Abstract
The occurrence of systemic inflammatory response syndrome (SIRS) remains a major problem in intensive care units with high morbidity and mortality. The differentiation between noninfectious and infectious etiologies of this disorder is challenging in routine clinical practice. Many biomarkers have been suggested for this purpose; however, sensitivity and specificity even of high-ranking biomarkers remain insufficient. Recently, metabolic profiling has attracted interest for biomarker discovery. The objective of this study was to identify metabolic biomarkers for differentiation of SIRS/sepsis. A total of 186 meta-bolites comprising six analyte classes were determined in 143 patients (74 SIRS, 69 sepsis) by LC-MS/MS. Two markers (C10:1 and PCaaC32:0) revealed significantly higher concentrations in sepsis. A classification model comprising these markers resulted in 80% and 70% correct classifications in a training set and a test set, respectively.This study demonstrates that acylcarnitines and glycerophosphatidylcholines may be helpful for differentiation of infectious from noninfectious systemic inflammation due to their significantly higher concentration in sepsis patients. Considering the well known pathophysiological relevance of lipid induction by bacterial components, metabolites as identified in this study are promising biomarker candidates in the differential diagnosis of SIRS and sepsis.
Keywords: SIRS, severe sepsis, inflammation, metabolomics, C10:1 acylcarnitine, PCaaC32:0 phosphatidylcholine
The development of systemic inflammatory response syndrome (SIRS) associated with multiple organ dysfunction is accompanied by high morbidity and mortality in critically ill patients (1, 2). In contrast to the progress made in the symptomatic supportive therapy of organ dysfunction, diagnosis of the underlying infectious-induced etiology of SIRS is often delayed because clinical symptoms are broad and unspecific (3, 4). Because delays in the administration of antimicrobial therapy increase mortality (5), biomarkers indicating infection in the critically ill are vital. Furthermore, avoiding antibiotics in patients with SIRS without infection reduces the risk for developing antibiotic resistance (6).
Many efforts have been made to find biomarkers that allow discrimination of noninfectious SIRS and sepsis at an early stage of the disease. Most of those studies addressed the transcriptome or proteome for biomarker identification (7–9). Even though 178 different biomarkers for sepsis have been proposed (7), most of them have only prognostic and not discriminative value. Only few have been suggested for differentiation of noninfectious induced SIRS from sepsis, and the majority have not been validated for clinical routine use (9). The only marker widely accepted in the clinical setting is procalcitonin. However, procalcitonin remains controversial for differentiation of SIRS/sepsis because it has been described to be elevated after major surgery, a common cause of SIRS without underlying infection (10–14).
Technical advances in mass spectrometry opened a new field to this research. In a previous study, we identified a peptide biomarker by proteome analysis that discriminates noninfectious induced SIRS from sepsis, which is being validated in other patient groups (15). Because SIRS and sepsis are accompanied by severe metabolic alterations (16–20), we hypothesize that a systematic analysis of the metabolome may lead to the identification of new disease-specific biomarkers. Although there are several studies addressing single metabolites or metabolite groups in sepsis, no study has investigated the differential metabolom of SIRS patients with and without infection. Because LC-MS/MS allows simultaneous detection and quantification of up to several hundred metabolites in one sample in a relatively short time, we chose to determine 186 metabolites comprising six analyte classes in these patients.
MATERIALS AND METHODS
Patients and sample collection
Patients admitted to the intensive care unit (ICU) of the Department of Anesthesiology and Intensive Care Medicine, Jena University Hospital, from September 2002 until September 2003 were screened for eligibility. Patients were enrolled if they fulfilled the sepsis criteria according to the ACCP/SCCM consensus conference or the criteria for SIRS of noninfectious origin with acute organ dysfunction, respectively (ICD-10-GM code) (3, 4). In patients with SIRS of noninfectious origin, blood samples were obtained within 24 h after ICU admission or at the onset of symptoms. In patients with sepsis, samples were obtained within 24 h after onset of the first sepsis-induced organ dysfunction. Sepsis patients had to have a microbiologically documented infection or a clinically suspected infection as evidenced by one or more of the following: white cells in a normally sterile body fluid, perforated viscus, radiographic evidence of pneumonia in association with the production of purulent sputum, and a syndrome associated with a high risk of infection (e.g., ascending cholangitis). Patients admitted to the ICU after major cardiac or vascular surgery who did not fulfill SIRS criteria served as a control group (ICU controls). Patients less than 18 years old, pregnant, or lacking informed consent by their representatives were excluded. The local ethics committee approved this study.
