our finding that genes involved in the generation of energy from fatty acids are in the gene coexpression subnetwork of Ehhadh, we reasoned that fasting would reveal a phenotype in Ehhadh KO mice. We fasted WT and Ehhadh KO and analyzed metabolites after 0 (fed), 6 (postabsorptive), and 20 h (fasted) of fasting. Blood glucose and plasma insulin decreased with time of fasting, whereas blood β-hydroxybutyrate levels increased (Fig. 2A). Although blood glucose and β-hydroxybutyrate levels were slightly lower in Ehhadh KO animals, the ketotic response upon fasting appeared adequate.
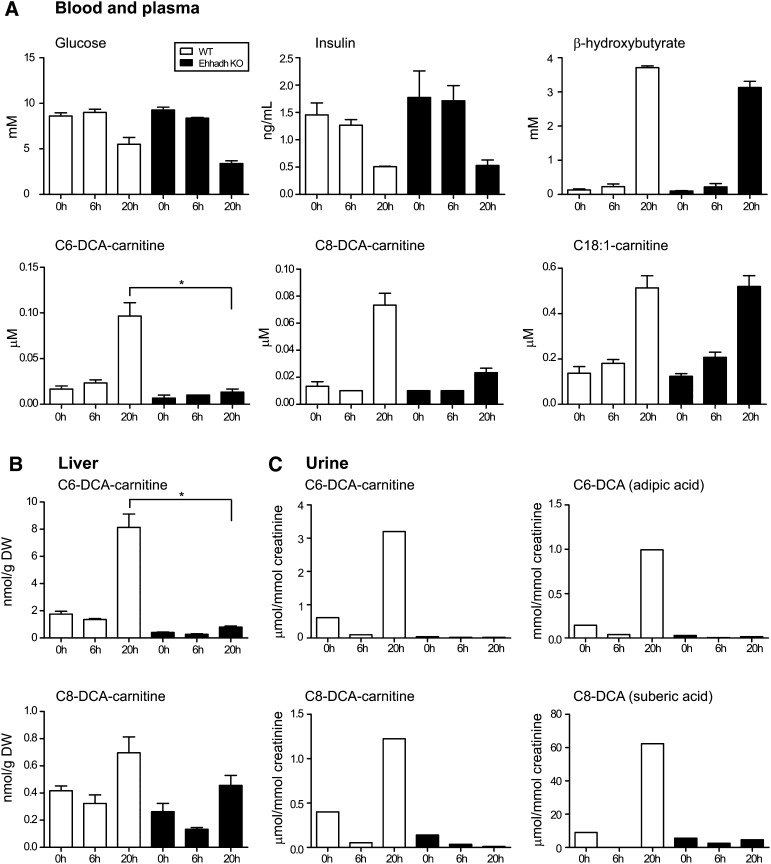
Fig.2.
Metabolites in WT and Ehhadh KO mice. A: Blood and plasma metabolites. B: Liver C6-DCA and C8-DCA acylcarnitine. C: Urinary C6-DCA and C8-DCA and their acylcarnitine esters. Values were obtained in pooled urine samples and determined in duplicate. Statistical significance in the Dunn's multiple comparison test is indicated as follows: * P < 0.05. The error bars indicate the SEM.
Next we determined the plasma acylcarnitine profile. Changes in the plasma acylcarnitine profile are thought to reflect tissue acyl-CoA levels. The induction of ω-oxidation after an overnight fast is reflected in an increase in plasma C6-DCA- and C8-DCA-carnitine in WT mice. In contrast, this increase was virtually absent in Ehhadh KO mice. Other long-chain acylcarnitines, such as C18:1-carnitine, increased upon fasting in WT and Ehhadh KO mice (Fig. 2A). Acylcarnitine analysis in tissues of WT mice revealed that liver, but not heart (data not shown), displayed an increase in C6-DCA- and C8-DCA-carnitine upon fasting. Again, this increase was virtually absent in Ehhadh KO mice (Fig. 2B). Urinary acylcarnitine analysis revealed a similar picture, with pronounced C6-DCA- and C8-DCA-carnitine excretion upon fasting in WT mice and a deficiency in this response in Ehhadh KO mice (Fig. 2C).
