SUMMARY
Lafora disease (LD) is a rare, fatal neurodegenerative disorder characterized by the accumulation of glycogen-like inclusions in the cytoplasm of cells from most tissues of affected patients. 100 years since the first description of these inclusions, the molecular bases underlying the processes involved in LD physiopathology are finally being elucidated. The main cause for the disease relies on the activity of two proteins, the dual specificity phosphatase laforin and the E3-ubiquitin ligase malin, that form a functional complex. Laforin is unique in humans since it is comprised of a carbohydrate binding module attached to a cysteine-based catalytic dual specificity phosphatase domain. Laforin directly dephosphorylates glycogen, but other proteinaceous substrates, if existent, have remained elusive. Recently, an emerging set of laforin binding partners apart from malin have been described, suggestive of laforin roles unrelated to its catalytic activity. Further investigations based on different transgenic mice models have shown that the laforin-malin complex is also involved in other cellular processes such as response to ER stress and misfolded proteins clearance by the lysosomal pathway. However, controversial data and some missing links still make difficult to assess the concrete relationship between glycogen deregulation and neuronal damage leading to the fatal symptoms observed in LD patients, such as myoclonic seizures and epilepsy. Consequently, clinical treatments are far from being achieved. In the present review, we focus on the knowledge of laforin biology not only as a glucan phosphatase, but also as an adaptor protein involved in several physiological pathways.
Keywords: Laforin, malin, glucan phosphatase, Lafora disease, Lafora bodies, glycogen, autophagy, ER stress
INTRODUCTION
The gene encoding the glucan phosphatase laforin is mutated in Lafora progressive myoclonus epilepsy (LD, OMIM 254780). LD is a fatal autosomal recessive neurodegenerative disorder characterized by the presence of progressive neurological deterioration, myoclonus and epilepsy (see [1] and [2] for review). LD initially manifests during adolescence with generalized tonic-clonic seizures, myoclonus, absences, drop attacks and visual hallucinations. As the disease proceeds, patients enter into a vegetative state and eventually die, usually within the first decade from onset of the first symptoms ([1], [3]).
A hallmark of LD is the accumulation of insoluble glucans (i.e. carbohydrates) called Lafora bodies (LBs) ([4], [5]). LBs form in the cytoplasm of cells from most tissues. LBs, like normal glycogen, are composed of glucose residues joined by α-1,4-glycosidic linkages with branches occurring via α-1,6-glycosidic linkages (reviewed in [2]). However, branches in LBs occur less frequently compared to glycogen, and LBs contain increased amounts of phosphate. These properties are shared with amylopectin, the major component of plant starch, and are the reason why LBs and plant starch are water insoluble. LD patients exhibit increased neuronal cell death, number of seizures, and LB accumulations as they age; thus, it is hypothesized that the LBs trigger these symptoms and ultimately the death of the patient [6].
Mutations causing LD have been identified in two genes, EPM2A ([7], [8]) and EPM2B (NHLRC1) [9], and there is evidence for a third locus [10]. EPM2A encodes the glucan phosphatase laforin, a type of dual specificity phosphatase, and EPM2B encodes malin, an E3-ubiquitin ligase ([9], [11], [12]). Laforin prevents Lafora disease by at least two mechanisms: 1) it avoids hyperphosphorylation of glycogen by dephosphorylating it, likely thereby allowing proper glycogen formation, and 2) laforin is an adapter protein and targets proteins to be ubiquitinated by the E3 ubiquitin ligase activity of malin.
Lafora disease was described over 100 years ago [4]. It took almost 90 years to identify the two genes mutated in LD, and 96 years to define biologically relevant substrates of laforin and malin. Our understanding of laforin’s multiple functions sheds insights into the mechanisms causing LD. These advances allow us to now postulate ideas to treat this devastating disease.
EPM2A gene
Laforin is encoded by the 130 Kb four-exon gene EPM2A on chromosome 6q24 of the human genome. It is ubiquitously expressed in all tissues, although brain, skeletal muscle, heart and liver have higher levels of expression [8]. In the brain, laforin is expressed predominantly in cerebellum, hippocampus, frontal cortex and olfactory bulb [13]. Laforin expression increases after birth, reaching a maximum during the adulthood [13].
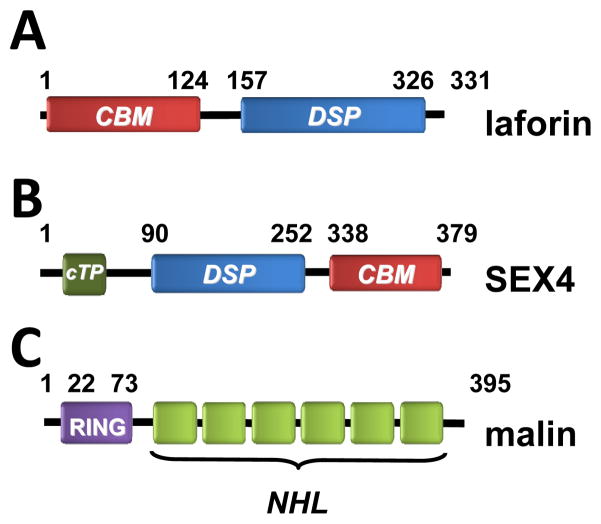
EPM2A encodes a 331 amino acid bi-modular protein with an amino-terminal carbohydrate binding module (CBM, residues 1–124) and a carboxy-terminal dual specificity phosphatase domain (DSP, residues 157–326) (Fig. 1A). Loss-of-function point mutations in either domain result in LD, demonstrating the essential nature of a functional CBM and DSP domains.(a comprehensive meta-analysis of reported mutations can be found in ref. [14]).
Fig. 1.
Schematic depicting of the domains present in human laforin (A), Arabidopsis SEX4 (B) and human malin (C). CBM, carbohydrate binding module; DSP, dual specificity phosphatase domain; cTP, chloroplast targeting peptide; RING, zinc-finger domain involved in E3-ubiquitin ligase activity; NHL, domains involved in protein-protein interaction.
EPM2A alternative splicing results in two laforin isoforms that are identical from amino acid 1–309, but contain a divergent C-terminal domain. Isoform laforin-331 is the most abundant form, possesses phosphatase activity, and when overexpressed in cell culture localizes to the cytoplasm and ER [15]. The minor isoform laforin-317 lacks phosphatase activity and localizes to the cytoplasm and nucleus [15]. Interestingly, Ganesh and colleagues found that the isoforms form heterodimers and that the heterodimers also lack phosphatase activity [15]. These results suggest that laforin-317 may modulate laforin activity by binding laforin-331, then functioning as a dominant negative. A recent study reported three additional isoforms of varying lengths, although the physiological role of these isoforms is still unclear [16].