According to a standardized protocol, 10 ml of blood was drawn into EDTA tubes (Sarstedt, Nuembrecht, Germany) immediately followed by the addition of 800 μl of a protease inhibitor cocktail (Complete Mini, Roche). Tubes were gently mixed and centrifuged at 3,750 rpm (2516 g at the middle of the tube) and 4°C for 10 min. Plasma aliquots were immediately frozen and stored at −80°C.
Determination of metabolite concentration
PITC, ammonium acetate, and HPLC water were purchased from Merck; methanol (HPLC grade) and acetonitril (LC-MS grade) were purchased from Roth; and formic acid was purchased from Sigma Aldrich. Metabolite concentrations were obtained using the AbsoluteIDQ kit p180 (Biocrates Life Science AG, Austria) according to manufacturer's instructions on an API4000™ LC/MS/MS System (AB SCIEX, USA) equipped with an electrospray ionization source, an Agilent G1367B autosampler, and the Analyst 1.51 software (AB SCIEX, USA). Samples (10 μl) were pipetted onto the spots of the kit plate. Spots were dried at room temperature in a nitrogen evaporator drying unit for 30 min. Twenty microliters of 5% PITC reagent was pipetted onto the spots and incubated for 20 min at RT. The plate was dried under the nitrogen evaporator for 60 min. Ammonium acetate (300 μl of 5 mM) in methanol was added to each well and incubated on a shaker (450 rpm) for 30 min. The plate was centrifuged at 100 g for 2 min, receiving about 250 μl sample in plate 1 (flow injection analysis [FIA] plate). The upper plate was removed, and 150 μl of each sample was transferred into a second plate (LC-MS plate). HPLC water (150 μl) was added to the LC-MS plate, and 500 μl of MS running solvent (Biocrates solvent diluted in methanol) was added to the FIA plate. The LC-MS plate was measured first by scheduled multiple reaction monitoring, and the FIA plate was stored at 4°C. Sample (10 μl) was injected onto a Zorbax Eclipse XDB C18, 3 × 100 mm column (Agilent) coupled to a C18, 4 × 3 mm security guard precolumn (Phenomenex) and eluted with solvent A (HPLC water + 0.2% formic acid) and solvent B (acetonitrile + 0.2% formic acid). Peaks were integrated, and concentrations were obtained (in comparison to internal standards) with Analyst 1.51. Evaluation of calibration curves, blanks, quality controls, and samples was accomplished in the MetIQ software (an integral part of the kit). The FIA plate was measured by FIA, and detection of fragments was performed in multiple reaction monitoring mode. Sample (20 μl) was injected directly into the MS at a flow of 30 μl/min with MS running solvent. Concentrations were calculated and evaluated in the Analyst/MetIQ software by comparing measured analytes in a defined extracted ion count section to those of specific labeled internal standards or nonlabeled, nonphysiological standards (semiquantitative) provided by the kit plate.
The nomenclature of phospholipids comprises a substance class of the same mass rather than a single substance. It includes the number of carbon atoms and the number of double bonds of both fatty acids linked to C1 and C2 of glycerophosphatidylcholine. The nature of fatty acid linkage is expressed as aa for diacyl or ae for acyl-alkyl.
Statistics
Statistical analysis was performed via SPSS Statistics 19 (IBM, USA). The dataset was divided into a training set and a test set. Normal distribution of the metabolite concentrations was tested. Depending on the outcome, Student's t-test (normal distribution) or the Mann-Whitney U test (no normal distribution) was chosen for determination of statistical significance. The level of statistical significance (0.05) was corrected with the Bonferroni method and then defined as 2.7 × 10−4. ROC curves were obtained by plotting sensitivity against 1-specificity. The obtained area under the curve (AUC) values are a measure for the predictive power of the analyte: the higher or lower the AUC (converges 1 or 0), the better is the classification by the corresponding analyte. For the combination of analytes, a binary logistic regression analysis was performed for the training set and applied to the test set. The obtained predicted probabilities for correct classification of each value can be plotted as ROC-curve. The proportion of correct classification (sensitivity and specificity) for the training set and the test set were obtained.
RESULTS
Samples of 143 patients were divided into a training set (30 sepsis, 33 noninfectious SIRS) for marker identification and establishment of a model for discrimination of SIRS and sepsis and a test set (39 sepsis, 41 noninfectious SIRS) for validation of that model. Metabolite concentrations of 16 ICU control subjects were determined. Clinical characteristics of the patients are depicted in Table 1.
