Abstract
Enterocyte cholesterol homeostasis reflects aggregated rates of sterol synthesis, efflux, and uptake from plasma and gut lumen. Cholesterol synthesis and LDL uptake are coordinately regulated by sterol regulatory element-binding proteins (SREBP), whereas sterol efflux is regulated by liver X receptors (LXR). How these processes are coordinately regulated in enterocytes, the site of cholesterol absorption, is not well understood. Here, we treat mice with ezetimibe to investigate the effect of blocking cholesterol absorption on intestinal SREBPs, LXRs, and their effectors. Ezetimibe increased nuclear SREBP-2 8-fold. HMG-CoA reductase (HMGR) and LDL receptor (LDLR) mRNA levels increased less than 3-fold, whereas their protein levels increased 30- and 10-fold, respectively. Expression of inducible degrader of LDLR (IDOL), an LXR-regulated gene that degrades LDLRs, was reduced 50% by ezetimibe. Coadministration of ezetimibe with the LXR agonist T0901317 abolished the reduction in IDOL and prevented the increase in LDLR protein. Ezetimibe-stimulated LDLR expression was independent of proprotein convertase subtilisin/kexin type 9 (PSCK9), a protein that degrades LDLRs. To maintain cholesterol homeostasis in the face of ezetimibe, enterocytes boost LDL uptake by increasing LDLR number, and they boost sterol synthesis by increasing HMGR and other cholesterologenic genes. These studies reveal a hitherto undescribed homeostatic network in enterocytes triggered by blockade of cholesterol absorption.
Keywords: cholesterol/absorption, cholesterol/biosynthesis, LDL/metabolism, lipoproteins/receptors, nuclear receptors, HMG-CoA reductase
The small intestine is central to the regulation of whole-body cholesterol balance in mammals. Enterocytes are unusual in that they have three, rather than two, sources of cholesterol. Enterocytes uniquely absorb free cholesterol from the gut lumen, and they also share two ubiquitous sources with other cells: endogenous synthesis of sterols de novo and uptake of LDL-derived cholesterol from plasma. The small intestine is one of the major sites of cholesterol synthesis in mammals. It is second only to liver in mice, and it may be quantitatively the single most important organ in other species, such as rabbit and guinea pig (1, 2). Likewise, the small intestine is second only to the liver in terms of overall LDL clearance in multiple species (1, 3, 4).
The transcriptional regulation of de novo sterol synthesis and LDL receptor-mediated sterol uptake in cultured cells and in liver has been well characterized (5). Transcription of the LDL receptor (LDLR) and of every gene of the sterol biosynthetic pathway is regulated by sterol regulatory element-binding protein-2 (SREBP-2), one of three SREBP family members (6). SREBPs are synthesized as transcriptionally inactive precursor molecules that are integral membrane proteins of the endoplasmic reticulum (ER). To become transcriptionally active, they must move to the Golgi complex, where they are proteolyzed to liberate the soluble N-terminal domain, which can enter the nucleus to activate transcription (7). ER-to-Golgi transport is accomplished by the action of Scap, a polytopic ER membrane protein that interacts with SREBPs and mediates their incorporation into COPII-coated vesicles (8, 9). Feedback inhibition of SREBP proteolysis is mediated by Insig proteins, membrane proteins that retain the Scap/SREBP complex in the ER when the cholesterol content of the ER membranes rises (10–12). Insigs also have a second action in feedback regulation of cholesterol synthesis; namely, they mediate sterol-regulated proteolytic degradation of HMG-CoA reductase (HMGR), a key enzyme required for mevalonate and cholesterol synthesis (13, 14).
Recent studies have shown that two factors affect posttranscriptional regulation of the LDLR. The first factor is proprotein convertase subtilisin/kexin type 9 (PSCK9), a secreted protein that degrades LDLRs and is a SREBP-2 target gene regulated coordinately with the LDLR and other SREBP-2-responsive genes (15, 16). PCSK9 potently regulates hepatic LDLRs (17), but its effects in other tissues are less well defined. PCSK9 does not function to degrade LDLRs equally well in all tissues. For instance, it does not appear to degrade LDLRs in adrenal glands (17). Overexpression of hepatic PCSK9 reduced LDLRs in the ileum by 33% (18). The second factor is inducible degrader of LDLR (IDOL), a recently described E3 ubiquitin ligase that mediates lysosomal degradation of polyubiquitinated LDLRs and is a transcriptional target of liver X receptors (LXR) (19–21), which are nuclear hormone receptors that are activated by oxysterols (22). Like PCSK9, IDOL's propensity to degrade LDLRs is tissue selective; administration of LXR agonists induced IDOL and degraded LDLRs in macrophages and intestine, but not in liver (19).
Sterol depletion, through activation of SREBP-2 processing, increases the transcription of both LDLR and PCSK9 (23, 24). In livers of mice, these two opposing effects counteract each other so that LDLR protein levels do not increase in response to sterol depletion with statin treatment (23). On the other hand, sterol excess, as can be caused by cholesterol feeding, inhibits hepatic SREBP processing and reduces the expression of target genes, such as LDLR, PCSK9, and others (12, 25). Furthermore, cholesterol feeding induces LXR activity and increases the expression of its target genes (26, 27). Whether pharmacologic sterol depletion modulates PCSK9 and IDOL expression in intestine in the same fashion as in liver remains an open question.
In the current studies, we explore the role of SREBPs and LXRs in the transcriptional and posttranscriptional regulation of intestinal cholesterol homeostasis. We induce a state of sterol depletion in enterocytes though the administration of ezetimibe (EZE), a widely used drug that blocks sterol absorption in enterocytes by inhibiting the transport function of Niemann-Pick C 1-like 1 protein (NPC1L1) (28). We characterize the effect of ezetimibe on SREBP processing and steady-state levels of HMGR and LDLR, the two key proteins that control de novo sterol synthesis and LDL uptake. The data show that blocking one of the three sources of enterocyte cholesterol, namely, lumenal cholesterol uptake, causes enterocytes to respond by upregulating the molecules needed for the other two sources, namely, LDL uptake and de novo sterol synthesis. This response is mediated by a complex network involving SREBP-2, LXR, PCSK9, and IDOL.
MATERIALS AND METHODS
Animals and diets
C57BL/6J mice were obtained from Jackson Laboratories. Mice carrying null PCSK9 alleles or age-matched wild-type mice were as described (23). Mice were housed in colony cages with a 12 h light/12 h dark cycle and fed a standard chow diet containing 6% fat (Teklad Mouse/Rat Diet 7002; Harlan Teklad Premier Laboratory Diets). Ezetimibe (#SRP04000e; Sequoia Research Products), cholesterol (#101382; MP Biomedical), and T0901317 (#71810; Cayman Chemicals) were mixed as dry powders into ground chow diet at the indicated concentration and fed ad libitum for the indicated duration. All mice were euthanized at the end of the dark cycle. All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee of the University of Texas Southwestern Medical Center at Dallas.