In addition to the acylcarnitine profiles, we also measured the organic acids in urine because the urinary excretion of free C6-DCA and C8-DCA is quantitatively greater than their excretion as acylcarnitines. As expected, urinary C6-DCA and C8-DCA levels were higher in fasted WT mice. In Ehhadh KO mice, however, C6-DCA and C8-DCA levels did not increase after fasting (Fig. 2C). We conclude that the increase in the formation of medium-chain DCAs upon fasting is virtually absent in Ehhadh KO mice suggesting that the peroxisomal β-oxidation of long-chain DCAs is disturbed.
Fasting induces PPARalpha targets in WT as well as Ehhadh KO mice
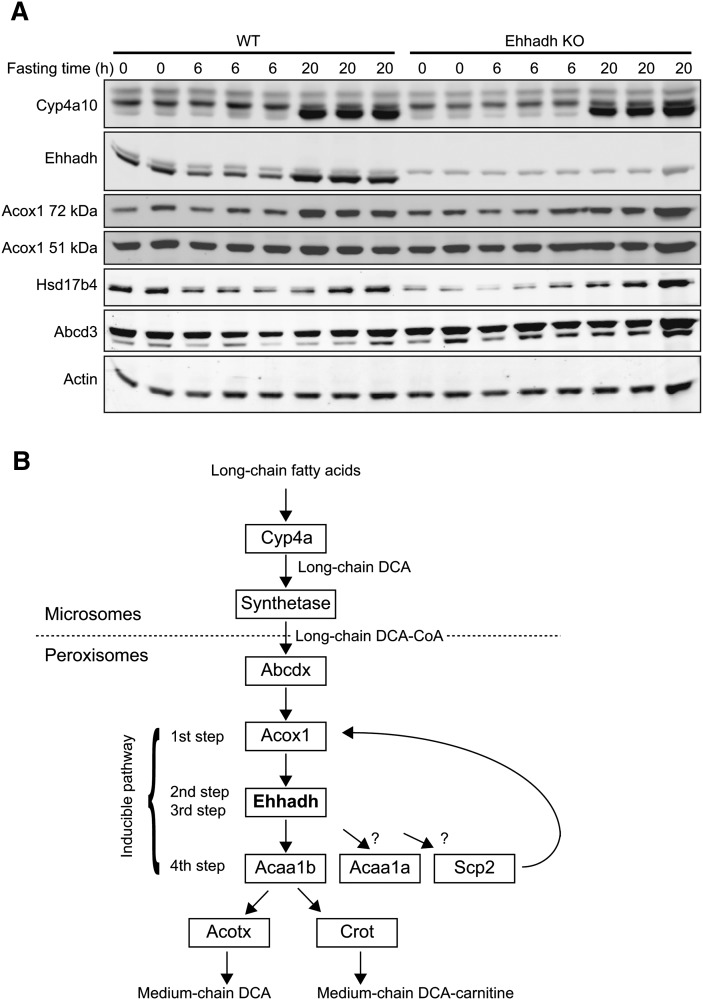
To verify whether the defect in the production of medium-chain DCAs in Ehhadh KO mice was not caused by a failure to induce genes involved in ω-oxidation, we analyzed expression of selected PPARα target genes using immunoblot analysis (Fig. 3A). Fasting induced a marked increase in the expression of Cyp4a10 in WT and Ehhadh KO mice. Ehhadh protein was also highly induced upon fasting in WT mice and, as expected, was absent in the Ehhadh KO mouse. Hsd17b4, which theoretically could compensate for Ehhadh deficiency, was slightly induced by fasting in WT and Ehhadh KO mice, which is consistent with its induction by PPARα agonists (35). An induction was also observed for Acox1, most notably for the 72 kDa full-length protein. Abcd3 (Pmp70) levels did not respond to fasting. Thus, the PPARα response upon fasting does not seem to be disturbed in Ehhadh KO mice.
Fig.3.
Immunoblots of ω- and β-oxidation proteins in WT and Ehhadh KO mice. A: Approximately 10 μg of total liver protein was used for electrophoresis and subsequently blotted onto nitrocellulose. B: Schematic representation of long-chain fatty acid ω-oxidation and subsequent peroxisomal long-chain DCA β-oxidation.
