Abstract
A disadvantage of 3D isotropic acquisition in whole-heart coronary MRI is the prolonged data acquisition time. Isotropic 3D radial trajectories allow undersampling of k-space data in all three spatial dimensions, enabling accelerated acquisition of the volumetric data. Compressed sensing (CS) reconstruction can provide further acceleration in the acquisition by removing the incoherent artifacts due to undersampling and improving the image quality. However, the heavy computational overhead of the CS reconstruction has been a limiting factor for its application. In this paper, a parallelized implementation of an iterative CS reconstruction method for 3D radial acquisitions using a commercial graphics processing unit (GPU) is presented. The execution time of the GPU-implemented CS reconstruction was compared with that of the C++ implementation and the efficacy of the undersampled 3D radial acquisition with CS reconstruction was investigated in both phantom and whole-heart coronary data sets. Subsequently, the efficacy of CS in suppressing streaking artifacts in 3D whole-heart coronary MRI with 3D radial imaging and its convergence properties were studied. The CS reconstruction provides improved image quality (in terms of vessel sharpness and suppression of noise-like artifacts) compared with the conventional 3D gridding algorithm and the GPU implementation greatly reduces the execution time of CS reconstruction yielding 34–54 times speed-up compared with C++ implementation.
Keywords: compressed sensing, accelerated imaging, GPU implementation, 3D radial acquisition, cardiac MR
INTRODUCTION
Cardiac MR (CMR) data is typically acquired using multiple two-dimensional (2D) slices. Imaging using a single large three-dimensional (3D) slab covering the whole heart from the base to the apex can significantly simplify image prescription. Whole-heart coronary MRI, analogous to coronary multi-detector CT, has replaced multiple small-slab targeted acquisitions for the individual coronary arteries (1–3). A single breath-hold accelerated 3D cine scan has been previously investigated for evaluation of cardiac function (4–6). Free-breathing, 3D late gadolinium enhancement (LGE) imaging has been used to identify fibrosis/scar with improved spatial resolution or coverage (7,8). Recently, 3D perfusion has also been applied to improve spatial coverage (9,10). The advantages of a 3D acquisition include superior spatial resolution, especially through-plane, ease of image prescription, superior signal-to-noise ratio (SNR) and easy reformatting of the image in any desired plane. However, one major disadvantage of 3D imaging is the long data acquisition time. For coronary MRI, a longer scan time usually makes the scan more susceptible to respiratory motion. For LGE, it results in imaging artifacts due to changes in optimal inversion time as the contrast washes out. For cine and perfusion, it typically results in lower temporal or spatial resolution. Therefore, methods to reduce scan time in 3D imaging could significantly improve the clinical utilization of 3D CMR.
3D whole-heart data is commonly acquired using Cartesian k-space sampling, however non-Cartesian sampling schemes, e.g. radial or spiral, have better data acquisition efficiency (11,12). Both 3D stack-of-radials and 3D radial (kooshball) acquisitions with isotropic spatial resolution have been previously used in 3D CMR (11,13,14). In these sampling schemes, a Nyquist sampling rate is not necessary because undersampling does not yield distinct fold-over artifacts; instead, it typically results in streaking artifacts. This allows high undersampling rates with less pronounced imaging artifacts compared to Cartesian acquisitions at the same sampling density. These potential benefits have been previously exploited to achieve whole-heart coronary MRI with isotropic spatial resolution (15). It has also been extensively investigated to improve dynamic imaging such as phase contrast, MR angiography, and cine imaging (16–18).