Real-time RT-PCR
Total RNA was prepared from mouse tissues using RNA STAT-60 kit (TEL-TEST Inc.). Equal aliquots from four or five mice were pooled and subjected to quantitative real-time PCR. Details of primers and PCR conditions are as described (29, 30), with the following additions: PCSK9 (forward) CAGGCGGC-CAGTGTCTATG, (reverse) GCTCCTTGATTTTGCATTCCA; NPC1L1 (forward) TGGACTGGAAGGACCATTTCC, (reverse) GCGCCCCGTAGTCAGCTAT; IDOL (forward) CGAGCCATCACCGAAACAC, (reverse) CGCGACTGTACTGCATCATGA.
Immunoblot analyses of intestine and liver nuclear extracts and crude membrane fractions
Membrane and nuclear fractions were prepared from the tissues of four or five mice (approximately 125 mg wet weight of enterocytes or 75 mg liver per mouse), and equal aliquots from each mouse were pooled and subjected to SDS-PAGE and immunoblot analysis (12, 23, 31, 32). For enterocytes, polytron homogenization was performed at 50% power for 10 s. NPC1L1 antibody was a kind gift of Helen Hobbs and Jonathan Cohen (UT-Southwestern Medical Center). Quantitative densitometry was performed using ImageJ software (National Institutes of Health).
Isolated intestine epithelial cell preparation
A crude intestine epithelial preparation was performed as described (33–35). Briefly, the intestine was dissected, and attached mesentery was completely removed. The lumen was flushed with normal saline. The first ∼6 cm segment, terminating just distal to common bile duct insertion, was divided and termed duodenum. The remaining small intestine was divided into two sections of equal length, the proximal termed jejunum and distal half termed ileum. In some experiments, jejunum and ileum were divided again into two segments of equal length, termed the first and second segments of jejunum or ileum, respectively. Intestine segments were opened length-wise and cut into 5–6 cm lengths so that the mucosa was completely effaced, and then placed into plastic tubes containing 15 ml of KRBE (Krebs-Ringer bicarbonate buffer containing 120 mM sodium chloride, 4.6 mM potassium chloride, 0.5 mM magnesium chloride, 10 mM D-glucose, 0.7 mM sodium phosphate dibasic, 1.5 mM sodium phosphate monobasic, 15 mM sodium bicarbonate, 1 mM DTT, and 1.5 mM EDTA, pH 7.4) in a shaking 37°C water bath for 20 min. For colonic segments, 10 mM EDTA was used. The intestine remnants were removed, and the cell suspension was put on ice and passed through a 100 μM cell strainer (#352360; BD Falcon). The cells were collected by centrifugation at 1000 g for 5 min. Cell pellets were washed with ice-cold KRBE buffer, collected by centrifugation at 1000 g for 5 min, and immediately frozen at −80°C for protein or biochemical analysis or immediately homogenized in RNA STAT-60 for mRNA analysis.
Isolation of enterocytes in a gradient along the crypt/villus axis was performed by the method previously described (36). Briefly, after dissection, washed jejunal segments were placed in tubes containing 20 ml of citrate buffer (27 mM sodium citrate, 96 mM sodium chloride, 1.5 mM potassium chloride, 8 mM potassium phosphate dibasic, 5.6 mM sodium phosphate dibasic, 1 mM DTT, pH 7.3) at 37°C for 5 min. Jejunal segments were then transferred into tubes contained 15 cc prewarmed PBSE buffer (Ca2+ and Mg2+-free PBS with 1.5 mM EDTA and 1 mM DTT, pH 7.3) in a shaking 37°C water bath. Cell-containing supernatants from eight sequential fractions were collected after incubation for 11, 17, 23, 29, 35, 45, 60, and 80 min. After each time point, the jejunal pieces were transferred into new tubes with 15 ml of fresh, prewarmed PBSE buffer and put back into shaking water bath. Cell suspensions were strained, pelleted, and frozen as described above. Alkaline phosphatase activity from the cell pellets was measured in triplicate by the manufacturer's protocol (#72146; Anaspec). Fractions were divided into tertiles of alkaline phosphatase activity and pooled into upper (fractions 1–3), mid (fractions 4 and 5), and lower (fractions 6–8) villus/crypt specimens.
Histology
Jejuna were fixed in 4% paraformaldehye in PBS overnight at 4°C. The fixed tissues were embedded in paraffin, sectioned at 5 μM, and immunohistochemistry was performed using anti-HMGR rabbit polyclonal antibody or preimmune antiserum at 6 μg/ml as previously described (37).
RESULTS
Blockade of lumenal cholesterol transport with ezetimibe upregulates intestinal SREBP-2 and its key targets, HMGR and LDLR
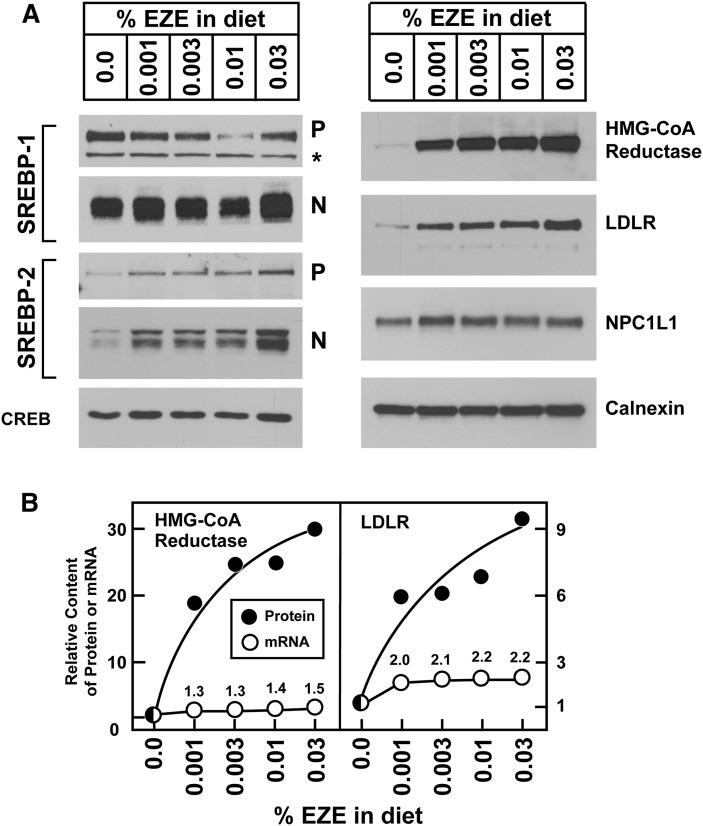
To study the effects of sterol deprivation on SREBP-mediated sterol homeostasis in enterocytes, mice were fed with ezetimibe, and intestinal epithelial cells were prepared by EDTA chelation. We studied the epithelial cells rather than intact intestines because enterocytes selectively express the target of ezetimibe action, the cholesterol transporter NPC1L1 (38). Inasmuch as the jejunum is the predominant organ responsible for cholesterol absorption, initial studies focused upon cells isolated from the jejunum. Fig. 1A shows immunoblot analysis of pooled enterocyte membrane fractions and nuclear extracts from mice fed with the indicated concentrations of ezetimibe for five days. In prior studies of mice, ezetimibe produced a 92% reduction in fractional cholesterol absorption at a dose approximating the 0.01% w/w group in Fig. 1 (39). We observed a dose-dependent increase in the content of precursor and nuclear SREBP-2, the SREBP isoform that predominantly drives the expression of genes involved in cholesterol synthesis and uptake (6). At the highest dose of ezetimibe, there was an 8-fold increase in active, nuclear SREBP-2. In contrast, ezetimibe had less effect on the content of precursor and nuclear SREBP-1. The protein levels of NPC1L1 and calnexin were not affected by ezetimibe administration.