Domains, biochemical properties, & phylogeny
Carbohydrate binding module (CBM)
CBMs are non-catalytic domains classified into sixty-four families based on evolutionary relationships, polypeptide folds, and substrate preferences according to the Carbohydrate-Active Enzymes (CAZY) database [17]. Proteins containing a CBM utilize the domain to bind carbohydrates and enzymatically modify the carbohydrates with a second domain (e.g. a hydrolase domain) [18]. The laforin CBM belongs to the CBM20 family [19]. CBM20 domains are 90–130 amino acids long. They are one of the most well characterized CBM families, and are characteristic of glucosylhydrolases and glucotransferases from bacteria, fungi, and plants ([18], [20], [21]). CBM20 modules are highly heterogeneous at the amino acid level and lack invariant residues, but contain moderately well-conserved aromatic residues that coordinate ligand binding. The CBM20 module typically consists of seven β-strands that form an open-sided distorted β-barrel with aromatic residues interacting with glucan chains rather than the starch crystalline surface as seen with other CBM families [22]. In the case of laforin, the CBM allows laforin to bind glycogen and LBs as well as plant amylopectin ([19], [23], [24]). Of note, laforin is the only human phosphatase with a CBM present in the same polypeptide chain as the catalytic domain ([25], [26]).
Since no crystal structure of laforin is yet available, we generated a homology model of the carbohydrate binding module of laforin (residues 1–116) using the best available structure [Geobacillus stearothermophilus cyclodextrin glycosyltransferase (PDB: 1CYG)] [27]. The homology model suggests that the laforin CBM folds into the characteristic two β-sheets fold, with the N- and C-termini pointing towards opposite ends of the longest axis of the molecule [28]. Conserved aromatic residues involved in carbohydrate binding are readily observable in the laforin CBM structure: W32, W85 and W99. In the model, these residues form a compact, rigid and surface-exposed hydrophobic site containing inter-ring spacing appropriate for binding to α(1,4)-linked glucoses, as is the case for glycoamylase [28].
Dual specificity phosphatase domain (DSP)
Laforin contains a carboxy-terminal dual specificity phosphatase (DSP) domain. The DSPs belong to the larger protein tyrosine phosphatase (PTP) superfamily of cysteine-dependent phosphatases that encompass around 107 human genes ([25], [26]). PTPs utilize a conserved CX5R motif to hydrolyze phosphoester bonds [26]. The DSP family includes phosphatases that dephosphorylate proteinaceous and/or non-proteinaceous substrates ([26], [29]). Similar to other DSPs, recombinant laforin dephosphorylates the artificial substrates para-nitrophenylphosphate (pNPP) and 3-O-methyl fluorescein phosphate (OMFP) ([30], [31], [32]). As a cysteine-based enzyme, laforin requires a reduced environment to be active and is reversibly inactivated under oxidative conditions [33].
The endogenous substrate for laforin remained elusive for some time, with several laboratories searching for it by targeted approaches and unbiased screening methods. Since laforin contains a CBM and the hallmark of LD is aberrant glycogen, multiple laboratories systematically tested proteins involved in glycogen metabolism as possible laforin substrates. In the end, a multi-system approach revealed that laforin directly dephosphorylates glucans instead of proteins involved in glycogen metabolism and these data established laforin as a glucan phosphatase ([23], [34], [35]) (see below).
Although the DSPs share significantly less amino acid conservation than the classical PTPs, they still retain the characteristic αβα PTP fold. In addition, the DSPs share many of the conserved elements first described for the classical PTPs. These enzymes utilize a cysteine residue at the base of the active site cleft within the PTP loop to perform nucleophilic attack of the phosphorous atom of the substrate. An aspartic acid around 30 amino acids N-terminal of the catalytic cysteine acts as the general acid catalyst, enhancing catalysis. A key difference between the classical PTPs and DSPs is that the classical PTPs possess a deeper active site allowing access to only phospho-tyrosine, whereas the DSPs active site is more shallow to accommodate phospho-serine, -threonine, and –tyrosine ([26], [29]).
The laforin DSP domain was modeled by comparing it with that of the human DUSP22 phosphatase (PDB: 1WRM; [36]) [27]. This in silico approach suggests that the laforin DSP folds into the characteristic αβα PTP fold consisting of four to five β-sheets surrounded by α-helices [37]. In this structure, the PTP loop, containing the catalytic cysteine residue (Cys266), and the conserved Asp residue (Asp235) point towards the catalytic groove. Despite these efforts, a crystal structure of laforin is needed to determine how phospho-glucans are bound by the CBM20 and positioned into the laforin DSP active site.
Laforin dimerization
Recombinant laforin purified from bacteria, laforin from cell culture, and laforin from tissue all form dimers ([38], [39], [40]). However, the domain(s) involved in this event, the mechanism(s) driving dimerization, and the biological function of dimerization are poorly understood. One study reported that laforin forms SDS-resistant dimers both in vitro and in vivo [39]. In addition, it was reported that monomeric laforin is inactive and that all of the phosphatase activity is from dimeric laforin [39]. However, recent studies have challenged this finding by demonstrating that monomeric laforin is the most abundant form of the phosphatase under normal reduced conditions, and that laforin phosphatase activity is similar for both monomer and dimer species [33]. The discrepancy is likely due to the oxidative conditions in the experimental methods, since an oxidative environment drives laforin oligomerization and abolishes laforin phosphatase activity. In the study performed by Liu and colleagues they used little to no reducing agents, and for this reason they concluded that monomeric laforin was inactive [39]. Cumulatively, the new data establish that monomeric and dimeric laforin possess similar phosphatase activity and glucan binding ability, and that dimerization is enhanced by increased oxidation [33]. Despite these findings, no biological role was identified for laforin dimers. Dimerization did not affect phosphatase activity, glucan binding, or binding to other known interacting partners. Thus, a role for laforin-331 homodimers is currently unknown.