TABLE 1.
Patient clinical characteristics for the SIRS and the sepsis group for the training set and the test set
| SIRS without infection |
SIRS with infection (severe sepsis /septic shock) |
ICU controls | |||
| Training samples (n = 33) | Test samples (n = 41) | Training samples (n = 30) | Test samples (n = 39) | (n = 16) | |
| Age, years (mean±SD) | 66.76 ± 10.29 | 67.46 ± 9.9 | 63.83 ± 11.39 | 63.56 ± 15.27 | 67.06 ± 11.73 |
| Gender (f/m) | 8/25 | 14/27 | 6/24 | 15/24 | 4/12 |
| APACHE II score (mean±SD) | 18.12 ± 7.1 | 19.22 ± 7.41 | 22.7 ± 7.6 | 25.79 ± 8.63 | 10.63 ± 1.93 |
| SOFA score (mean±SD) | 7.97 ± 3.89 | 8.83 ± 3.36 | 9.43 ± 3.58 | 11.44 ± 3.88 | 4.87 ± 1.89 |
| Mechanical ventilation (%) | 45.5 | 65.9 | 83.3 | 94.9 | 5.9 |
| PCT procalcitonin ([ng/ml], mean±SD) | 14.04 ± 29.94 | 14.95 ± 25.12 | 26.58 ± 76.6 | 12.97 ± 27.05 | 0.3 ± 0.01 |
| Diagnosis (MDI/CDI) | 18/12 | 25/14 | |||
| Pathogen (gram+/gram−/fungal/ mixed infection)* | 11/5/1/1 | 4/18/1/2 | |||
| Recent surgical history (%) | 100 | 100 | 96.7 | 89.7 | 100 |
| Type of surgery (n) | |||||
| Abdominal surgery | 2 | 14 | |||
| Cardiothoracic surgery | 30 | 38 | 17 | 12 | 15 |
| Obstetrics | 2 | ||||
| Maxillofacial surgery | 1 | ||||
| Neurosurgery | 3 | 6 | 2 | ||
| Trauma surgery | 1 | ||||
| Urosurgery | 2 | ||||
| vVascular surgery | 3 | 3 | 2 | 1 | |
| No history of surgery | 1 | 4 | |||
| Focus (n) | |||||
| Intra-abdominal | 2 | ||||
| Primary bacteremia | 1 | ||||
| Catheter-associated | 4 | 3 | |||
| Wound infection | 1 | ||||
| Mediastinitis | 3 | ||||
| Meningitis | 1 | 1 | |||
| Other | 3 | ||||
| Pericarditis | 1 | ||||
| Peritonitis | 2 | 17 | |||
| Pleural empyema | 1 | 1 | |||
| Pneumonia | 13 | 14 | |||
| Urogenital | 1 | ||||
| No | 33 | 41 | 16 | ||
MDI/CDI, microbiologically documented /clinically documented infection.
Isolated pathogens of microbiologically documented sepsis patients are depicted in supplementary Table I.
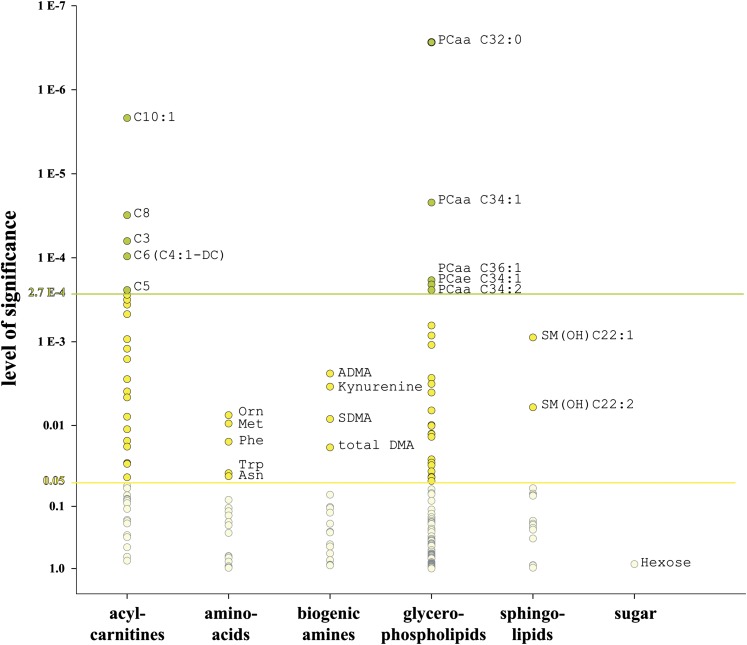
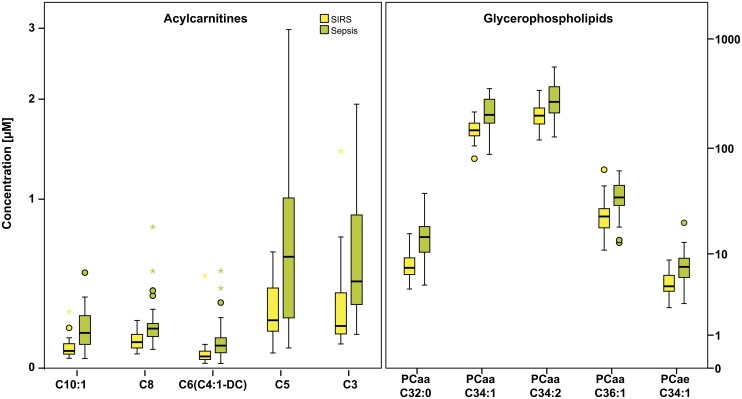