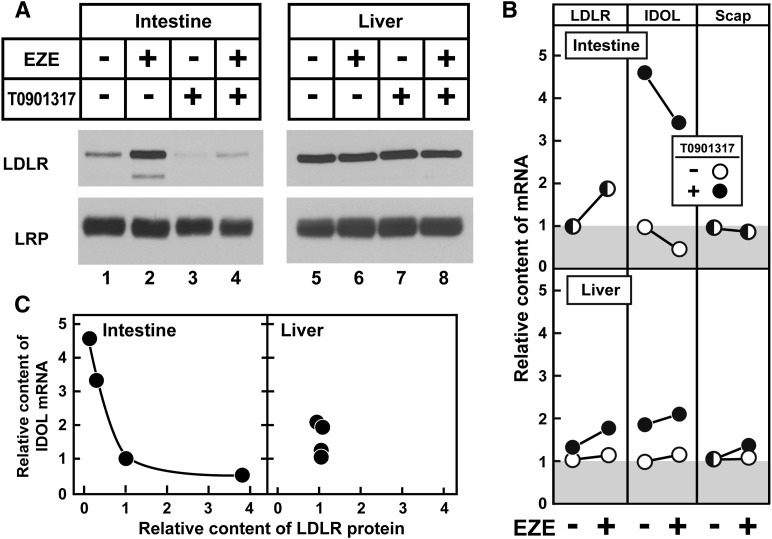
Fig. 1.
Ezetimibe upregulates SREBP-2 and its key effectors in jejunocytes. (A) Immunoblot analysis of nuclear extracts and membrane fractions from 11-week-old male C57BL/6J mice fed increasing concentrations of ezetimibe in the diet for three days. Jejunocytes (five mice per group) were separately fractionated, and equal amounts of protein from each mouse were pooled; 30 μg aliquots of the membrane and nuclear extract fractions were subjected to SDS-PAGE and immunoblot analysis. The precursor and nuclear forms of SREBPs are denoted as P and N, respectively. Asterisks denote nonspecific bands. Immunoblots of CREB and calnexin were used as loading controls for the nuclear extract and membrane fractions, respectively. (B) Comparison of relative mRNA levels and protein levels of HMG-CoA reductase and LDLR. Immunoblots from panel A were scanned and quantified by densitometry (filled circles). Total RNA from jejunocytes (five mice per group) was separately isolated; equal amounts of RNA from each mouse were pooled and subjected to quantitative real-time PCR using cyclophilin as the invariant control (open circles). Each value represents the relative mRNA level or protein level in ezetimibe-treated mice compared with control mice who did not receive ezetimibe (0.0%), which was arbitrarily defined as 1.0.
We used immunoblots and quantitative real-time PCR to measure the protein and mRNA levels of two key SREBP-2-regulated genes, HMGR, which catalyzes the rate-limiting step of cholesterol biosynthesis, and LDLR, which is responsible for the majority of LDL uptake. Both proteins increased markedly after ezetimibe administration (Fig. 1A). Posttranscriptional regulation of protein stability for both HMGR and LDLR has been well described in cultured cells and liver (14, 20, 40). Fig. 1B shows the relative changes in protein and mRNA levels for HMGR and LDLR. Whereas treatment with the highest dose of ezetimibe (0.03% w/w) caused a modest 1.5-fold increase in HMGR mRNA, a dramatic 30-fold increase in HMGR protein was observed. Similarly, this dose of ezetimibe provoked increases in LDLR mRNA and protein by 2.2-fold and 10-fold, respectively. The relative increases in protein were much greater than the increases in the respective mRNAs, which suggests that ezetimibe affects posttranscriptional modes of regulation as well as transcriptional ones.
Ezetimibe causes SREBP-2 and its targets to redistribute to middle and upper villus enterocytes of the jejunum
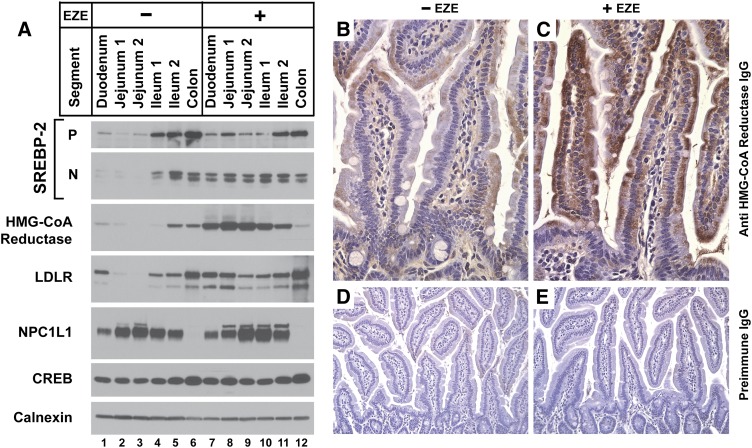
To determine whether the effect of ezetimibe on SREBP-2 and its downstream targets correlates with NPC1L1 expression, intestines from ezetimibe-treated and control mice were divided into anatomic segments proximally to distally from duodenum to colon, and enterocytes were isolated and subjected to immunoblot analysis (Fig. 2A). In agreement with previous studies, NPC1L1 expression was highest in the jejunum and absent in the colon (38). Ezetimibe treatment did not alter NPC1L1 levels in any intestinal segment. In animals not treated with ezetimibe, precursor and nuclear SREBP-2 was most abundant in the distal intestines, including ileum and colon (Fig. 2A, lanes 4–6). These data agree with prior studies in hamsters that reported higher levels of SREBP-2 and LDLR mRNA in distal versus proximal small intestine (41). In contrast, following ezetimibe administration, nuclear SREBP-2 was readily detectable in the proximal intestines from duodenum to distal jejunum, and its level in the proximal ileum increased as well (Fig. 2A, lanes 7–10). LDLR showed the same shift in distribution with enrichment in the proximal intestine after ezetimibe treatment, as was seen with nuclear SREBP-2, in agreement with the idea that SREBP-2 is controlling LDLR expression in the intestines. In the absence of ezetimibe, HMGR was most abundant in the distal ileum, in agreement with prior studies in mice that reported higher levels of de novo sterol synthesis in distal versus proximal small intestine (42). Following ezetimibe treatment, HMGR also increased greatly from duodenum through the ileum, with peak expression in the jejunum.
Fig. 2.