For single-phase anatomical imaging such as coronary MRI, a gridding algorithm is commonly used in the reconstruction of 3D radial acquisitions (19). Although the gridding algorithm can efficiently reconstruct data acquired using a 3D radial trajectory, its performance deteriorates significantly for highly undersampled data due to significant undersampling of outer k-space regions (20). Parallel imaging methods including SENSE (21) and GRAPPA (22) have been previously applied for 2D radial acquisitions to reduce streaking artifacts (23,24). Recently, compressed sensing (CS) has been applied to remove streaking artifacts for 2D radial acquisitions (20,25). In this approach, additional constraints based on image properties are used to improve the image reconstruction. The CS reconstruction techniques are usually implemented with iterative procedures that solve the optimization problem with relatively computationally cheap matrix-vector multiplications (26). The CS reconstruction for 3D radial acquisition has been recently demonstrated for imaging of the hand using 512 radial profiles on a matrix size of 1283 (27). The computational overhead of the iterative CS reconstruction increases as the size of the 3D k-space increases, resulting in prolonged reconstruction time.
Recently, graphics processing units (GPUs) have become available for high computation-intensive applications. Hardware manufacturers provide parallel computing architectures (such as CUDA and FireStream) that enable researchers to implement GPU programs using high-level programming languages without knowledge of the GPU hardware structure. Recent studies have shown that GPU-accelerated reconstructions can be used to achieve reduced and low-latency reconstruction times for various MR applications (28–30). GPU-accelerated reconstructions for 2D radial acquisitions were demonstrated with approximately 6–32 times speed-up in reconstruction time compared to central processing unit (CPU) implementations (28,31). GPU implementations have been shown to greatly accelerate CS reconstructions of 3D non-Cartesian trajectories such as stack-of-radials and stack-of-spirals (30,32), where the k-space samples are equidistantly spaced along one k-space dimension. However, GPU implementation for a true 3D non-Cartesian trajectory such as 3D radial sampling is not straightforward from those for stack-of-radials/spirals trajectories and has not been previously reported due to the large size of the 3D sampling data and GPU hardware limitations.
In this paper, we propose to implement and evaluate the performance of a GPU-accelerated reconstruction for 3D radial reconstruction using the latest commercially-available GPU hardware. Subsequently, we will investigate the efficacy of a 3D radial acquisition with CS reconstruction for whole-heart coronary MRI.
MATERIALS AND METHODS
All phantom and volunteer data were obtained using 1.5T Achieva magnet (Philips Healthcare, Best, The Netherlands) with a 5 channel phased-array coil. The acquired MR data were transferred to a stand-alone computer and the image reconstruction was performed off-line. All in vivo studies were approved by our institutional review board and all subjects provided consent prior to participation in the study.
3D Radial Acquisition and Reconstruction
In this section, we will review and present the formulation for 3D radial image acquisition and reconstruction using CS. The 3D radial sampling trajectory consists of Ni interleaves, where each interleaf has Np projection lines with Ns sample points (15). Each interleaf is the rotated version of the first interleaf around the kz-axis. The isotropy (or uniformity) of the sampling point distribution can be quantified by the standard deviation of the distance between adjacent sampling points on the k-space sphere, and is kept at less than 10% of the mean distance when the total number of projections Np×Ni is between 100 and 10,000 (33). The sampling density of a 3D radial acquisition is defined as the ratio of the total number of k-space samples of the 3D radial acquisition over that of a Nyquist-sampled 3D Cartesian acquisition with the same resolution and the same field-of-view (FOV). A gridding algorithm (19) is commonly used to reconstruct 3D radial data. In the conventional gridding algorithm, each data point is compensated for its non-uniform sampling density by the density compensation function (DCF) which is calculated based on the sampling trajectory (34–36). The data point is convolved with a gridding kernel and re-sampled onto the Cartesian grid. The re-gridded k-space samples are then inverse Fourier transformed to obtain the desired image. De-apodization is performed after the inverse Fourier transform by dividing the image by the apodization function which is given by the Fourier transform of the gridding kernel function (19). Since all of the operations are linear, this procedure can be expressed in a matrix-vector format:
| [1] |
where x̂ is the reconstructed image, y is the measured 3D radial k-space data, P is a diagonal matrix performing the density compensation, S denotes the convolution matrix for the gridding operator, F* denotes the inverse fast Fourier transform (IFFT), and D is a diagonal matrix performing the de-apodization. We note that all the voxels of the 3D image are represented in a single column vector x̂ for mathematical convenience.