Concentrations of 186 metabolites belonging to six analyte classes (acylcarnitines, amino acids, biogenic amines, glycerophospholipids, sphingolipids, and carbohydrates) were determined by LC-MS/MS. Analytes and their respective P values for these classes in the trainings set are shown in Fig. 1. At a significance level of 0.05, analytes from all classes but the carbohydrates were significantly different. After Bonferroni correction, the level of significance was defined as 2.7 × 10−4. At this level, only acylcarnitines and the glycerophospholipids were significantly higher in sepsis samples. Boxplots of the respective markers are shown in Fig. 2.
Fig. 1.
Level of significance. All 186 metabolites and their respective P values are shown for the six analyte classes. At a level of 0.05, representatives of all classes but the sugars are significant (yellow circles). After Bonferroni correction, the level of significance is 2.7 E-4. Only metabolites of the acylcarnitines and glycerophosphatidylcholines are significant at this level (green circles).
Fig. 2.
Box plots of the significant acylcarnitines and glycerophospholipids. The box represents the 25th–75th percentiles. Whiskers indicate the lowest and highest values of the respective quartile, dots present mild outliers, and asterisks extreme outliers. All significant metabolites, both acylcarnitines and glycerophospholipids, have significant higher concentrations in sepsis.
Metabolite concentrations for most of the 186 analytes in ICU controls were at the SIRS level (data not shown).
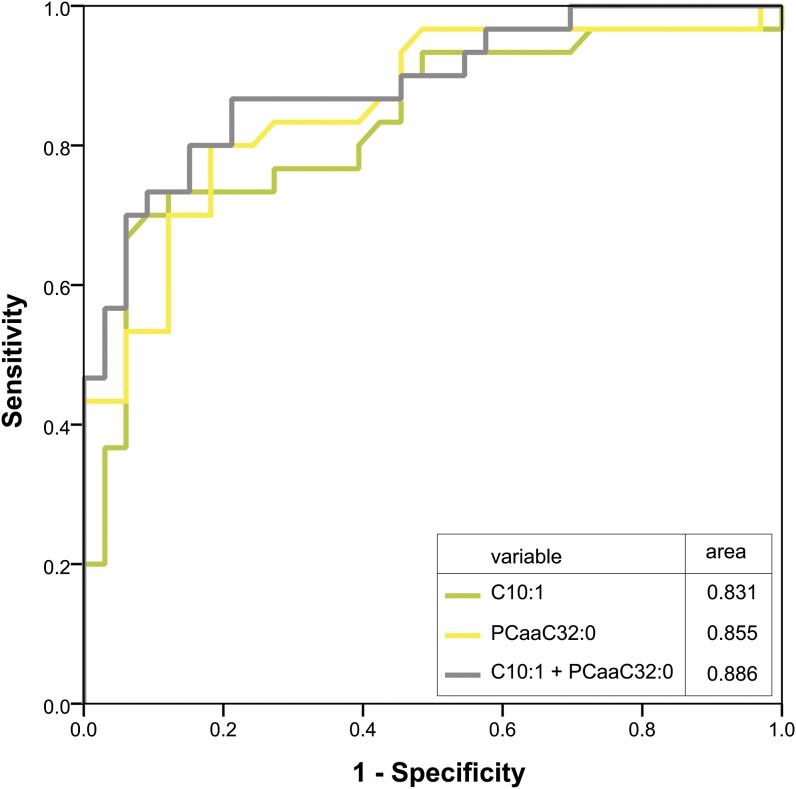
To analyze the diagnostic value of these markers, we included all significant markers of the training set after Bonferroni correction into a model for classification of samples in the SIRS and the sepsis group. For validation, we applied this model to a test set. The AUC values for the single markers and for the combination of markers as well as the percentages of correctly classified patients for the test set and the training set are depicted in Table 2. About 80% of patients in the training set were correctly classified by the two markers (C10:1 + PCaaC32:0), and these results were reproducible in the test set (70% correct classified samples). Moreover, the combination of more than two markers does not yield improvement in classification, which can be traced back to a strong correlation of markers within one substance class (Fig. 3).