Anatomic distribution of ezetimibe's effect on SREBP-2 and its key effectors in the intestines. (A–E) Ages 12- to 13-week-old male C57BL/6J mice were fed chow diets with or without 0.01% ezetimibe in the diet for five days. (A) Immunoblot analysis of nuclear extracts and membrane fractions from enterocytes from sequential segments of intestine. Intestines were divided into five anatomic segments from duodenum to colon (see Materials and Methods) arranged in proximal to distal order. Enterocytes (four mice per group) were fractionated; 20 μg aliquots of the membrane and nuclear extract fractions were subjected to SDS-PAGE and immunoblot analysis. Immunoblots of CREB and calnexin were used as loading controls for the nuclear extract and membrane fractions, respectively. (B–E) Immunohistochemical analysis of HMG-CoA reductase in jejuna of untreated mice (B) or those who received ezetimibe (C), magnification 40×. Jejuna from untreated mice (D) and ezetimibe-treated mice (E), probed with preimmune antisera, magnification 20×.
Fig. 2B–E shows immunohistochemical analysis for HMGR in mouse jejunal villi following ezetimibe treatment. Ezetimibe treatment had no apparent effect on overall villus structure by light microscopy (Fig. 2D, E). Without ezetimibe, cytoplasmic HMGR staining in enterocytes and stromal cells could be detected at low levels in patches along the villus and in the crypts (Fig. 2B). Following ezetimibe administration, dense cytoplasmic HMGR staining was visualized in enterocytes, along both the apical and basolateral aspects of the enterocytes, without obvious enrichment in the stromal cells (Fig. 2C). This enrichment in HMGR staining was most dramatic in the mid and upper villus.
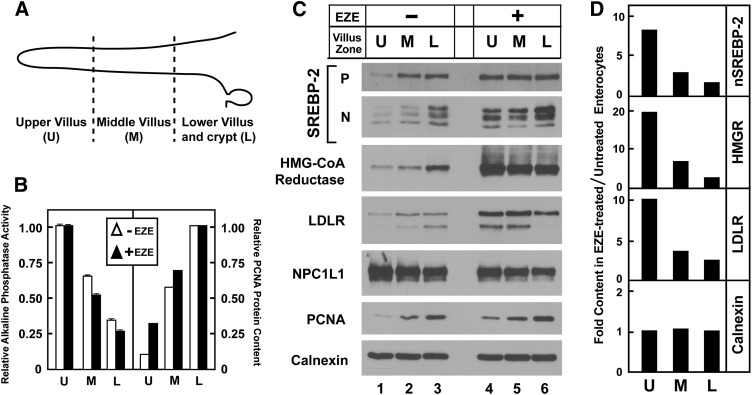
To better define the effect of ezetimibe on the anatomic distribution of SREBP-2 and its effectors along the crypt/villus axis, enterocytes from ezetimibe-treated and control mice were isolated in a gradient from villus tip to crypt. The tissue fractionation was validated by measuring markers of DNA synthesis and absorptive cell differentiation: proliferating cell nuclear antigen (PCNA) protein levels and alkaline phosphatase (AP) activity, respectively. Cells were isolated from tip to crypt in eight sequential fractions and pooled into three groups by virtue of decreasing AP activity. Fig. 3B shows that irrespective of ezetimibe treatment, cells in the early fractions (“upper villus”) had the highest AP activity and lowest PCNA levels, cells in the middle fractions (“middle villus”) had intermediate AP activity and PCNA levels, and cells in the late fractions (“low villus and crypt”) had the lowest AP activity and highest PCNA levels, as has been described previously (43). In the absence of ezetimibe, levels of precursor and nuclear SREBP-2, HMGR, and the LDLR were highest in the low villus fractions (Fig. 3C), in agreement with previous reports that sterol synthesis, LDL uptake, and mRNAs for SREBP-2 and LDLR are highest in the low villus and crypt (44, 45). Ezetimibe provoked a redistribution of these factors, such that their levels in the upper villus were increased. The levels of a control protein, calnexin, were equal in all fractions. The ratio of protein levels in ezetimibe-treated compared with untreated specimens is plotted in Fig. 3D. The ratio of HMGR protein in the upper villus was 20-fold, and it declined to 2.5-fold in the lower villus and crypt, owing primarily to increased HMGR levels in the lower villus and crypt of untreated mice. Similar trends for nuclear SREBP-2 and LDLR were noted, whereas the ratio for calnexin was 1.0 and invariant across the crypt axis.
Fig. 3.
Modulation of SREBP-2 and its key effector molecules along the crypt/villus axis. Thirteen-week-old male C57BL/6J mice were fed chow diets with or without 0.01% ezetimibe in the diet for 5 days. (A) Schematic of crypt/villus axis. U, upper villus zone; M, middle villus zone; L, lower villus/crypt zone. Jejunocytes (four mice per group) were isolated along the crypt-villus axis in timed aliquots and pooled into three groups based on alkaline phosphatase activity (early fractions with highest AP activity defined as upper villus; middle fractions with intermediate activity defined as middle villus; latest fractions with lowest activity defined as lower villus/crypt). (B) Relative alkaline phosphatase activity and PCNA protein levels from pooled jejunocytes as described in panel A. Alkaline phosphatase activity from pooled jejunocytes was measured in triplicate. Data are mean ± SEM; the upper villus groups arbitrarily set as 1.0. PCNA immunoblot from panel C was quantified by densitometry, and the lower villus groups arbitrarily defined at 1.0. (C) Immunoblot analysis of nuclear extracts and membrane fractions from pooled jejunocytes isolated as described in panel A. (D) Ratio of protein levels in ezetimibe-treated compared with untreated jejunocytes. Immunoblots from panel C were quantified by densitometry, and the ratio of expression for each crypt zone was calculated by dividing the value of the ezetimibe group by that of the untreated group.
Ezetimibe suppresses IDOL, an LXR-responsive degrader of LDLR
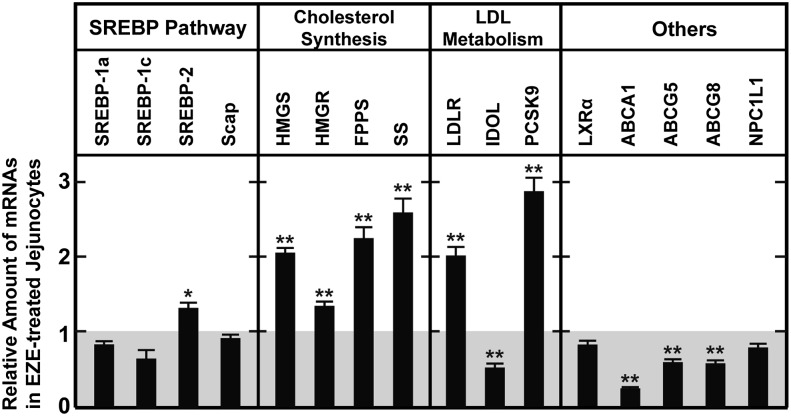
The above data are consistent with the hypothesis that ezetimibe blocks apical cholesterol influx by antagonizing NPC1L1, which depletes enterocyte ER membrane cholesterol to induce SREBP-2 processing and expression of genes required for cholesterol uptake from the plasma and for endogenous sterol synthesis. To verify that the entire genetic program required for cholesterol biosynthesis becomes activated in response to ezetimibe and to uncover clues to explain the apparent discrepancy between the relative increases in mRNA and protein for LDLR and HMGR, microarray analysis of jejunal mRNA was performed (supplementary Fig. I) on mice treated with ezetimibe and compared with untreated controls. As expected, the mRNA for nearly every gene that encodes an enzyme required for conversion of acetyl-CoA to cholesterol, which are known to be SREBP-2 target genes, was increased (46). Seventeen out of 22 genes were upregulated by at least 2-fold, and an additional 3 genes were increased by at least 50%. Only 2 genes, sterol C5-desaturase and 24-dehydrocholesterol reductase (DHCR24) had unchanged expression. Of other known SREBP-2 target genes not directly involved in cholesterol biosynthesis (46), only 3 genes were increased by at least 2-fold: Insig1, PCSK9, and aldolase C.