DISCUSSION
In this study, we combined prior knowledge with gene coexpression subnetworks and mouse experiments to elucidate the role of Ehhadh in metabolism. Ehhadh belongs to the so-called classical peroxisomal β-oxidation pathway, together with Acox1 and Acaa1b. In the liver, this pathway is induced by peroxisome proliferators in a mechanism that depends on PPARα. A similar response is elicited by fasting, a stress that leads to physiologic PPARα activation. In addition to peroxisomal β-oxidation, PPARα induces mitochondrial β-oxidation, ketogenesis, and microsomal ω-oxidation, among many other pathways (36). We found that hepatic Ehhadh coexpression subnetworks were composed of genes involved in the tricarboxylic acid cycle and mitochondrial and peroxisomal fatty acid oxidation, which is consistent with a function in the generation of energy from fatty acids. We were able to demonstrate that, during fasting, Ehhadh plays an indispensable role in the formation of the medium-chain DCAs adipic and suberic acid and their carnitine esters. Our results offer an explanation for the coordinated induction of mitochondrial and peroxisomal oxidative pathways during fasting.
Medium-chain DCAs are the final products of microsomal ω-oxidation of long-chain fatty acids and the subsequent peroxisomal β-oxidation of long-chain DCAs (Fig. 3B). The Ehhadh coexpression subnetwork contained at least three genes involved in this process: Cyp4a10, Cyp4a14, and Aldh3a2 (Fig. 1; Supplementary Fig. IIA). Previous biochemical studies have indicated that peroxisomes are important for the β-oxidation of long-chain DCAs (37–42). This was confirmed in vitro using peroxisome-deficient cells. Oxidation of hexadecanedioic acid (C16-DCA) was strongly reduced in fibroblasts of patients with a peroxisome biogenesis disorder (18). The use of a KO mouse model revealed a similar picture with deficient C14-DCA oxidation in hepatocytes devoid of Pex5 (17). Before peroxisomal β-oxidation, long-chain DCAs are activated to their CoA ester by a microsomal acyl-CoA synthetase (42, 43), the molecular identity of which is unknown. Acsl1, which is present in the gene coexpression subnetwork, is a candidate gene for this function (44). The resulting long-chain DCA-CoA is subsequently transported across the peroxisomal membrane probably by one of the three peroxisomal ABC half-transporters (Fig. 3B). Abcd3 has been hypothesized to perform this role based on preliminary reports on the Abcd3 KO mice (45). Indeed, Abcd3 was present in our gene coexpression subnetwork.
For the individual β-oxidation steps of long-chain DCAs, it has been demonstrated that ACOX1 is crucial for the first step in humans (18). The last step in the β-oxidation of DCAs can be performed by multiple thiolases (Acaa1a, Acaa1b, and Scp2). Based on our results, it seems reasonable to speculate that the inducible Acaa1b is crucial. Indeed, Acaa1b was present in the gene coexpression subnetwork. On the other hand, fibroblasts of patients with rhizomelic chondrodysplasia punctata type 1, in which the mature 41 kDa form of ACAA1 is absent, have normal β-oxidation of long-chain DCAs, suggesting that, at least in humans, ACAA1 in not crucial (18). The role of Acaa1b in the β-oxidation of long-chain DCAs could be investigated in the Acaa1b KO mouse model during fasting (46). Fibroblasts from patients with HSD17B4 deficiency had normal oxidation of C16-DCA, suggesting that Ehhadh has a role in performing the second and third step of long-chain DCA β-oxidation (18). Indeed, recombinant Hsd17b4 and Ehhadh handle long-chain DCA substrates with comparable efficiency (18). Moreover, oxidation of C14-DCA was reduced in Ehhadh-deficient hepatocytes, whereas it was elevated in Hsd17b4 KO hepatocytes (17). The fact that the oxidation of C14-DCA was more deficient in Pex5 KO hepatocytes when compared with Ehhadh KO hepatocytes hindered concluding that Ehhadh is essential for oxidation of long-chain DCAs. In addition, Hsd17b4 is inducible by PPARα (35) and thus could theoretically compensate for Ehhadh deficiency also during fasting. Moreover, Hsd17b4 expression correlated significantly with Ehhadh in the B6BTBRF2 and BHF2 crosses (data not shown). Indeed, we observed a small increase in the protein levels of Hsd17b4 upon fasting in Ehhadh KO mice, but production of medium-chain DCAs was deficient, from which we concluded that Hsd17b4 is unable to compensate for Ehhadh deficiency. Therefore, we provide the first in vivo evidence that Ehhadh is essential for the generation of medium-chain DCAs.