TABLE 2.
AUCs and model quality
| Six significant analytes | Five significant analytes | Four significant analytes | Three significant analytes | Two significant analytes | |
| Training set | |||||
| Correct classifications (%) | |||||
| SIRS | 81.8 | 87.9 | 84.8 | 84.8 | 84.8 |
| Sepsis | 80.0 | 76.7 | 76.7 | 73.3 | 73.3 |
| Total | 81.0 | 82.5 | 81.0 | 79.4 | 79.4 |
| AUC | |||||
| PCaaC32:0 | 0.855 | 0.855 | 0.855 | 0.855 | 0.855 |
| C10:1 | 0.831 | 0.831 | 0.831 | 0.831 | 0.831 |
| PCaaC34:1 | 0.800 | 0.800 | 0.800 | 0.800 | |
| C8 | 0.795 | 0.795 | 0.795 | ||
| C3 | 0.784 | 0.784 | |||
| C6 (C4:1-DC) | 0.778 | ||||
| Binary logistic regression | 0.899 | 0.894 | 0.894 | 0.897 | 0.886 |
| Test set | |||||
| Correct classifications (%) | |||||
| SIRS | 75.6 | 75.6 | 75.6 | 80.5 | 80.5 |
| Sepsis | 64.1 | 69.2 | 69.2 | 64.1 | 66.7 |
| Total | 70.0 | 72.5 | 72.5 | 72.5 | 73.8 |
| AUC | |||||
| PCaaC32:0 | 0.752 | 0.752 | 0.752 | 0.752 | 0.752 |
| C10:1 | 0.758 | 0.758 | 0.758 | 0.758 | 0.758 |
| PCaaC34:1 | 0.713 | 0.713 | 0.713 | 0.713 | |
| C8 | 0.706 | 0.706 | 0.706 | ||
| C3 | 0.725 | 0.725 | |||
| C6 (C4:1-DC) | 0.764 | ||||
| Binary logistic regression | 0.781 | 0.792 | 0.801 | 0.806 | 0.805 |
Shown are the AUCs for the single markers and their combination in a binary logistic regression analysis and the correct classifications in percent for the combination of two to six markers. The combination of more than two markers does not yield further improvement, probably due to strong correlation between markers within one analyte group.
The training set mainly consisted of gram-positive sepsis cases, whereas the test set contained more gram-negative cases (Table 1 and Supplementary Table I). The discrimination of SIRS and sepsis by selected analyte concentrations worked well in both sets (Table 2), suggesting that the observed increase of analyte concentrations is a general host response to infection independent on the gram status of bacteria. However, only 11 out of 23 g negative sepsis cases compared with 13 out of 15 g positive sepsis cases (training set and test set) were classified correctly (Supplementary Table II). This might suggest that C10:1 + PCaaC32:0 are more suitable for the detection of gram-positive sepsis, but this hypothesis needs to be confirmed in a larger patient cohort.
Fig. 3.
ROC curve of C10:1, PCaaC32:0, and the combination of both markers. ROC curves were created for the best markers of the acylcarnitines (green) and glycerophosphatidylcholines (yellow) in the training set. Both markers were included in a binary logistic regression model and the obtained predicted probabilities plotted as ROC curve representing the combination of those markers (gray). Respective AUCs are shown for the single markers and the combination.