As an alternative approach, the acquired k-space signals can be formulated in an encoding matrix format as y= Ax, where A denotes the encoding matrix and x denotes the actual image. A can be considered as taking the reverse steps of the conventional gridding algorithm without the density compensation:
| [2] |
where D is a diagonal matrix performing the de-apodization, F denotes the fast Fourier transform (FFT) matrix, and S* denotes convolution matrix from Cartesian to radial sample points. x is de-apodized and Fourier transformed into the k-space, then the Cartesian k-space samples are re-gridded onto the 3D radial sample points using the gridding kernel. Unlike the conventional gridding algorithm, the density compensation is not required before the re-gridding because the density of the Cartesian grid is uniform (37). Equation [2] holds regardless of the Nyquist criterion, but the encoding matrix is not invertible for undersampled data, since Eq. [2] is underdetermined and there are multiple solutions that will satisfy the system equation. CS reconstruction utilizes the sparsity of the image to reconstruct the undersampled data using a constrained minimization problem:
| [3] |
where λ is a regularization parameter which determines the tradeoff between the data consistency and the sparsity level of the image, ||·||p denotes the lp norm of the vector which is defined by , and Ψ is a sparsifying transform matrix such as a wavelet transform or total variation (TV) operator.
To solve Eq. [3], we adopt an iterative method which alternately enforces the data consistency and sparsity of the image estimate at each iteration (38). The image update at the (t+1)-th iteration is given by solving the following two sub-problems:
| [4] |
and
| [5] |
Equation [4] is called the data consistency step as the solution tends to decrease the l2-norm error between the measured data and the k-space of the image estimate. For any unitary sparsifying transform Ψ, Eq. [5] can be re-expressed with respect to the transform domain vector zt= Ψxt as
| [6] |
Equation [6] can be solved by a simple coefficient-wise thresholding function as:
| [7] |
where and denote the i-th coefficient of the transform domain vector zt and wt= Ψut of the solution of the first sub problem in Eq. [4], respectively. The second sub problem is called the thresholding step. For αt, we adopt the step size from (39), where αt is determined so that αtI approximates the Hessian of the data consistency term as below:
| [8] |
The overall iterative reconstruction procedure is summarized in Figure 1. The reconstruction starts from an initial image estimate, which in our experiments was chosen to be the gridding reconstruction. The image is de-apodized, Fourier transformed into k-space, and then re-gridded onto the radial sample points. The estimated radial samples are subtracted from the actual measurement data, convolved onto the Cartesian k-space grid, inverse Fourier transformed and an image estimate is obtained after de-apodization. The image estimate is combined with the intermediate image from the previous iteration. The combined image is then thresholded in the transform domain to produce a new image estimate, and the intermediate image is updated. The final image estimate is obtained as the result of the iterative procedures.
Figure 1.
3D radial reconstruction using compressed sensing. The iterative process consists of two steps of data consistency and thresholding. The image is updated to reduce the l2-norm error between the measured data and the k-space of the image estimate in the data consistency step, and to enforce the sparsity of the image estimate in the transform domain in the thresholding step. The final image is obtained as the result of the iterative process.
GPU-Accelerated CS Reconstruction for 3D Radial Trajectory
The computational burden of a 3D radial trajectory with CS reconstruction is a major drawback and its feasibility has not been studied in the literature. In this section, we will present our implementation of a GPU-based reconstruction of a 3D radial acquisition that allows us to further explore the utility of this reconstruction for 3D whole-heart CMR.
The reconstruction algorithm in this paper was implemented using an NVIDIA (Santa Clara, CA) graphics card and parallel computing architecture, CUDA (Compute Unified Device Architecture). The CUDA program consists of two parts: host code that is executed on the CPU and device code that is executed on the GPU. The code which has little or no parallelism in computation is written in host code using ANSI C language, and the code which has a large amount of parallelism in computation is written in device code using a slightly modified C-like language. The functions written in the device code are called kernels, and each kernel generates a large number of threads as a result of data parallelism once the kernel is invoked. All the threads generated by a kernel invocation are called a grid. The threads in a grid are grouped into blocks, which are the basic allocation unit for the execution resources on the hardware. All the blocks in the same grid must have the same number of threads.