The final steps in the peroxisomal metabolism of DCAs are poorly understood. The thiolytic cleavage of the CoA esters is achieved by acyl-CoA thioesterases (Fig. 3B). In mice and humans, Acot4 or Acot8 are most likely involved in the metabolism of medium-chain DCAs (47), and, indeed, Acot4 and cytosolic Acot1 are present in our Ehhadh coexpression subnetworks. Alternatively, the CoA ester can be converted into a carnitine ester by carnitine octanoyltransferase (Crot). Whether a DCA is converted to an organic acid or an acylcarnitine probably depends on the relative expression levels of Crot versus the thioesterases (48). Expression of Crot in liver is high (48), which is in accordance with the fact that we could measure the formation of C6-DCA-carnitine in liver but not in heart.
ω-Oxidation contributes significantly to the oxidation of fatty acids. Increased medium-chain DC aciduria is observed during conditions characterized by increased fatty acid oxidation, such as fasting and diabetic ketosis. In addition, ω-oxidation is increased in patients with an inborn error of mitochondrial β-oxidation. In these cases, ω-oxidation followed by peroxisomal β-oxidation seems to function as a sink for fatty acids overflowing from the mitochondrial □-oxidation system. We hypothesize that this function is the explanation for the coordinate induction of mitochondrial and peroxisomal β-oxidation after PPARα activation. The Ehhadh KO mouse is the first model with a clear deficiency in the peroxisomal β-oxidation of long-chain DCAs and can therefore be used to understand the physiological role of this pathway. We did not find evidence for additional defects in peroxisomal functions upon fasting. After the overnight fast, there was no accumulation of bile acid precursors or of very long-chain fatty acids (data not shown).
Besides this role in the peroxisomal β-oxidation of DCAs, recent studies suggest additional functions for Ehhadh in humans. A systematic genetic analysis of cytochrome P450 enzyme activities in human liver revealed EHHADH as a novel regulatory gene for the P450 system (49). In a bioinformatics study, EHHADH was selected as a candidate gene for nonalcoholic fatty liver disease. Subsequent genetic analysis revealed SNPs in EHHADH that associated with fasting insulin levels (50). This coincides with our observation that in the B6BTBRF2 cross, hepatic Ehhadh expression significantly and negatively correlated with fasting plasma glucose, suggesting that variation in the expression of Ehhadh and its partners in the coexpression network may affect glucose metabolism. Moreover, when compared with steatosis-sensitive C57BL/6 mice, peroxisomal biogenesis and β-oxidation were increased in steatosis-resistant A/J mice upon high-fat feeding (51). We observed that fasting-induced steatosis in Ehhadh KO mice was not more pronounced when compared with WT mice (data not shown). Therefore, the role of Ehhadh in diet-induced obesity and related traits warrants more investigation.
Another interesting feature of Ehhadh is that it is a trifunctional enzyme harboring Δ3,Δ2-enoyl-CoA isomerase activity (52). Although peroxisomes have a second Δ3,Δ2-enoyl-CoA isomerase (53), we hypothesize that the isomerase activity of Ehhadh is crucial to efficiently metabolize the DCAs of unsaturated fatty acids such as cis-9-octadecenedioic acid, the DCA of oleic acid.
In summary, we demonstrate that Ehhadh is involved in the fasting-induced production of medium-chain DCAs in vivo.
Supplementary Material
Acknowledgments
The authors thank Wilma Smit for organic acid analysis.
Footnotes
Abbreviations:
- Crot
- carnitine octanoyltransferase
- DCA
- dicarboxylic acid
- Ehhadh
- L-bifunctional enzyme
- GO
- Gene Ontology
- KO
- knockout
- PPARα
- peroxisome proliferator-activated receptor α
- Scp
- sterol carrier protein
This work was supported by The Netherlands Organisation for Scientific Research VIDI grant 016.086.336 (S.M.H.) and the by National Institutes of Health grant DK083163 (J.K.R.).