DISCUSSION
Sepsis is accompanied by severe metabolic changes, such as an increase of plasma lipids, including triglycerides (TGs), phospholipids, and free fatty acids (FAs). In animals, plasma lipids can be induced by lipopolysaccharides (LPSs) and lipoteichoic acid, a component of the cell wall of gram-positive bacteria, due to increased lipolysis and impaired lipid catabolism (16). This seems to be the result of a coordinated, predominantly down-regulation of many proteins taking part in FA transport , FA oxidation (carnitine palmitoyl transferase, medium chain acyl CoA dehydrogenase [MCAD]), and the breakdown of triglycerides (LPL) (17–20). A well characterized enzyme taking part in FA oxidation that is suppressed in sepsis is MCAD (17, 19). This mitochondrial enzyme processes FAs bound to CoA (acyl-CoA) with a chain length of C5–C14. Inhibition of oxidation by MCAD leads to accumulation of medium-chain acyl-CoA in the mitochondria, which is quite toxic for the cell. As a consequence, medium-chain FA intermediates are transferred to carnitine and are capable of leaving the mitochondria and the cell as acylcarnitine. This is supported by our observation that most of the acylcarnitines increased in sepsis samples belong to medium-chain acylcarnitines (Figs. 1 and 2). Suppression of MCAD has also been shown in zymosan- and turpentine-induced inflammation and therefore has been suggested as a general response in inflammation (21), but we could not confirm this in our patients because the intense rise of MCAD intermediates seems to be sepsis specific and does not occur in noninfectious SIRS compared with ICU control subjects. However, these models might not properly reflect the conditions in our SIRS patients.
In plasma, most lipids are bound to albumin, chylomicrons, and lipoproteins. The latter particles, specifically LDL and VLDL, have also been shown to be highly elevated in sepsis (22–24). We suppose that the elevated levels of phosphatidylcholines (PCs) observed in sepsis patients are attributed to high concentrations of lipoproteins. Because PCs with two fatty acids of C16–C20 are major components of mammalian lipoproteins, we observed PCaaC32–PCaaC36 as the most significant PCs in our samples. Furthermore Drobnik et al. (25) have shown decreased lysophosphatidylcholine/PC and increased ceramide/sphingomyelin ratios in septic patients compared with healthy control subjects. However, although comparison of sepsis patients with healthy control subjects might not be appropriate regarding our hypothesis, this is still in concordance with our results. Again, highly increased PCs seem to be sepsis specific because they are not detectable in SIRS samples without infection compared with ICU control subjects.
Recent data suggest that lipemia in sepsis is not only a reaction of the host to provide energy but is also an integral part of the innate immunity to neutralize bacterial toxins (26) by which the lipid-A moiety of LPS is embedded in the phospholipid layer of lipoproteins. This neutralizing effect has been shown for all lipoproteins in vitro and in vivo (27, 28) and has been attributed to the PC content of lipoproteins.
Together, these data indicate that lipemia in systemic inflammation, particularly the increase in PCs, is a specific host response to infection-induced inflammation (bacteria and their toxins). Because it has been shown that this effect occurs within 2 h after LPS administration and is sustained for at least 24 h (29, 30), medium-chain acylcarnitines and phospholipids might be appropriate biomarkers for differentiation of sepsis and noninfectious SIRS in the early stage of the disease.
It is known that medication can alter metabolism. Therefore, we gathered information on blood products (antithrombin III, packed red blood cells, fresh frozen plasma, prothrombin complex concentrate, and platelet concentrate) and medications such as glucocorticoids (hydrocortisone, methylprednisolone, prednisolone), insulin, and propofol (a hypnotic agent often used in general anesthesia that has been described to influence metabolism), and tested for correlation with C10:1 and PCaaC32:0. None of the aforementioned parameters had a significant influence on those markers (Supplementary Table III and Supplementary Figs. II and III). Also, clinical data that might have confounded the results were tested for correlation with metabolic markers and differences in falsely and correctly classified patients (Supplementary Table II and Supplementary Fig. I). No significant confounders could be detected.
In conclusion, we were able to demonstrate that SIRS and sepsis patients can be distinguished with high sensitivity and specificity by using two metabolic markers (C10:1 and PCaaC32:0). These markers belong to the MCAD intermediates and the most common glycerophospholipids, respectively. In animal models, they have been shown to be responsive to cell wall components of gram-negative and gram-positive bacteria, indicating that they are specific for infection, which we could confirm in our patient cohort. However, the study has several limitations. Concentrations of phosphatidylcholines were determined semiquantitatively to nonphysiological standards. Another limitation is that the sample size is rather small. Although we could exclude that differences in mechanical ventilation, severity of disease, age, or gender confound the findings (data not shown), we cannot out rule that there are other factors that might influence the results. Thus, validation of our markers in independent, prospectively collected larger sample cohorts is mandatory. Considering that mass spectrometry, with analysis times of 5–10 min for determination of 186 metabolites in parallel, is fairly quick, metabolic markers or marker classes as identified in this study may serve as very promising candidates in the differential diagnosis of sepsis and noninfectious SIRS in the clinical routine setting.