Insights from phylogeny
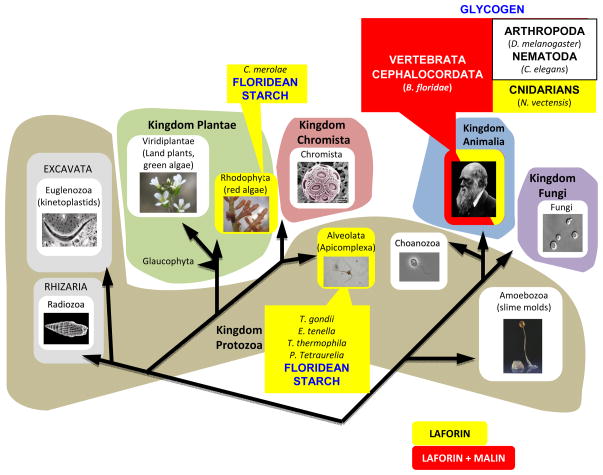
The laforin gene is conserved in all vertebrate genomes, but it is absent from genomes of most non-vertebrate organisms including the standard model organisms yeast, flies, and worms. ([2], [23], [41]) (Fig. 2). Surprisingly, laforin is conserved in the cephalochordate Branchiostoma floridae and in the cnidarian Nematostella vectensis as well as in the following five protozoans Cyanidioschyzon merolae, Toxoplasma gondii, Eimeria tenella, Tetrahymena thermophila, and Plasmodium tetraurelia ([23], [41]). Thus, laforin possesses an ancient and unique evolutionary lineage. Laforin conservation in these five protozoa was a surprising and fortuitous finding. These five organisms all undergo a type of hibernation at some point in their life-cycle and when they “hibernate” they form an insoluble glucan (floridean starch) that resembles a Lafora body. This result provided an additional link to insoluble glucans and thus offered an insight into the biological substrate of laforin.
Fig. 2.
Laforin and malin phylogeny. Schematic view of the presence of laforin (yellow background) or laforin and malin (red background) in the different kingdoms of the eukaryotic tree of life. Groups that do not contain either laforin or malin orthologs are displayed on a white background. In groups containing laforin, malin or both of them, the corresponding organisms are indicated, as well as the type of polyglucosan used as energy source. Note that malin orthologs are only present in organisms that also contain laforin. Modified from [88].
A phylogenetic study of malin was also recently performed. It indicates that malin is present in all vertebrate species and a cephalochordate [42]. When the species distribution of malin was compared with that described for laforin ([23], [41]), it was observed that laforin and malin do not correlate in species distribution (Fig. 2). Since laforin is present in the genome of more evolutionarily basal organisms than malin, these results suggest that laforin may perform additional functions independent of malin. It is possible that these functions are conserved from red algae to humans, but these results indicate that at least in lower eukaryotes laforin possesses malin-independent functions, likely glucan dephosphorylation [42].
Biological function
Glucan phosphatase activity
A single experiment based on unforeseen findings in the literature revealed that laforin is the founding member of a unique class of phosphatases that dephosphorylate phospho-glucans, the glucan phosphatases [34]. In the 1960s, Yokoi, Sakai and colleagues purified and biochemically characterized LBs from brains of LD patients ([6], [43]). They utilized electron-probe microanalysis, by focusing a 1 micron beam of electrons on a LB and they analyzed the wavelength of excited X-rays to determine specific elements within the LB. In a small table as part of a 33 page study, they reported that LBs possess 2–3 fold higher amounts of phosphate compared to glycogen, while other elements were equal in both samples ([6], [43]). While they were unaware of laforin, they did postulate that ester-linked phosphate might explain why amylolytic enzymes are largely unable to degrade LBs [43]). In addition, they surmised that LBs are biochemically more like plant amylopectin than animal glycogen.
Work by Zeeman, Smith and colleagues and the Moorhead lab using Arabidopsis also provided an intriguing clue to the function of laforin. These groups identified a protein similar to laforin in Arabidopsis that contains both a CBM and DSP domain, but the domains are in the opposite orientation as laforin ([44], [45]) (Fig. 1B). In addition, the Zeeman and Smith labs demonstrated that mutation of the gene results in an accumulation of starch and designated the protein as SEX4 (Starch EXcess phenotype 4) [45]). Prior to these data, multiple laboratories had identified glucan water dikinase (GWD) and phosphoglucan water dikinase (PWD) as two Arabidopsis dikinases that phosphorylate the C6 and C3 position of glucose moieties on starch, respectively ([46], [47], [48], [49]).
These findings, along with the finding of laforin in protozoan models, provided the impetus to test laforin as a glucan phosphatase. Initially, laforin was shown to dephosphorylate amylopectin from plant starch ([23], [34]). This result prompted the hypothesis that laforin removes phosphate monoesters from glycogen, allowing glycogen metabolism to proceed normally. In the absence of this activity, glycogen would accumulate more phosphate residues and longer unit chains, due to inhibited branching by the phosphates, and eventually would form an insoluble LB that biochemically resembles amylopectin. The presence of phosphate groups in glycogen was demonstrated back in the 1980s and 1990s ([50], [51]) but up to now, no report on how these phosphates were removed was known. Roach and colleagues confirmed the in vitro dephosphorylation of amylopectin by laforin and also showed that mammalian glycogen was a substrate of this phosphatase [35]. In addition, they demonstrated that glycogen isolated from laforin knockout mice was hyperphosphorylated and developed an abnormal structure ([35], [52]). Cumulatively, these data established laforin as a glucan phosphatase and provide one mechanism for LB formation. A recent paper from the Roach and DePaoli-Roach labs completed the circle by identifying the source of glycogen phosphate. They found that glycogen synthase incorporates the β-phosphate of UDP-glucose (its substrate) at a rate of 1 phosphate/10,000 glucose moieties as C2- and C3-linked monoesters [53]. Thus, one function for laforin is to prevent the enzymatic error mediated by glycogen synthase leading to the phosphorylation of glycogen.