REFERENCES
- 1.Poirier Y., Antonenkov V. D., Glumoff T., Hiltunen J. K. 2006. Peroxisomal beta-oxidation–a metabolic pathway with multiple functions. Biochim. Biophys. Acta. 1763: 1413–1426 [DOI] [PubMed] [Google Scholar]
- 2.Van Veldhoven P. P. 2010. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51: 2863–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wanders R. J., Waterham H. R. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75: 295–332 [DOI] [PubMed] [Google Scholar]
- 4.Van Veldhoven P. P., Vanhove G., Assselberghs S., Eyssen H. J., Mannaerts G. P. 1992. Substrate specificities of rat liver peroxisomal acyl-CoA oxidases: palmitoyl-CoA oxidase (inducible acyl-CoA oxidase), pristanoyl-CoA oxidase (non-inducible acyl-CoA oxidase), and trihydroxycoprostanoyl-CoA oxidase. J. Biol. Chem. 267: 20065–20074 [PubMed] [Google Scholar]
- 5.Antonenkov V. D., Van Veldhoven P. P., Waelkens E., Mannaerts G. P. 1997. Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase purified from normal rat liver peroxisomes. Sterol carrier protein 2/3-oxoacyl-CoA thiolase is involved in the metabolism of 2-methyl-branched fatty acids and bile acid intermediates. J. Biol. Chem. 272: 26023–26031 [DOI] [PubMed] [Google Scholar]
- 6.Antonenkov V. D., Van Veldhoven P. P., Waelkens E., Mannaerts G. P. 1999. Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B from rat liver. Biochim. Biophys. Acta. 1437: 136–141 [DOI] [PubMed] [Google Scholar]
- 7.Qi C., Zhu Y., Pan J., Usuda N., Maeda N., Yeldandi A. V., Rao M. S., Hashimoto T., Reddy J. K. 1999. Absence of spontaneous peroxisome proliferation in enoyl-CoA Hydratase/L-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver. Further support for the role of fatty acyl CoA oxidase in PPARalpha ligand metabolism. J. Biol. Chem. 274: 15775–15780 [DOI] [PubMed] [Google Scholar]
- 8.Dieuaide-Noubhani M., Novikov D., Baumgart E., Vanhooren J. C., Fransen M., Goethals M., Vandekerckhove J., Van Veldhoven P. P., Mannaerts G. P. 1996. Further characterization of the peroxisomal 3-hydroxyacyl-CoA dehydrogenases from rat liver. Relationship between the different dehydrogenases and evidence that fatty acids and the C27 bile acids di- and tri-hydroxycoprostanic acids are metabolized by separate multifunctional proteins. Eur. J. Biochem. 240: 660–666 [DOI] [PubMed] [Google Scholar]
- 9.Dieuaide-Noubhani M., Asselberghs S., Mannaerts G. P., Van Veldhoven P. P. 1997. Evidence that multifunctional protein 2, and not multifunctional protein 1, is involved in the peroxisomal beta-oxidation of pristanic acid. Biochem. J. 325: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieuaide-Noubhani M., Novikov D., Vandekerckhove J., Van Veldhoven P. P., Mannaerts G. P. 1997. Identification and characterization of the 2-enoyl-CoA hydratases involved in peroxisomal beta-oxidation in rat liver. Biochem. J. 321: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin Y. M., Haapalainen A. M., Conry D., Cuebas D. A., Hiltunen J. K., Novikov D. K. 1997. Recombinant 2-enoyl-CoA hydratase derived from rat peroxisomal multifunctional enzyme 2: role of the hydratase reaction in bile acid synthesis. Biochem. J. 328: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y. M., Poutanen M. H., Helander H. M., Kvist A. P., Siivari K. M., Schmitz W., Conzelmann E., Hellman U., Hiltunen J. K. 1997. Peroxisomal multifunctional enzyme of beta-oxidation metabolizing D-3-hydroxyacyl-CoA esters in rat liver: molecular cloning, expression and characterization. Biochem. J. 321: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baes M., Huyghe S., Carmeliet P., Declercq P. E., Collen D., Mannaerts G. P., Van Veldhoven P. P. 2000. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J. Biol. Chem. 275: 16329–16336 [DOI] [PubMed] [Google Scholar]
- 14.Ferdinandusse S., Denis S., Mooyer P. A., Dekker C., Duran M., Soorani-Lunsing R. J., Boltshauser E., Macaya A., Gartner J., Majoie C. B., et al. 2006. Clinical and biochemical spectrum of D-bifunctional protein deficiency. Ann. Neurol. 59: 92–104 [DOI] [PubMed] [Google Scholar]