The microarray analysis also revealed that the expression levels of several LXR-responsive genes, such as ABCA1, were decreased. This finding agrees with prior studies that reported decreases in the intestinal expression of ABCA1, ABCG5, and ABCG8 in response to ezetimibe in multiple species (47, 48). Although the mechanism for this effect is unclear, the reduced sterol flux associated with NPC1L1 blockade may reduce the availability of endogenous LXR ligands. Of these LXR-responsive genes, the expression of IDOL was decreased by 70%. These gene expression changes were confirmed by quantitative real-time PCR (Fig. 4).
Fig. 4.
Ezetimibe-mediated changes in jejunal expression of key cholesterol homeostatic genes. Analysis of pooled data from four independent studies is presented. Ages 11- to 14-week-old male C57BL/6J mice were fed chow diets with or without 0.003% ezetimibe in the diet for three or four days. Total RNA from jejunocytes (four or five mice per group in each of four studies) was separately isolated; equal amounts of RNA from each mouse was pooled and subjected to quantitative real-time PCR using cyclophilin as the invariant control. Each value represents the relative mRNA level or protein level in ezetimibe-treated mice compared with control mice who did not receive ezetimibe, which was normalized to 1.0. Each bar represents the mean ± SEM from the pooled data from four studies; overall n = 18 mice. The levels of statistical significance between the control and ezetimibe-treated groups are indicated (paired Student t-test; *P < 0.05, **P < 0.01).
Comprehensive studies by Tontonoz and colleagues (19, 20) have established that IDOL, an E3 ubiquitin ligase, promotes the degradation of LDLR. This raises the possibility that the ezetimibe-induced increase in intestinal LDLR protein levels is caused by a decrease in IDOL. To test this hypothesis, mice were treated with ezetimibe either alone or in combination with an exogenous LXR agonist, T0901317. In the intestine, ezetimibe alone produced the expected increase in LDLR protein and decrease in IDOL mRNA (Fig. 5A, lane 2, and 5B). No change in LRP, an LDLR family member resistant to IDOL-dependent degradation (19, 49), was observed. T0901317 increased IDOL expression and decreased jejunal LDLR protein without affecting LDLR mRNA (Fig. 5A, lane 3, and 5B). Coadministration of ezetimibe with T0901317 largely prevented both the ezetimibe-induced decrease in IDOL mRNA and the increase in LDLR protein levels (Fig. 5A, lane 4). Importantly, the LDLR mRNA level in the intestine was induced equally by ezetimibe with or without T0901317 (Fig. 5B). Scap mRNA levels were not altered by ezetimibe or T0901317 administration. Hepatic levels of LDLR and LRP protein were not altered by ezetimibe or T0901317 (Fig. 5A, lanes 5–8). In Fig. 5C, the level of LDLR protein content is plotted as a function of IDOL mRNA level for the four experimental groups from Fig. 5A, B. Because antibodies capable of detecting endogenous IDOL are not readily available, IDOL mRNA was used here as a surrogate for its activity. A negative relation between IDOL and LDLR protein level was noted in intestine but not in liver. These data show that ezetimibe's effect to increase LDLR number can be abrogated by maintaining an artificially high level of IDOL.
Fig. 5.
Exogenous LXR agonism abrogates ezetimibe's increase in jejunal LDLR via increased IDOL expression. (A) Immunoblot analysis of jejunocyte and liver membrane fractions from 15- to 18-week-old female C57BL/6J mice fed diets with or without 0.01% ezetimibe (for five days) and 0.015% T0901317 (for two days) either singly or in combination. All mice were euthanized on the same day, and the T0901317 dosing was delayed so that it was given the two days preceding euthanasia. Jejunocytes or livers (five mice per group) were fractionated and subjected to SDS-PAGE and immunoblot analysis. LRP served as loading control for the membrane fraction. (B) Comparison of relative hepatic and jejunocyte mRNA levels. Total RNA from jejunocytes or livers (five mice per group) was isolated and subjected to quantitative real-time PCR using cyclophilin as the invariant control. Animals who did not receive T1317 are indicated by open circles, whereas those who received T0901317 are indicated by filled circles. Ezetimibe administration is as indicated. mRNA levels from mice that did not receive either drug were normalized to 1.0. (C) Relationship between IDOL expression and LDLR protein levels in jejunum and liver. LDLR immunoblots from panel A were quantified by densitometry and plotted against IDOL mRNA from panel B; protein and mRNA levels of mice that did not receive either drug were normalized to 1.0.
Balance of PCSK9 and IDOL regulates jejunal LDLR content
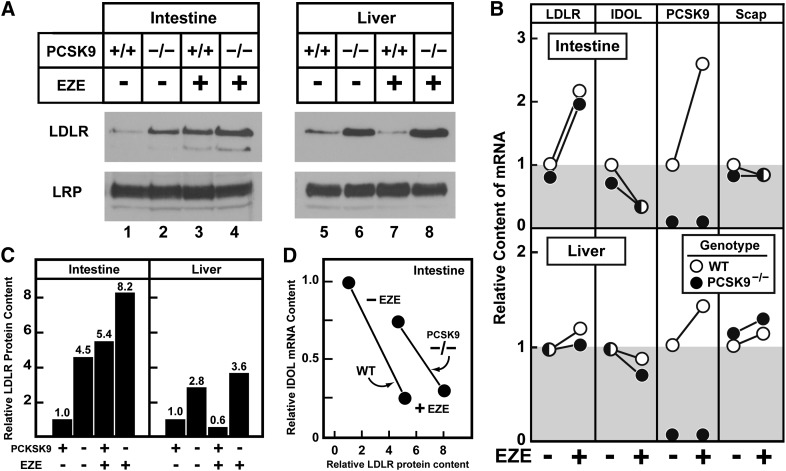
The above results present an apparent paradox in that ezetimibe, which dramatically increases LDLR protein levels, differentially affects the expression of two negative regulators of LDLR, namely PCSK9 and IDOL. Ezetimibe reduces the expression of IDOL, which would be predicted to increase LDLR protein, but ezetimibe also increases the expression of PCSK9, which would be predicted to reduce LDLR protein. To assess whether the ezetimibe-induced increase in intestinal PCSK9 mRNA attenuates the increase in intestinal LDLR protein, ezetimibe was administered to mice with a germline ablation of PCSK9 and to matched wild-type mice (23). In the intestine, the loss of PCSK9 was associated with a 4.5-fold increase in LDLR (Fig. 6A, lane 2). Ezetimibe treatment was associated with a 5.4-fold increase in LDLR (Fig. 6A, lane 3). Ezetimibe administration to PCSK9 knockout mice was associated with an 8.2-fold increase in the jejunal LDLR (Fig. 6A, lane 4), which is close to the predicted response if the individual fold increases in LDLR from PCSK9 ablation or from ezetimibe treatment were summed arithmetically (i.e., 8.2-fold observed versus 8.9-fold expected). In liver, PCSK9 ablation was associated with a ∼3-fold increase in LDLR levels, which was not altered by ezetimibe administration (Fig. 6A, lanes 5–8). Levels of the control receptor LRP were equal across groups in both liver and intestine.