Adapter protein of enzymes involved in glycogen synthesis
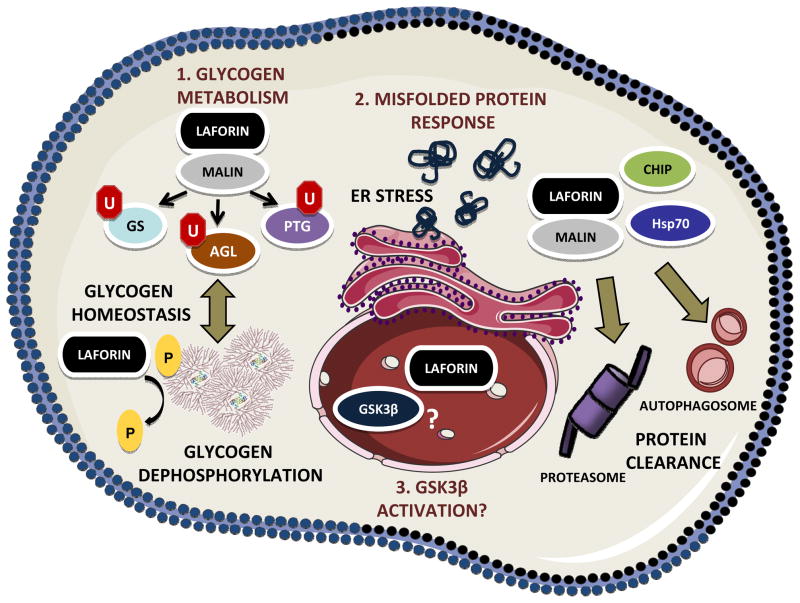
Malin is an E3-ubiquitin ligase that contains an amino-terminal RING domain and six carboxy-terminal NHL domains that are predicted to form a β-propeller type protein interaction domain ([11], [12]) (Fig. 1C). Multiple labs have demonstrated that laforin and malin form a complex and that laforin recruits substrates to be ubiquitinated by malin. These substrates are ubiquitinated by malin in a laforin-dependent manner and many of the substrates are enzymes involved in glycogen synthesis. The laforin-malin complex binds and ubiquitinates the muscle isoform of glycogen synthase [54] and PTG, the glycogen targeting subunit of protein phosphatase type 1 (PP1) ([55], [56]). In these experiments, the laforin-malin complex ubiquitinates the substrate, decreases the substrate protein levels, and downregulates glycogen levels (Fig. 3). The role of laforin as an adapter protein is uncoupled from its role as a glucan phosphatase since a catalytically inactive phosphatase mutant (C266S) still recruits malin and targets it to glycogen related enzymes ([55], [56]). In addition to PTG and glycogen synthase, one report suggests that malin ubiquitinates glycogen debranching enzyme (GDE/AGL) [57]. This report shows that AGL ubiquitination is increased in a wild-type malin-dependent manner when both proteins are overexpressed. Additionally, these investigators demonstrated that increased levels of cAMP increases the interaction between malin and AGL as measured by co-immunoprecipitation and subcellular localization. New information on the contribution of the laforin-malin complex to glycogen regulation has been reported very recently. Jana and colleagues reported that the laforin-malin complex interacts with neuronatin, an 81 amino acid protein that stimulates glycogenesis. The laforin-malin complex ubiquitinates and promotes the proteasomal degradation of neuronatin; therefore, they proposed that in the presence of an inactive laforin-malin complex, neuronatin accumulates and hyperstimulates glycogen synthesis [58].
Fig. 3.
Laforin functions. Schematic view of the different functions of laforin in cell physiology. GS, glycogen synthase; AGL, glycogen debranching enzyme; PTG, protein targeting to glycogen; U, ubiquitin; P, phosphate. See text for details. Figure was produced using Server Medical Art platform.
Many of the conclusions regarding glycogen synthase, PTG and other glycogen related proteins were based on cell culture experiments overexpressing malin, laforin and/or the putative substrate. However, multiple laboratories have recently tested these initial findings under more biologically relevant conditions and they have found conflicting results. In contrast with the cell culture data, 3-month old mice lacking laforin did not show increased levels of glycogen synthase or PTG in muscle or brain extracts [52]. Additionally, two groups found no increase in glycogen synthase, PTG or AGL in malin-deficient mice of 3 to 6 months of age ([59], [60]). However, a third group found dramatically higher levels of glycogen synthase in brain extracts from 11-month old malin-deficient mice compared to wild-type mice [61].
Although protein ubiquitination was first described as a mechanism for targeting proteins for rapid proteasomal degradation, in recent years other functions of ubiquitination have been delineated that are driven by different types of ubiquitination, e.g. monoubiquitination, multi- versus polyubiquitination, and different chain topology ([62], [63]). A recent study reported that the laforin-malin complex produces K63-linked poly-ubiquitin chains in PTG, AMPKα and AMPKβ [64]. These results suggest that the modification introduced by the laforin-malin complex may play a different role from targeting substrates for degradation by the proteasome.
It is possible that under certain circumstances the laforin-malin complex could also promote the formation of K48-linked ubiquitins. This possibility has also been described for parkin, an E3-ubiquitin ligase involved in Parkinson disease that modifies synphilin-1 with both K63- and K48-linked ubiquitin chains [65]. Perhaps this is the reason why the overexpression of laforin and malin promote the proteasomal degradation of PTG [55]. This possibility would reconcile the results obtained in mouse models lacking either laforin or malin, where it has been described that, in spite of having increased levels of glycogen in different tissues (skeletal muscle and brain), there are no differences in either the activity or the protein levels of glycogen synthase or PTG ([35], [59], [60]). Although, the most recent report on this matter indicates that in the brain of 11 month old mice lacking malin, there is an increase in the levels of the muscle glycogen synthase [61]. These results indicate that the age of the mice may prove important, as the previous studies examined younger mice. These latest results suggest that the laforin-malin complex indeed has a role in vivo in downregulating proteins involved in glycogen homeostasis, but more work is needed to define the mechanism of these events. Supporting the possible role of the laforin-malin complex in the regulation of PTG levels, we found that PTG protein levels were also increased in primary fibroblasts from LD patients [66]. Further bolstering PTG as a bona fide substrate is the ability to recapitulate in vitro ubiquitination of PTG by malin in a laforin-dependent manner using purified components [56].
Adapter protein in ER-stress and protein clearance
The laforin-malin complex also plays a role in protecting cells from ER-stress conditions. In cell culture models depleted of laforin there is enhanced sensitivity to agents that trigger ER-stress, e.g. thapsigargin and tunicamicin [67]. In laforin-depleted cells, there is a decrease in proteasome activity and an increase in apoptosis upon drug treatment, as compared to control cells [67]. Similar results regarding protein aggregation induced cell death in malin-depleted cells were also reported [68]. Therefore in the absence of either laforin or malin, there is increased ER-stress response that eventually leads to decreased proteasome function and increased apoptosis, which could be important factors in the development of LD. Strong corroborating evidence for these cell models is that tissue from mice lacking laforin and human LD patients have increased ER stress markers [67] (Fig. 3).
In addition to loss of laforin resulting in enhanced sensitivity to ER stress, laforin itself seems to contribute to ER stress. Overexpressed laforin is prone to aggregate and these aggregates localize in perinuclear aggresome structures that co-localize with ubiquitin, ER-chaperones, and proteasome subunits [69]. We have also observed co-localization of the autophagy maker p62 with these structures, which suggests that they might be labeled for degradation by autophagy (unpublished results). Laforin aggregation is enhanced when some LD mutant forms of laforin are overexpressed and these aggregates also contribute to increased ER-stress response and increased apoptosis [70].