- 15.Lalwani N. D., Reddy M. K., Mangkornkanok-Mark M., Reddy J. K. 1981. Induction, immunochemical identity and immunofluorescence localization of an 80 000-molecular-weight peroxisome-proliferation-associated polypeptide (polypeptide PPA-80) and peroxisomal enoyl-CoA hydratase of mouse liver and renal cortex. Biochem. J. 198: 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osumi T., Hashimoto T. 1979. Peroxisomal beta oxidation system of rat liver. Copurification of enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase. Biochem. Biophys. Res. Commun. 89: 580–584 [DOI] [PubMed] [Google Scholar]
- 17.Dirkx R., Meyhi E., Asselberghs S., Reddy J., Baes M., Van Veldhoven P. P. 2007. Beta-oxidation in hepatocyte cultures from mice with peroxisomal gene knockouts. Biochem. Biophys. Res. Commun. 357: 718–723 [DOI] [PubMed] [Google Scholar]
- 18.Ferdinandusse S., Denis S., van Roermund C. W., Wanders R. J., Dacremont G. 2004. Identification of the peroxisomal beta-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J. Lipid Res. 45: 1104–1111 [DOI] [PubMed] [Google Scholar]
- 19.Nguyen S. D., Baes M., Van Veldhoven P. P. 2008. Degradation of very long chain dicarboxylic polyunsaturated fatty acids in mouse hepatocytes, a peroxisomal process. Biochim. Biophys. Acta. 1781: 400–405 [DOI] [PubMed] [Google Scholar]
- 20.Reddy J. K., Goel S. K., Nemali M. R., Carrino J. J., Laffler T. G., Reddy M. K., Sperbeck S. J., Osumi T., Hashimoto T., Lalwani N. D., et al. 1986. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc. Natl. Acad. Sci. USA. 83: 1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone T. C., Weinheimer C. J., Kelly D. P. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA. 96: 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesler E. J., Lu L., Wang J., Williams R. W., Manly K. F. 2004. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 7: 485–486 [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Williams R. W., Manly K. F. 2003. WebQTL: web-based complex trait analysis. Neuroinformatics. 1: 299–308 [DOI] [PubMed] [Google Scholar]
- 25.Lan H., Chen M., Flowers J. B., Yandell B. S., Stapleton D. S., Mata C. M., Mui E. T., Flowers M. T., Schueler K. L., Manly K. F. 2006. Combined expression trait correlations and expression quantitative trait locus mapping. PLoS Genet. 2: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatti D., Maki A., Chesler E. J., Kirova R., Kosyk O., Lu L., Manly K. F., Williams R. W., Perkins A., Langston M. A., et al. 2007. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 46: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatti D. M., Zhao N., Chesler E. J., Bradford B. U., Shabalin A. A., Yordanova R., Lu L., Rusyn I. 2010. Sex-specific gene expression in the BXD mouse liver. Physiol. Genomics. 42: 456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., Schadt E. E., Wang S., Wang H., Arnold A. P., Ingram-Drake L., Drake T. A., Lusis A. J. 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maere S., Heymans K., Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- 31.van Vlies N., Tian L., Overmars H., Bootsma A. H., Kulik W., Wanders R. J., Wood P. A., Vaz F. M. 2005. Characterization of carnitine and fatty acid metabolism in the long-chain acyl-CoA dehydrogenase-deficient mouse. Biochem. J. 387: 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vreken P., van Lint A. E., Bootsma A. H., Overmars H., Wanders R. J., van Gennip A. H. 1999. Quantitative plasma acylcarnitine analysis using electrospray tandem mass spectrometry for the diagnosis of organic acidaemias and fatty acid oxidation defects. J. Inherit. Metab. Dis. 22: 302–306 [DOI] [PubMed] [Google Scholar]
- 33.Marcus S. L., Miyata K. S., Zhang B., Subramani S., Rachubinski R. A., Capone J. P. 1993. Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression. Proc. Natl. Acad. Sci. USA. 90: 5723–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B., Marcus S. L., Miyata K. S., Subramani S., Capone J. P., Rachubinski R. A. 1993. Characterization of protein-DNA interactions within the peroxisome proliferator-responsive element of the rat hydratase-dehydrogenase gene. J. Biol. Chem. 268: 12939–12945 [PubMed] [Google Scholar]
- 35.Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273: 5678–5684 [DOI] [PubMed] [Google Scholar]