Supplementary Material
Acknowledgments
The authors thank the study nurses A. Braune, U. Redlich, and P. Bloos, Dept. of Anesthesiology and Intensive Care Medicine, Jena-University Hospital for sample drawing and collection of clinical data; Dirk Osterloh SIRS-Lab GmbH, Jena, Germany for sample drawing; and J. Koehler, C. Richert, and K. Stoetzer, Department of Clinical Chemistry and Laboratory Medicine, Jena University Hospital, for excellent technical assistance.
Footnotes
Abbreviations:
- AUC
- area under the curve
- FIA
- flow injection analysis
- LPS
- lipopolysaccharide
- MCAD
- medium chain acyl CoA dehydrogenase
- PC
- phosphatidylcholine
- SIRS
- systemic inflammatory response syndrome
The research was supported by the Thuringia Government (grant B-309-00014) and by the Ministry of Thuringia (ProExcellence; PE 108-2); the Thuringian Foundation for Technology, Innovation and Research (STIFT); the German Sepsis Society (GSS); the German Center for Sepsis Control & Care (CSCC); and by the Ministry of Education and Research (BMBF), Grant No. 01 E0 1002.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables and three figures.
REFERENCES
- 1.Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., Pinsky M. R. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29: 1303–1310 [DOI] [PubMed] [Google Scholar]
- 2.Brun-Buisson C. 2000. The epidemiology of the systemic inflammatory response. Intensive Care Med. 26(Suppl 1): S64–S74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone R. C., Balk R. A., Cerra F. B., Dellinger R. P., Fein A. M., Knaus W. A., Schein R. M., Sibbald W. J. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 101: 1644–1655 [DOI] [PubMed] [Google Scholar]
- 4.Reinhart K., Brunkhorst F., Bone H., Gerlach H., Grundling M., Kreymann G., Kujath P., Marggraf G., Mayer K., Meier-Hellmann A., et al. 2006. Diagnosis and therapy of sepsis: guidelines of the German Sepsis Society Inc. and the German Interdisciplinary Society for Intensive and Emergency Medicine (in German) Anaesthesist. 55(Suppl 1): 43–56 [PubMed] [Google Scholar]
- 5.Kumar A., Roberts D., Wood K. E., Light B., Parrillo J. E., Sharma S., Suppes R., Feinstein D., Zanotti S., Taiberg L., et al. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34: 1589–1596 [DOI] [PubMed] [Google Scholar]
- 6.Singh N., Rogers P., Atwood C. W., Wagener M. M., Yu V. L. 2000. Short-course empiric antibiotic therapy for patients with pulmonary infiltrates in the intensive care unit. A proposed solution for indiscriminate antibiotic prescription. Am. J. Respir. Crit. Care Med. 162: 505–511 [DOI] [PubMed] [Google Scholar]
- 7.Pierrakos C., Vincent J. L. 2010. Sepsis biomarkers: a review. Crit. Care. 14: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland A., Thomas M., Brandon R. A., Brandon R. B., Lipman J., Tang B., McLean A., Pascoe R., Price G., Nguyen T., et al. 2011. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit. Care. 15: R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall T. C., Bilku D. K., Al-Leswas D., Horst C., Dennison A. R. 2011. Biomarkers for the differentiation of sepsis and SIRS: the need for the standardisation of diagnostic studies. Ir. J. Med. Sci. 180: 793–798 [DOI] [PubMed] [Google Scholar]
- 10.Meisner M., Tschaikowsky K., Hutzler A., Schick C., Schuttler J. 1998. Postoperative plasma concentrations of procalcitonin after different types of surgery. Intensive Care Med. 24: 680–684 [DOI] [PubMed] [Google Scholar]
- 11.Mimoz O., Benoist J. F., Edouard A. R., Assicot M., Bohuon C., Samii K. 1998. Procalcitonin and C-reactive protein during the early posttraumatic systemic inflammatory response syndrome. Intensive Care Med. 24: 185–188 [DOI] [PubMed] [Google Scholar]
- 12.Mitaka C. 2005. Clinical laboratory differentiation of infectious versus non-infectious systemic inflammatory response syndrome. Clin. Chim. Acta. 351: 17–29 [DOI] [PubMed] [Google Scholar]