The laforin-malin complex has also been implicated in suppressing cytotoxicity produced by the accumulation of misfolded proteins. Ganesh and coworkers overexpressed aggregate prone proteins and demonstrated that laforin-malin in conjunction with HSP70 degrade the aggregates and protect against cytotoxicity [71]. The laforin-malin complex interacts with misfolded proteins and targets them for degradation by the proteasome. Follow up studies demonstrated that laforin and malin co-immunoprecipitate with the co-chaperone protein CHIP and showed that CHIP stabilizes malin’s tertiary structure ([72], [73]).
Finally, laforin has also been described as a positive regulator of autophagy. In both cellular and mouse models lacking laforin there is decreased autophagy. This decrease is due to impaired formation of autophagosomes that leads to decreased content of autophagic vesicles and lower levels of the LC3-II autophagic marker. The molecular basis of this defect is not known, although it seems that in cells lacking laforin the mTOR pathway is overactivated. The changes in autophagy mediated by the absence of laforin may lead to the accumulation of diverse autophagy substrates that would contribute to cell stress and may contribute to cell death [74] (Fig. 3). Similar defects in autophagy have been recently described in a mouse model lacking malin (Epm2b−/−) [75]. Therefore, autophagy dysfunction is observed in both mouse models of Lafora disease (Epm2a−/− and Emp2b−/−).
Unexpected role: laforin as a tumor suppressor
An unexpected proposed function of laforin is as tumor suppressor. One line of mice expressing SV40 large T antigen and a rearranged T-cell receptor (TCR) developed T cell lymphoma with almost 100% penetrance ([76], [77]). The Zheng laboratory later discovered that in these mice, the TCR disrupted exon 1 of one EPM2A locus and the second locus underwent epigenetic silencing and concluded that laforin could act as a tumor suppressor [77]. The tumor suppression is associated with laforin phosphatase activity, since mice injected with a T lymphoma cell line transduced with a wild type laforin lentiviral vector displayed protection against tumor formation, whereas mice injected with a T-lymphoma cell line transduced with a catalytically inactive laforin (C266S) were not protected [77]. The authors proposed that laforin dephosphorylated p-Ser9-GSK3β and in the absence of laforin GSK3β would accumulate in its inactive phosphorylated form. As GSK3β is a key regulator of the Wnt signaling, the inactivation of GSK3β would lead to the accumulation of β-catenin inside the nucleus, producing an increase in tumorigenesis [77]. In a follow-up study, the authors reported that laforin negatively regulates the cell cycle through dephosphorylation of GSK3β and its regulation of cyclin D1. Lack of laforin results in increased levels of cyclin D1, which promotes cell cycle progression [78] (Fig. 3). Despite convincing data demonstrating that laforin suppresses tumor growth in immunocompromised mice, which strongly links laforin with cell cycle progression, the data that supports laforin as a direct GSK3β phosphatase is controversial. Other laboratories tested GSK3β as a laforin substrate using in vivo and in vitro methods during targeted searches for a substrate, before the glucan phosphatase activity was discovered, and did not observe dephosphorylation of GSK3β by laforin ([34], [35], [79]). Thus, the link between laforin and tumor suppression in immunocompromised mice remains to be elucidated.
Et alii
Although laforin has been definitively shown to be a glucan phosphatase, multiple studies have also found that laforin directly and/or indirectly interacts with many proteins. Multiple techniques have been utilized to identify possible interaction partners and/or putative substrates and these results are summarized in Table I. Many of these interactions have been discussed above, but, apart from malin, it is unclear at this time what the physiological relevance of some of these interactions may be.
Table I.
Laforin interaction partners. Proteins reported to interact with laforin were indicated, along with their biological function and the corresponding identification method. Co-IP, co-immunoprecipitation.
| PROTEIN | FUNCTION | IDENTIFICATION METHOD | REFERENCE |
|---|---|---|---|
| Malin | E3-ubiquitin ligase | Yeast two-hybrid screening | [38] |
| PTG | PPP1R3C Regulatory subunit | Yeast two-hybrid screening | [38] |
| GL | PPP1R3B Regulatory subunit | Co-IP | [34] |
| R6 | PPP1R3D Regulatory subunit | Functional interaction | [56] |
| HIRIP5 | Possibly involved in iron homeostasis | Yeast two-hybrid screening | [85] |
| EPM2AIP1 | Unknown | Yeast two-hybrid screening | [86] |
| GS | Glycogen synthase | Co-IP | [34] |
| GSK3β | Involved in Wnt pathway regulation | Mammalian two-hybrid; Co-IP | [11] |
| AMPKα/β subunits | Cell energy sensor | Yeast two-hybrid Co-IP | [55] |
| TAU | Microtubule-associated protein | Pull-down | [87] |
Controlling/Regulating laforin
While the list of interactive proteins and putative roles of laforin continues to expand, to date only four mechanisms have been described on the regulation of laforin. The first discovery of how laforin is regulated was both surprising and perplexing. Using cell culture models we found that malin binds, ubiquitinates, and targets laforin for degradation; and we were able to recapitulate the ubiquitination using purified components in vitro [12]. This result is surprising given that laforin and malin activity are both necessary to inhibit Lafora body formation and LD. However, malin-directed degradation of laforin has been verified in multiple cell culture systems, mouse models, and data from LD patient tissue ([54], [55], [59], [60], [61], [67]).
An additional mechanism regulating laforin protein levels is directly tied to glycogen stores. Roach and coworkers examined mouse models that accumulate higher or lower levels of glycogen and found that laforin protein levels directly correlate with the amount of glycogen [80]. While this link has been described, a mechanism regulating this fluctuation is currently unknown.
Recently, we demonstrated that laforin physically interacts with the AMPKα and AMPKβ subunits of the heterotrimeric AMP-activated protein kinase (AMPK), a key cellular energy sensor [55]. We found that AMPK is a positive regulator of the laforin-malin complex, since the interaction between laforin and malin is enhanced under conditions of AMPK activation [55]. In a follow-up study, we demonstrated that AMPK phosphorylates laforin at Ser25 both in vivo and in vitro [27]. We found that Ser25 is critical for both laforin phosphatase activity and for its ability to interact with established binding partners, e.g. dimerization with itself, malin, and PTG [27]. These results suggest that laforin-Ser25 phosphorylation by AMPK modulates the laforin-malin interaction and provides a means to regulate their role in glycogen metabolism. However, these data, as with many in the laforin field, are also controversial. Roach and colleagues investigated the levels of PTG in exercised mice, which activate AMPK, and they saw no change in PTG levels [59]. Thus, additional work must be done to determine the role of AMPK in controlling the laforin-malin complex.