Fig. 6.
PCSK9 regulates basal intestinal LDLR protein levels in jejunum. (A) Immunoblot analysis of jejunocyte and liver membrane fractions from 13- to 16-week-old male wild-type or PCSK9 knockout mice fed diets with or without 0.01% ezetimibe for five days. Jejunocytes or livers (four mice per group) were fractionated and subjected to SDS-PAGE and immunoblot analysis as described in Fig. 1. (B) Comparison of relative hepatic and jejunocyte mRNA levels. Total RNA from jejunocytes or livers (four mice per group) was isolated and subjected to quantitative real-time PCR as described in Fig. 1. Wild-type animals are indicated by open circles, whereas PCSK9 knockout mice are indicated by filled circles. Ezetimibe administration is as indicated. mRNA levels from wild-type mice that did not receive ezetimibe were normalized to 1.0. (C) Quantitation of LDLR protein levels. Immunoblots from panel A were quantified by densitometry. Each bar represents the relative LDLR protein level from each group as described in panel A; protein levels from wild-type mice that did not receive ezetimibe were normalized to 1.0. (D) Relationship between IDOL expression and LDLR protein levels in jejunum of wild-type and PCSK9 knockout mice. Relative LDLR protein levels from panels A and C are plotted against IDOL mRNA from panel B. Wild-type mice that did not receive ezetimibe were normalized to 1.0 for protein and mRNA levels.
Importantly, the levels of intestinal LDLR and IDOL mRNA were similar between the wild-type and PCSK9 knockout mice with or without ezetimibe exposure (Fig. 6B, top panel). Scap mRNA levels were not altered by ezetimibe administration or by PCSK9 ablation. In Fig. 6D, the level of jejunal LDLR protein content is plotted as a function of IDOL mRNA level for the four experimental groups from Fig. 6A, B. Wild-type and PCSK9 knockout mice are plotted as separate curves. The slope of the relationship between LDLR and IDOL jejunal expression is similar for mice of both genotypes, but the curve is shifted rightward in the PCSK9 knockout mice. This indicates that the basal level of the LDLR is increased in PCSK9 knockout mice, but that the reduction by IDOL is the same in wild-type and knockout mice. Taken together, these data suggest that PCSK9 reduces basal LDLR levels in the intestine, and that the effect of ezetimibe is not dependent upon changes in PCSK9.
Although the jejunal PCSK9 mRNA was consistently elevated following ezetimibe administration, driven by the increase in nuclear SREBP-2 (Figs. 4 and 6B), plasma PCSK9 levels were not altered in the ezetimibe-treated mice under these dosing conditions (data not shown). Jejunal PCSK9 mRNA levels were 20% of those seen in liver, and in the current studies, jejunal PCSK9 protein was not detected with antibodies that can detect endogenous PCSK9 in mouse liver (23, 50). As such, the ezetimibe-induced increase in intestinal PCSK9 mRNA is not associated with a detectable increase in plasma or intestinal PCSK9 protein.
DISCUSSION
The current results confirm that, to maintain sterol homeostasis in the face of ezetimibe-induced sterol depletion, enterocytes activate LDL uptake by increasing LDLR number and boost sterol synthesis by increasing the expression of HMGR and nearly all other genes needed for sterol synthesis. Both of these effects are mediated in part via activation of SREBP-2 processing. The increases in the levels of LDLR and HMGR proteins are more dramatic than would have been expected for the relatively modest increases in their cognate mRNAs, which suggests that posttranscriptional mechanisms are affected. These effects are concentrated in the enterocytes of the middle and upper villus of the jejunum, which contain the enterocyte population that is the most active in lipoprotein metabolism and presumably cholesterol absorption (51–53). Whereas cholesterol synthesis and LDL uptake are most active in the lower villus and crypt (44), ezetimibe redistributes these processes to the middle and upper villus. These data indicate that the low rates of cholesterol synthesis and LDL uptake in the upper villus are attributable to constant cholesterol uptake into these cells that is mediated by NPC1L1. When NPC1L1 is blocked by ezetimibe, this repression is relieved and the cells respond by obtaining cholesterol from endogenous sources. It is significant that SREBP-2 targets in the upper villus are repressed even when mice ingest a low-cholesterol diet. It is likely that this repression is mediated by the reabsorption of cholesterol from biliary secretions and from sloughing of the gut epithelium.
The current studies confirm and extend prior studies that report increased LDLR and HMGR mRNAs in the intestines of miniature pigs, hamsters, and mice in response to ezetimibe, as well other reports that in vivo sterol synthesis rates in the intestine are increased by ezetimibe in hamsters and mice (39, 47, 54). Ezetimibe has also been shown to increase whole-body fractional LDL clearance as estimated by apoB kinetics in miniature pigs and humans (54, 55). On the basis of our current data, we speculate that the increased LDL uptake occurs largely in the jejunum rather than in the liver.
The mechanism for the ezetimibe-induced increase in HMGR protein remains unclear. This degree of elevation is reminiscent to what is seen in the tissues of mice that lack Insig-1 and Insig-2 (11, 56), which bind to HMGR in a sterol-dependent fashion to mediate its rapid proteosomal degradation. Alternatively, deficiencies in function of GP78 or other factors required for regulated proteosomal degradation of HMGR could be present (57). We note that enterocyte GP78 levels are not affected by ezetimibe as measured by immunoblotting (data not shown). To test the hypothesis that altered Insig function or abundance contributes to the upregulation of HMGR abundance in enterocytes, further studies in mice with intestine-specific overexpression or ablation of Insig proteins are underway and are beyond the scope of the current studies.