- 13.Reith H. B., Mittelkotter U., Debus E. S., Kussner C., Thiede A. 1998. Procalcitonin in early detection of postoperative complications. Dig. Surg. 15: 260–265 [DOI] [PubMed] [Google Scholar]
- 14.Tang B. M., Eslick G. D., Craig J. C., McLean A. S. 2007. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect. Dis. 7: 210–217 [DOI] [PubMed] [Google Scholar]
- 15.Kiehntopf M., Schmerler D., Brunkhorst F. M., Winkler R., Ludewig K., Osterloh D., Bloos F., Reinhart K., Deufel T. 2011. Mass spectometry-based protein patterns in the diagnosis of sepsis/systemic inflammatory response syndrome. Shock. 36: 560–569 [DOI] [PubMed] [Google Scholar]
- 16.Khovidhunkit W., Kim M. S., Memon R. A., Shigenaga J. K., Moser A. H., Feingold K. R., Grunfeld C. 2004. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 45: 1169–1196 [DOI] [PubMed] [Google Scholar]
- 17.Feingold K. R., Moser A., Patzek S. M., Shigenaga J. K., Grunfeld C. 2009. Infection decreases fatty acid oxidation and nuclear hormone receptors in the diaphragm. J. Lipid Res. 50: 2055–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feingold K. R., Wang Y., Moser A., Shigenaga J. K., Grunfeld C. 2008. LPS decreases fatty acid oxidation and nuclear hormone receptors in the kidney. J. Lipid Res. 49: 2179–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maitra U., Chang S., Singh N., Li L. 2009. Molecular mechanism underlying the suppression of lipid oxidation during endotoxemia. Mol. Immunol. 47: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer J. J. 1979. Lipid metabolism in endotoxic shock. Circ. Shock Suppl. 1: 69–79 [PubMed] [Google Scholar]
- 21.Kim M. S., Shigenaga J. K., Moser A. H., Feingold K. R., Grunfeld C. 2005. Suppression of estrogen-related receptor alpha and medium-chain acyl-coenzyme A dehydrogenase in the acute-phase response. J. Lipid Res. 46: 2282–2288 [DOI] [PubMed] [Google Scholar]
- 22.Samra J. S., Summers L. K., Frayn K. N. 1996. Sepsis and fat metabolism. Br. J. Surg. 83: 1186–1196 [PubMed] [Google Scholar]
- 23.Tripp R. J., Tabares A., Wang H., Lanza-Jacoby S. 1993. Altered hepatic production of apolipoproteins B and E in the fasted septic rat: factors in the development of hypertriglyceridemia. J. Surg. Res. 55: 465–472 [DOI] [PubMed] [Google Scholar]
- 24.Wolfe R. R., Shaw J. H., Durkot M. J. 1985. Effect of sepsis on VLDL kinetics: responses in basal state and during glucose infusion. Am. J. Physiol. 248: E732–E740 [DOI] [PubMed] [Google Scholar]
- 25.Drobnik W., Liebisch G., Audebert F. X., Frohlich D., Gluck T., Vogel P., Rothe G., Schmitz G. 2003. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 44: 754–761 [DOI] [PubMed] [Google Scholar]
- 26.Harris H. W., Gosnell J. E., Kumwenda Z. L. 2000. The lipemia of sepsis: triglyceride-rich lipoproteins as agents of innate immunity. J. Endotoxin Res. 6: 421–430 [PubMed] [Google Scholar]
- 27.Cue J. I., DiPiro J. T., Brunner L. J., Doran J. E., Blankenship M. E., Mansberger A. R., Hawkins M. L. 1994. Reconstituted high density lipoprotein inhibits physiologic and tumor necrosis factor alpha responses to lipopolysaccharide in rabbits. Arch. Surg. 129: 193–197 [DOI] [PubMed] [Google Scholar]
- 28.Parker T. S., Levine D. M., Chang J. C., Laxer J., Coffin C. C., Rubin A. L. 1995. Reconstituted high-density lipoprotein neutralizes gram-negative bacterial lipopolysaccharides in human whole blood. Infect. Immun. 63: 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feingold K. R., Grunfeld C. 1987. Tumor necrosis factor-alpha stimulates hepatic lipogenesis in the rat in vivo. J. Clin. Invest. 80: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feingold K. R., Staprans I., Memon R. A., Moser A. H., Shigenaga J. K., Doerrler W., Dinarello C. A., Grunfeld C. 1992. Endotoxin rapidly induces changes in lipid metabolism that produce hypertriglyceridemia: low doses stimulate hepatic triglyceride production while high doses inhibit clearance. J. Lipid Res. 33: 1765–1776 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.