The last reported means of regulating laforin activity is via heterodimerization of different splice variants. Ganesh and colleagues characterized different laforin isoforms and found that they display distinct subcellular localization in cell culture ([15], [16]). In addition, they reported that heterodimerization between truncated isoforms and full-length laforin results in a phosphatase inactive complex [15], thus offering a mechanism to regulate laforin function.
LD causes & possible therapeutics
Given the ever-expanding reports of putative laforin functions and laforin-interacting proteins, it is clear that multiple mechanisms drive the progression of Lafora disease. The different lines of data strongly suggest that glycogen phosphorylation, interactions with glycogen metabolism enzymes, and cellular stresses are all intimately involved in disease progression. However, deciphering the intercalated pathways driving these mechanisms will likely take many more years.
One definitive function of laforin is to remove phosphate from glycogen. Failure to remove covalently attached phosphate from glycogen disrupts glycogen organization and results in LB formation. Laforin clearly functions as a glucan phosphatase, but all of the other proposed functions include some level of controversy. It seems likely that laforin participates in other aspects of glycogen metabolism via functioning as an adapter protein for malin-directed ubiquitination of some glycogen metabolism enzymes. If both of these functions are correct then LB formation would result from either lack of laforin glucan phosphatase activity or lack of laforin’s scaffolding ability, with either resulting in LB formation. Once a LB begins to nucleate, it seems probable that the cell would sense a disturbance in its homeostasis and respond with increased unfolded protein response (UPR), ubiquitination, and autophagy (Fig. 3). Since the reoccurring theme in LD is glycogen, it is not surprising that a link between laforin, energy metabolism, and cell cycle progression has been uncovered. How these pathways impact Lafora disease remains to be determined. Despite a lack of consensus regarding many of the proposed pathways that laforin impacts and the proposed laforin-interacting proteins, laforin and LD researchers have made significant strides. These results are allowing researchers to propose and test putative therapeutic paths.
Both the glucan phosphatase activity of laforin and the ability of laforin to act as a scaffold impinge on glycogen metabolism. In addition, if either of these functions falters then one would predict that inhibiting glycogen synthesis might prevent LB formation and if LBs are the causative agent of LD then preventing glycogen synthesis should relieve neuronal cell death and epilepsy. An elegant collaborative study found that depletion of PTG in mice lacking laforin resulted in a down-regulation of glycogen synthesis with near-complete disappearance of Lafora bodies as well as decreased neuronal cell death and myoclonic epilepsy [81], supporting a role for glycogen dysregulation in LD pathogenesis. Thus, removing PTG, an activator of glycogen synthase and inhibitor of glycogen phosphorylase, dramatically reduced the hallmark features of LD and caused no obvious harm to the mice. This study opens the possibility of utilizing chemical inhibitors to disrupt the PTG-glycogen synthase interaction and/or the PTG-glycogen phosphorylase interaction as a means to inhibit glycogen accumulation and disease progression.
A similar line of thinking could be utilized to explore therapeutic options focused on the UPR and autophagy. Since autophagy is impaired in the absence of laforin, the use of different strategies aimed to enhance autophagy could be an interesting therapeutic possibility. Similarly, one could attempt to enhance protein folding through the upregulation of chaperone proteins and/or increase proteasomal activity as a means to turnover misfolded proteins.
Additional experimental therapeutics are still in their infancy. The use of gene therapy to express the EPM2A gene or treatment with Trojan horse liposomes (also called PEGylated immunoposomes) containing the gene of interest are both putative options [82]. In the cases where the disease is produced by nonsense mutations in the EPM2A gene (i.e, R241X, the most frequent mutation in Mediterranean countries), treatment with gentamycin or other aminoglycoside antibiotics that produce read-through of stop codons, could be potentially relevant. The use of these antibiotics would be clinically justified for compassionate use of this fatal disorder similar to its use of cystic fibrosis patients ([1], [83], [84])
In summary, 100 years after the first clinical description of LD, the molecular bases of the disease are beginning to be understood. However, more work is still needed to fully decipher the functions of the two main players in the disease, laforin and malin. With this knowledge, rational therapeutic designs will be proposed that could offer a window of hope to patients suffering from this devastating disease.
Acknowledgments
C.R-M. is supported by a contract from la Fundació La Marató de TV3. This work was supported by a grant from the Spanish Ministry of Education and Science (SAF2011-27442), a grant from la Fundació La Marato de TV3 (ref. 100130) and a grant from Generalitat Valenciana (Prometeo 2009/051) to P.S. and the National Institutes of Health grants R00NS061803, P20RR020171, R01NS070899 and University of Kentucky College of Medicine startup funds to M.S.G.