- 36.Mandard S., Muller M., Kersten S. 2004. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 61: 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergseth S., Poisson J. P., Bremer J. 1990. Metabolism of dicarboxylic acids in rat hepatocytes. Biochim. Biophys. Acta. 1042: 182–187 [DOI] [PubMed] [Google Scholar]
- 38.Kolvraa S., Gregersen N. 1986. In vitro studies on the oxidation of medium-chain dicarboxylic acids in rat liver. Biochim. Biophys. Acta. 876: 515–525 [DOI] [PubMed] [Google Scholar]
- 39.Pourfarzam M., Bartlett K. 1991. Products and intermediates of the beta-oxidation of [U-14C]hexadecanedionoyl-mono-CoA by rat liver peroxisomes and mitochondria. Biochem. J. 273: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki H., Yamada J., Watanabe T., Suga T. 1989. Compartmentation of dicarboxylic acid beta-oxidation in rat liver: importance of peroxisomes in the metabolism of dicarboxylic acids. Biochim. Biophys. Acta. 990: 25–30 [DOI] [PubMed] [Google Scholar]
- 41.Vamecq J., Draye J. P. 1989. Peroxisomal and mitochondrial beta-oxidation of monocarboxylyl-CoA, omega-hydroxymonocarboxylyl-CoA and dicarboxylyl-CoA esters in tissues from untreated and clofibrate-treated rats. J. Biochem. 106: 216–222 [DOI] [PubMed] [Google Scholar]
- 42.Vamecq J., Draye J. P., Brison J. 1989. Rat liver metabolism of dicarboxylic acids. Am. J. Physiol. 256: G680–G688 [DOI] [PubMed] [Google Scholar]
- 43.Vamecq J., de Hoffmann E., Van Hoof F. 1985. The microsomal dicarboxylyl-CoA synthetase. Biochem. J. 230: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watkins P. A., Howard A. E., Gould S. J., Avigan J., Mihalik S. J. 1996. Phytanic acid activation in rat liver peroxisomes is catalyzed by long-chain acyl-CoA synthetase. J. Lipid Res. 37: 2288–2295 [PubMed] [Google Scholar]
- 45.Jimenez-Sanchez G., Hebron K. J., Silva-Zolezzi I., Mihalik S. J., Watkins P. A., Espeel M., Moser A., Thomas G., Roels F., Valle D. 2000 Fasting fuel homeostasis triggered by defective phytanic and pristanic acids metabolism in the 70kDa peroxisomal membrane protein (PMP70) deficient mice. Annual Meeting American Soc Human Genet, October 3–7, 2000, Philadelphia, PA. Abstract no. 282. [Google Scholar]
- 46.Chevillard G., Clemencet M. C., Latruffe N., Nicolas-Frances V. 2004. Targeted disruption of the peroxisomal thiolase B gene in mouse: a new model to study disorders related to peroxisomal lipid metabolism. Biochimie. 86: 849–856 [DOI] [PubMed] [Google Scholar]
- 47.Westin M. A., Hunt M. C., Alexson S. E. 2005. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. J. Biol. Chem. 280: 38125–38132 [DOI] [PubMed] [Google Scholar]
- 48.Westin M. A., Hunt M. C., Alexson S. E. 2008. Short- and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of beta-oxidation products out of peroxisomes. Cell. Mol. Life Sci. 65: 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Zhang B., Molony C., Chudin E., Hao K., Zhu J., Gaedigk A., Suver C., Zhong H., Leeder J. S., et al. 2010. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 20: 1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banasik K., Justesen J. M., Hornbak M., Krarup N. T., Gjesing A. P., Sandholt C. H., Jensen T. S., Grarup N., Andersson A., Jorgensen T., et al. 2011. Bioinformatics-driven identification and examination of candidate genes for non-alcoholic fatty liver disease. PLoS ONE. 6: e16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall D., Poussin C., Velagapudi V. R., Empsen C., Joffraud M., Beckmann J. S., Geerts A. E., Ravussin Y., Ibberson M., Oresic M., et al. 2010. Peroxisomal and microsomal lipid pathways associated with resistance to hepatic steatosis and reduced pro-inflammatory state. J. Biol. Chem. 285: 31011–31023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palosaari P. M., Hiltunen J. K. 1990. Peroxisomal bifunctional protein from rat liver is a trifunctional enzyme possessing 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and delta 3, delta 2-enoyl-CoA isomerase activities. J. Biol. Chem. 265: 2446–2449 [PubMed] [Google Scholar]
- 53.Geisbrecht B. V., Zhang D., Schulz H., Gould S. J. 1999. Characterization of PECI, a novel monofunctional Delta(3), Delta(2)-enoyl-CoA isomerase of mammalian peroxisomes. J. Biol. Chem. 274: 21797–21803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.