The current studies highlight a previously undescribed connection between NPC1L1, LXRs, and LDLR through IDOL in the intestine. The intestinal mRNAs for several LXR-responsive genes, such as IDOL, ABCG5, and ABCG8, were reduced by ezetimibe. Lumenal sterols are likely acting to provide sterol ligands that activate LXRs in enterocytes, and the reduced sterol flux caused by ezetimibe reduces the concentrations of these ligands. Inasmuch as IDOL is an LXR-responsive gene, its expression is reduced in response to ezetimibe in the intestine. The increase in LDLR mRNA mediated by increased SREBP-2 processing, in concert with reduced IDOL expression mediated presumably through reduced LXR activity, led to a dramatic increase in steady-state LDLR protein levels. However, coincident with the increase in SREBP-2 processing was an increase in PCSK9 expression, which would be predicted to degrade intestinal LDLRs in response to ezetimibe. This situation is analogous to that of statin-treated mouse liver, where LDLR protein does not increase despite an increase in LDLR mRNA, which is attributable to an increase in hepatic and plasma PCSK9 levels (23). In contrast to liver, the increased jejunal PCSK9 expression does not delimit the increase in LDLR protein in response to ezetimibe (Fig. 6C). We speculate that this failure is due to the lower overall expression of PCSK9 in the intestine compared with the liver or that the increase in enterocyte PCSK9 mRNA does not provoke an increase in plasma PCSK9. Under any circumstance, in the current studies, IDOL trumps PCSK9 in the regulation of jejunal LDLRs.
With respect to the use of ezetimibe as a hypocholesterolemic agent in humans, these findings are a mix of good and bad. The increase in HMGR and nearly all genes required for sterol synthesis is predicted to lessen the LDL-lowering capacity of ezetimibe, whereas an increase in intestinal LDL uptake is predicted to improve it. If the ameliorative effects of NPC1L1 inhibition (blockage of bulk cholesterol influx and increased LDL clearance) could be uncoupled from the pejorative effects (increased de novo sterol synthesis and possibly increased PCSK9), a more potent LDL-lowering agent could be achieved. Further studies into whether the regulatory sterol pools that affect each arm of this complex network, namely, SREBP processing, LXR activation, HMG-CoA degradation, and others, can be dissected from one another are warranted.
Supplementary Material
Acknowledgments
The authors thank Michael Brown and Joseph Goldstein for their mentorship and support; Jay Horton and Joyce Repa for critical review of the manuscript; Stephen Turley for helpful discussions; Isis Desoto, Monica Mendoza, Norma Anderson, and Tuyet Dang for technical assistance; Jeff Cormier for help with RT-PCR; the UT-Southwestern Microarray Core Facility for help with the microarray studies; and the UT-Southwestern Richardson Molecular Pathology Core Facility for tissue sectioning.
Footnotes
Abbreviations:
- AP
- alkaline phosphatase
- CREB
- cAMP response element binding
- ER
- endoplasmic reticulum
- EZE
- ezetimibe
- FPPS
- farnesyl pyrophosphate synthase
- HMGR
- HMG-CoA reductase
- HMGS
- HMG-CoA synthase
- IDOL
- inducible degrader of LDLR
- LDLR
- LDL receptor
- LRP
- LDL receptor-related protein
- LXR
- liver X receptor
- NPC1L1
- Niemann-Pick C1-like 1
- PCNA
- proliferating cell nuclear antigen
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- SREBP
- sterol regulatory element-binding protein
- nSREBP
- nuclear SREBP
- SS
- squalene synthase
This work was supported by grants from the National Institutes of Health (HL-20948) and the Moss Heart Foundation. L. J. Engelking was supported by National Institutes of Health Institutional Training Grant 2T32-DK-007745-16. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Xie C., Turley S. D., Dietschy J. M. 2009. ABCA1 plays no role in the centripetal movement of cholesterol from peripheral tissues to the liver and intestine in the mouse. J. Lipid Res. 50: 1316–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spady D. K., Dietschy J. M. 1983. Sterol synthesis in vivo in 18 tissues of the squirrel monkey, guinea pig, rabbit, hamster, and rat. J. Lipid Res. 24: 303–315 [PubMed] [Google Scholar]
- 3.Spady D. K., Bilheimer D. W., Dietschy J. M. 1983. Rates of receptor-dependent and -independent low density lipoprotein uptake in the hamster. Proc. Natl. Acad. Sci. USA. 80: 3499–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spady D. K., Turley S. D., Dietschy J. M. 1985. Receptor-independent low density lipoprotein transport in the rat in vivo. Quantitation, characterization, and metabolic consequences. J. Clin. Invest. 76: 1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M. S., Goldstein J. L. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340 [DOI] [PubMed] [Google Scholar]
- 6.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nohturfft A., DeBose-Boyd R. A., Scheek S., Goldstein J. L., Brown M. S. 1999. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. USA. 96: 11235–11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102: 315–323 [DOI] [PubMed] [Google Scholar]
- 9.Espenshade P. J., Li W. P., Yabe D. 2002. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA. 99: 11694–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500 [DOI] [PubMed] [Google Scholar]
- 11.Engelking L. J., Liang G., Hammer R. E., Takaishi K., Kuriyama H., Evers B. M., Li W. P., Horton J. D., Goldstein J. L., Brown M. S. 2005. Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115: 2489–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelking L. J., Kuriyama H., Hammer R. E., Horton J. D., Brown M. S., Goldstein J. L., Liang G. 2004. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 113: 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 11: 25–33 [DOI] [PubMed] [Google Scholar]
- 14.DeBose-Boyd R. A. 2008. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 18: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton J. D., Cohen J. C., Hobbs H. H. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S. W., Moon Y. A., Horton J. D. 2004. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9 in mouse liver. J. Biol. Chem. 279: 50630–50638 [DOI] [PubMed] [Google Scholar]
- 17.Grefhorst A., McNutt M. C., Lagace T. A., Horton J. D. 2008. Plasma PCSK9 preferentially reduces liver LDL receptors in mice. J. Lipid Res. 49: 1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian Y. W., Schmidt R. J., Zhang Y. Y., Chu S. Y., Lin A. M., Wang H., Wang X. L., Beyer T. P., Bensch W. R., Li W. M., et al. 2007. Secreted PCSK9 downregulates low density lipoprotein receptor through receptor-mediated endocytosis. J. Lipid Res. 48: 1488–1498 [DOI] [PubMed] [Google Scholar]
- 19.Zelcer N., Hong C., Boyadjian R., Tontonoz P. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 325: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tontonoz P. 2011. Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors. Cold Spring Harb. Symp. Quant. Biol. Epub ahead of print. August 22, 2011; doi: 10.1101/sqb.2011.76.010702. [DOI] [PubMed] [Google Scholar]
- 21.Scotti E., Hong C., Yoshinaga Y., Tu Y., Hu Y., Zelcer N., Boyadjian R., de Jong P. J., Young S. G., Fong L. G., et al. 2011. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol. Cell. Biol. 31: 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tontonoz P., Mangelsdorf D. J. 2003. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 17: 985–993 [DOI] [PubMed] [Google Scholar]