References
- 1.Delgado-Escueta AV. Advances in lafora progressive myoclonus epilepsy. Curr Neurol Neurosci Reports. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 2.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. Recent advances in the molecular basis of Lafora’s progressive myoclonus epilepsy. J Hum Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
- 4.Lafora GR, Glueck B. Beitrag zur histogpathologie der myoklonischen epilepsie. Gesamte Neurol Psychiatr. 1911;6:1–14. [Google Scholar]
- 5.Collins GH, Cowden RR, Nevis AH. Myoclonus epilepsy with Lafora bodies. An ultrastruc- tural and cytochemical study. ArchPpathol. 1968;86:239–254. [PubMed] [Google Scholar]
- 6.Yokoi S, Austin J, Witmer F, Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol. 1968;19:15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 7.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC, 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 8.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 9.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 10.Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, Minassian BA. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004;63:565–567. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- 11.Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 12.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci U S A. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganesh S, Agarwala KL, Amano K, Suzuki T, Delgado-Escueta AV, Yamakawa K. Regional and developmental expression of Epm2a gene and its evolutionary conservation. Biochem Biophys Res Commun. 2001;283:1046–1053. doi: 10.1006/bbrc.2001.4914. [DOI] [PubMed] [Google Scholar]
- 14.Singh S, Ganesh S. Lafora progressive myoclonus epilepsy: a meta-analysis of reported mutations in the first decade following the discovery of the EPM2A and NHLRC1 genes. Human Mut. 2009;30:715–723. doi: 10.1002/humu.20954. [DOI] [PubMed] [Google Scholar]
- 15.Dubey D, Ganesh S. Modulation of functional properties of laforin phosphatase by alternative splicing reveals a novel mechanism for the EPM2A gene in Lafora progressive myoclonus epilepsy. Hum Mol Genet. 2008;17:3010–3020. doi: 10.1093/hmg/ddn199. [DOI] [PubMed] [Google Scholar]
- 16.Dubey D, Parihar R, Ganesh S. Identification and characterization of novel splice variants of the human EPM2A gene mutated in Lafora progressive myoclonus epilepsy. Genomics. 2011 doi: 10.1016/j.ygeno.2011.10.001. in press. [DOI] [PubMed] [Google Scholar]
- 17.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Stuckey JA, Wishart MJ, Dixon JE. A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J Biol Chem. 2002;277:2377–2380. doi: 10.1074/jbc.C100686200. [DOI] [PubMed] [Google Scholar]
- 20.Janecek S, Sevcik J. The evolution of starch-binding domain. FEBS Lett. 1999;456:119–125. doi: 10.1016/s0014-5793(99)00919-9. [DOI] [PubMed] [Google Scholar]
- 21.Janecek S, Svensson B, Macgregor EA. Structural and evolutionary aspects of two families of non-catalytic domains present in starch and glycogen binding proteins from microbes, plants and animals. Enz Microb Technol. 2011;49:429–440. doi: 10.1016/j.enzmictec.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Machovic M, Svensson B, MacGregor EA, Janecek S. A new clan of CBM families based on bioinformatics of starch-binding domains from families CBM20 and CBM21. FEBS J. 2005;272:5497–5513. doi: 10.1111/j.1742-4658.2005.04942.x. [DOI] [PubMed] [Google Scholar]
- 23.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Delgado-Escueta AV, Yamakawa K. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem Biophys Res Commun. 2004;313:1101–1109. doi: 10.1016/j.bbrc.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 25.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature reviews. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 27.Roma-Mateo C, Solaz-Fuster MC, Gimeno-Alcaniz JV, Dukhande V, Donderis J, Worby CA, Marina A, Criado O, Koller A, Rodriguez de Cordoba S, Gentry MS, Sanz P. Laforin, a dual specificity protein phosphatase involved in Lafora disease, is phosphorylated at Ser25 by AMP-activated protein kinase. Biochem J. 2011;439:265–275. doi: 10.1042/BJ20110150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machovic M, Janecek S. Starch-binding domains in the post-genome era. Cell Mol Life Sci. 2006;63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 30.Ganesh S, Agarwala KL, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado-Escueta AV, Yamakawa K. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum Mol Genet. 2000;9:2251–2261. doi: 10.1093/oxfordjournals.hmg.a018916. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Roach PJ. Glycogen and related polysaccharides inhibit the laforin dual-specificity protein phosphatase. Biochem Biophys Res Commun. 2004;325:726–730. doi: 10.1016/j.bbrc.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 32.Girard JM, Le KH, Lederer F. Molecular characterization of laforin, a dual-specificity protein phosphatase implicated in Lafora disease. Biochimie. 2006;88:1961–1971. doi: 10.1016/j.biochi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Dukhande VV, Rogers DM, Roma-Mateo C, Donderis J, Marina A, Taylor AO, Sanz P, Gentry MS. Laforin, a dual specificity phosphatase involved in lafora disease, is present mainly as monomeric form with full phosphatase activity. PloS one. 2011;6:e24040. doi: 10.1371/journal.pone.0024040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worby CA, Gentry MS, Dixon JE. Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci U S A. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokota T, Nara Y, Kashima A, Matsubara K, Misawa S, Kato R, Sugio S. Crystal structure of human dual specificity phosphatase, JNK stimulatory phosphatase-1, at 1.5 A resolution. Proteins. 2007;66:272–278. doi: 10.1002/prot.21152. [DOI] [PubMed] [Google Scholar]
- 37.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, Garcia-Fojeda B, Medrano-Fernandez I, Gomez-Garre P, Sanz P, Serratosa JM, Rodriguez de Cordoba S. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum Mol Genet. 2003;12:3161–3171. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Wang Y, Wu C, Liu Y, Zheng P. Dimerization of Laforin Is Required for Its Optimal Phosphatase Activity, Regulation of GSK3beta Phosphorylation, and Wnt Signaling. J Biol Chem. 2006;281:34768–34774. doi: 10.1074/jbc.M607778200. [DOI] [PubMed] [Google Scholar]
- 40.Castanheira P, Moreira S, Gama M, Faro C. Escherichia coli expression, refolding and characterization of human laforin. Protein Expr Purif. 2010;71:195–199. doi: 10.1016/j.pep.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glucan metabolism of the organism. BMC Evol Biol. 2009;9:138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roma-Mateo C, Moreno D, Vernia S, Rubio T, Bridges TM, Gentry MS, Sanz P. Lafora disease E3-ubiquitin ligase malin is related to TRIM32 at both the phylogenetic and functional level. BMC Evol Biol. 2011;11:225. doi: 10.1186/1471-2148-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20:160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 44.Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 2006;46:400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- 45.Niittyla T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, Zeeman SC, Smith AM. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 46.Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci U S A. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580:4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 48.Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137:242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 50.Fontana JD. The presence of phosphate in glycogen. FEBS Lett. 1980;109:85–92. doi: 10.1016/0014-5793(80)81317-2. [DOI] [PubMed] [Google Scholar]
- 51.Lomako J, Lomako WM, Whelan WJ, Marchase RB. Glycogen contains phosphodiester groups that can be introduced by UDPglucose: glycogen glucose 1-phosphotransferase. FEBS Lett. 1993;329:263–267. doi: 10.1016/0014-5793(93)80234-l. [DOI] [PubMed] [Google Scholar]
- 52.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, Garcia-Rocha M, Soriano E, Rodriguez de Cordoba S, Guinovart JJ. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nature neuroscience. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 55.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, Aguado C, Knecht E, Serratosa J, Guinovart JJ, Sanz P, Rodriguez de Cordoba S. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17:667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 56.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J Biol Chem. 2008;283:4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21:2399–2409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma J, Rao SN, Shankar SK, Satishchandra P, Jana NR. Lafora disease ubiquitin ligase malin promotes proteasomal degradation of neuronatin and regulates glycogen synthesis. Neurobiol Disease. 2011;44:133–141. doi: 10.1016/j.nbd.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 59.DePaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to lafora bodies and the accumulation of insoluble laforin. J Biol Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turnbull J, Wang P, Girard JM, Ruggieri A, Wang TJ, Draginov AG, Kameka AP, Pencea N, Zhao X, Ackerley CA, Minassian BA. Glycogen hyperphosphorylation underlies lafora body formation. Annals Neurology. 2010;68:925–933. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 61.Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, Gruart A, Guinovart JJ. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Molecular Medicine. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 63.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreno D, Towler MC, Hardie DG, Knecht E, Sanz P. The laforin-malin complex, involved in Lafora disease, promotes the incorporation of K63-linked ubiquitin chains into AMP-activated protein kinase beta subunits. Mol Biol Cell. 2010;21:2578–2588. doi: 10.1091/mbc.E10-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vernia S, Solaz-Fuster MC, Gimeno-Alcaniz JV, Rubio T, Garcia-Haro L, Foretz M, de Cordoba SR, Sanz P. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunit of the R5/PTG-protein phosphatase 1 holoenzyme, and accelerates its down-regulation by the laforin-malin complex. J Biol Chem. 2009;284:8247–8255. doi: 10.1074/jbc.M808492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vernia S, Rubio T, Heredia M, Rodriguez de Cordoba S, Sanz P. Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PloS one. 2009;4:e5907. doi: 10.1371/journal.pone.0005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao SN, Maity R, Sharma J, Dey P, Shankar SK, Satishchandra P, Jana NR. Sequestration of chaperones and proteasome into Lafora bodies and proteasomal dysfunction induced by Lafora disease-associated mutations of malin. Hum Mol Genet. 2010;19:4726–4734. doi: 10.1093/hmg/ddq407. [DOI] [PubMed] [Google Scholar]
- 69.Mittal S, Dubey D, Yamakawa K, Ganesh S. Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet. 2007;16:753–762. doi: 10.1093/hmg/ddm006. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Wang Y, Wu C, Liu Y, Zheng P. Deletions and missense mutations of EPM2A exacerbate unfolded protein response and apoptosis of neuronal cells induced by endoplasm reticulum stress. Hum Mol Genet. 2009;18:2622–2631. doi: 10.1093/hmg/ddp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garyali P, Siwach P, Singh PK, Puri R, Mittal S, Sengupta S, Parihar R, Ganesh S. The malin-laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system. Hum Mol Genet. 2009;18:688–700. doi: 10.1093/hmg/ddn398. [DOI] [PubMed] [Google Scholar]
- 72.Rao SN, Sharma J, Maity R, Jana NR. Co-chaperone CHIP stabilizes aggregate-prone malin, a ubiquitin ligase mutated in Lafora disease. J Biol Chem. 2010;285:1404–1413. doi: 10.1074/jbc.M109.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sengupta S, Badhwar I, Upadhyay M, Singh S, Ganesh S. Malin and laforin are essential components of a protein complex that protects cells from thermal stress. J Cell Sci. 2011;124:2277–2286. doi: 10.1242/jcs.082800. [DOI] [PubMed] [Google Scholar]
- 74.Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, Juana-Lopez L, Dominguez M, Navarro C, Serratosa JM, Sanchez M, Sanz P, Bovolenta P, Knecht E, Rodriguez de Cordoba S. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet. 2012 doi: 10.1093/hmg/ddr590. ahead of print. [DOI] [PubMed] [Google Scholar]
- 76.Geiger T, Gooding LR, Flavell RA. T-cell responsiveness to an oncogenic peripheral protein and spontaneous autoimmunity in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:2985–2989. doi: 10.1073/pnas.89.7.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Liu Y, Wu C, Zhang H, Zheng X, Zheng Z, Geiger TL, Nuovo GJ, Liu Y, Zheng P. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer Cell. 2006;10:179–190. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 78.Liu R, Wang L, Chen C, Liu Y, Zhou P, Wang Y, Wang X, Turnbull J, Minassian BA, Liu Y, Zheng P. Laforin negatively regulates cell cycle progression through glycogen synthase kinase 3beta-dependent mechanisms. Mol Cell Biol. 2008;28:7236–7244. doi: 10.1128/MCB.01334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang W, Lohi H, Skurat AV, DePaoli-Roach AA, Minassian BA, Roach PJ. Glycogen metabolism in tissues from a mouse model of Lafora disease. Arch Biochem Biophys. 2007;457:264–269. doi: 10.1016/j.abb.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Parker GE, Skurat AV, Raben N, DePaoli-Roach AA, Roach PJ. Relationship between glycogen accumulation and the laforin dual specificity phosphatase. Biochem Biophys Res Commun. 2006;350:588–592. doi: 10.1016/j.bbrc.2006.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turnbull J, DePaoli-Roach AA, Zhao X, Cortez MA, Pencea N, Tiberia E, Piliguian M, Roach PJ, Wang P, Ackerley CA, Minassian BA. PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS Genetics. 2011;7:e1002037. doi: 10.1371/journal.pgen.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pardridge WM. Preparation of Trojan horse liposomes (THLs) for gene transfer across the blood-brain barrier. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5407. pdb prot5407. [DOI] [PubMed] [Google Scholar]
- 83.Howard M, Frizzell RA, Bedwell DM. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat Med. 1996;2:467–469. doi: 10.1038/nm0496-467. [DOI] [PubMed] [Google Scholar]
- 84.Delgado-Escueta AV, Bourgeois BF. Debate: Does genetic information in humans help us treat patients? PRO--genetic information in humans helps us treat patients. CON--genetic information does not help at all. Epilepsia. 2008;49:13–24. doi: 10.1111/j.1528-1167.2008.01922.x. [DOI] [PubMed] [Google Scholar]
- 85.Ganesh S, Tsurutani N, Suzuki T, Ueda K, Agarwala KL, Osada H, Delgado-Escueta AV, Yamakawa K. The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Genet. 2003;12:2359–2368. doi: 10.1093/hmg/ddg253. [DOI] [PubMed] [Google Scholar]
- 86.Ianzano L, Zhao XC, Minassian BA, Scherer SW. Identification of a novel protein interacting with laforin, the EPM2a progressive myoclonus epilepsy gene product. Genomics. 2003;81:579–587. doi: 10.1016/s0888-7543(03)00094-6. [DOI] [PubMed] [Google Scholar]
- 87.Puri R, Suzuki T, Yamakawa K, Ganesh S. Hyperphosphorylation and aggregation of Tau in laforin-deficient mice, an animal model for Lafora disease. J Biol Chem. 2009;284:22657–22663. doi: 10.1074/jbc.M109.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cavalier-Smith T. Only six kingdoms of life. Proceedings. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]