- 23.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 102: 5374–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng Z., Otani H., Brown M. S., Goldstein J. L. 1995. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. USA. 92: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimomura I., Bashmakov Y., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. 1997. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. USA. 94: 12354–12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRa and LXRb. Genes Dev. 14: 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repa J. J., Berge K. E., Pomajzl C., Richardson J. A., Hobbs H., Mangelsdorf D. J. 2002. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277: 18793–18800 [DOI] [PubMed] [Google Scholar]
- 28.Davis H. R., Jr, Altmann S. W. 2009. Niemann-Pick C1 Like 1 (NPC1L1) an intestinal sterol transporter. Biochim. Biophys. Acta. 1791: 679–683 [DOI] [PubMed] [Google Scholar]
- 29.Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. 2002. Diminished hepatic response to fasting/refeeding and LXR agonists in mice with selective deficiency of SREBP-1c. J. Biol. Chem. 277: 9520–9528 [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Goldstein J. L., Hammer R. E., Moon Y. A., Brown M. S., Horton J. D. 2001. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl. Acad. Sci. USA. 98: 13607–13612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimano H., Shimomura I., Hammer R. E., Herz J., Goldstein J. L., Brown M. S., Horton J. D. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda M., Korn B. S., Hammer R. E., Moon Y. A., Komuro R., Horton J. D., Goldstein J. L., Brown M. S., Shimomura I. 2001. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Y., Nassir F., Luo J., Buhman K., Davidson N. O. 2003. Intestinal lipoprotein assembly in apobec-1−/− mice reveals subtle alterations in triglyceride secretion coupled with a shift to larger lipoproteins. Am. J. Physiol. Gastrointest. Liver Physiol. 285: G735–G746 [DOI] [PubMed] [Google Scholar]
- 34.Drover V. A., Ajmal M., Nassir F., Davidson N. O., Nauli A. M., Sahoo D., Tso P., Abumrad N. A. 2005. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J. Clin. Invest. 115: 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Wu G., Chapkin R. S., Lupton J. R. 1998. Energy metabolism of rat colonocytes changes during the tumorigenic process and is dependent on diet and carcinogen. J. Nutr. 128: 1262–1269 [DOI] [PubMed] [Google Scholar]
- 36.Weiser M. M. 1973. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J. Biol. Chem. 248: 2536–2541 [PubMed] [Google Scholar]
- 37.Evers B. M., Farooqi M. S., Shelton J. M., Richardson J. A., Goldstein J. L., Brown M. S., Liang G. 2010. Hair growth defects in Insig-deficient mice caused by cholesterol precursor accumulation and reversed by simvastatin. J. Invest. Dermatol. 130: 1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altmann S. W., Davis H. R., Zhu L. J., Yao X. R., Hoos L. M., Tetzloff G., Iyer S. P. N., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204 [DOI] [PubMed] [Google Scholar]
- 39.Repa J. J., Turley S. D., Quan G., Dietschy J. M. 2005. Delineation of molecular changes in intrahepatic cholesterol metabolism resulting from diminished cholesterol absorption. J. Lipid Res. 46: 779–789 [DOI] [PubMed] [Google Scholar]
- 40.Horton J. D., Cohen J. C., Hobbs H. H. 2009. PCSK9: a convertase that coordinates LDL catabolism. J. Lipid Res. 50(Suppl.):S172–S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Field F. J., Born E., Murthy S., Mathur S. N. 2001. Regulation of sterol regulatory element-binding proteins in hamster intestine by changes in cholesterol flux. J. Biol. Chem. 276: 17576–17583 [DOI] [PubMed] [Google Scholar]
- 42.Schwarz M., Russell D. W., Dietschy J. M., Turley S. D. 1998. Marked reduction in bile acid synthesis in cholesterol 7alpha-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J. Lipid Res. 39: 1833–1843 [PubMed] [Google Scholar]
- 43.Mariadason J. M., Nicholas C., L'Italien K. E., Zhuang M., Smartt H. J., Heerdt B. G., Yang W., Corner G. A., Wilson A. J., Klampfer L., et al. 2005. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 128: 1081–1088 [DOI] [PubMed] [Google Scholar]
- 44.Stange E. F., Dietschy J. M. 1983. Cholesterol synthesis and low density lipoprotein uptake are regulated independently in rat small intestinal epithelium. Proc. Natl. Acad. Sci. USA. 80: 5739–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Field F. J., Born E., Murthy S., Mathur S. N. 2001. Gene expression of sterol regulatory element-binding proteins in hamster small intestine. J. Lipid Res. 42: 1–8 [PubMed] [Google Scholar]
- 46.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valasek M. A., Repa J. J., Quan G., Dietschy J. M., Turley S. D. 2008. Inhibiting intestinal NPC1L1 activity prevents diet-induced increase in biliary cholesterol in Golden Syrian hamsters. Am. J. Physiol. Gastrointest. Liver Physiol. 295: G813–G822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valasek M. A., Weng J., Shaul P. W., Anderson R. G., Repa J. J. 2005. Caveolin-1 is not required for murine intestinal cholesterol transport. J. Biol. Chem. 280: 28103–28109 [DOI] [PubMed] [Google Scholar]
- 49.Hong C., Duit S., Jalonen P., Out R., Scheer L., Sorrentino V., Boyadjian R., Rodenburg K. W., Foley E., Korhonen L., et al. 2010. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J. Biol. Chem. 285: 19720–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. 2006. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 116: 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sparrow C. P., Patel S., Baffic J., Chao Y. S., Hernandez M., Lam M. H., Montenegro J., Wright S. D., Detmers P. A. 1999. A fluorescent cholesterol analog traces cholesterol absorption in hamsters and is esterified in vivo and in vitro. J. Lipid Res. 40: 1747–1757 [PubMed] [Google Scholar]
- 52.Suzuki T., Mochizuki K., Goda T. 2009. Localized expression of genes related to carbohydrate and lipid absorption along the crypt-villus axis of rat jejunum. Biochim. Biophys. Acta. 1790: 1624–1635 [DOI] [PubMed] [Google Scholar]
- 53.Stange E. F., Dietschy J. M. 1983. Absolute rates of cholesterol synthesis in rat intestine in vitro and in vivo: a comparison of different substrates in slices and isolated cells. J. Lipid Res. 24: 72–82 [PubMed] [Google Scholar]
- 54.Telford D. E., Sutherland B. G., Edwards J. Y., Andrews J. D., Barrett P. H., Huff M. W. 2007. The molecular mechanisms underlying the reduction of LDL apoB-100 by ezetimibe plus simvastatin. J. Lipid Res. 48: 699–708 [DOI] [PubMed] [Google Scholar]
- 55.Tremblay A. J., Lamarche B., Cohn J. S., Hogue J. C., Couture P. 2006. Effect of ezetimibe on the in vivo kinetics of apoB-48 and apoB-100 in men with primary hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 26: 1101–1106 [DOI] [PubMed] [Google Scholar]
- 56.Engelking L. J., Evers B. M., Richardson J. A., Goldstein J. L., Brown M. S., Liang G. 2006. Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo Y., Debose-Boyd R. A. 2010. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit. Rev. Biochem. Mol. Biol. 45: 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.