The gridding and re-gridding operations are the most computationally intensive part of the iterative CS reconstruction. Since the width of the convolution window is much smaller than the size of the entire k-space, the gridding/re-gridding can be performed in a parallel manner for each measured radial point, and are well-suited for CUDA implementation. In this paper, we assigned each 3D radial data point to one CUDA thread. Each projection line corresponds to one block, which consists of Ns threads. The grid has a 2D block structure (Np, Ni) to represent all the projection lines and interleaves of the 3D radial trajectory. Figure 2 shows a simplified example of a grid hierarchy and thread assignment of our implementation, where we have 8 sample points in one projection, 3 projection lines per interleaf, and 2 interleaves. In the gridding operation, contributions from adjacent radial samples are accumulated to a Cartesian sample point as illustrated in Figure 3(a), which results in cumulative memory writes during the parallelized execution. The cumulative memory writes can produce incorrect results if more than two threads try to access the same memory simultaneously. This is prevented using CUDA’s atomic operation, which is capable of reading and writing on a memory address without interruption by other threads, allowing concurrent threads to correctly perform the required memory access. The performance of atomic operation in CUDA is greatly improved on recent “Fermi”-based GPUs offered by NVIDIA, which provide up to 20 times faster atomic operation compared to the their former generation GPUs (40).
Figure 2.
CUDA grid hierarchy and thread assignment: A grid, which consists of multiple threads, is generated once the device kernel is invoked. Each projection line of the 3D radial trajectory is assigned to one block of threads. Each thread in a block corresponds to a 3D radial sample point in the same projection line. The total number of projections is equal to the total number of blocks. This example shows a thread assignment of a 3D radial trajectory with (Ns, Np, Ni) = (8, 3, 2).
Figure 3.
Thread assignment strategies for implementation of a gridding algorithm in CUDA programming: (a) radial point driven assignment, (b) Cartesian point driven assignment. Cumulative memory writes can be observed in the radial point driven assignment. The central grid point has a larger workload than the outer grid point in the Cartesian point driven assignment.
Besides the gridding/re-gridding operations, most of the CS reconstruction procedures including FFT/IFFT, wavelet/inverse-wavelet transforms, de-apodization and thresholding were parallelized and written in device code. cuFFT and cuBLAS packages were used for FFT/IFFT and other arithmetic operations. Due to the limited global memory size of current GPU hardware, we could not parallelize the reconstruction for the multiple coil elements. The reconstruction was performed sequentially for each coil and the final reconstructed image was obtained as the root-sum-square of the individual coil images. The CS reconstruction was also implemented in standard C++ environment for the comparison of the reconstruction time. The FFTw package (41) was used for FFT/IFFT operations. The GPU and C++ implementations of the CS reconstruction were based on single precision floating point arithmetic, and they were executed on a PC with Intel (Santa Clara, CA) Core2 Quad Q9400 CPU (2.66 GHz), 8.0 GB memory, and NVIDIA GeForce GTX 480 Graphics card (480 cores, 1.5 GB memory) running on a 64 bit Windows 7 operating system.
Phantom Study
Two experiments were performed for the phantom study. The first experiment is to demonstrate the capability of improving the reconstruction quality of 3D radial acquisitions using the CS reconstruction method. The second experiment is to investigate the convergence properties of the CS reconstruction method over different numbers of iterations.
For the first experiment, a high resolution phantom was scanned with a steady-state free precession (SSFP) sequence using 3D radial trajectories with Ns = 344 and Ni = 10 for five different sampling densities 7.5, 10, 20, 30, 40% and 100%, which correspond to the number of projections per interleaf Np of 221, 289, 576, 896, 1184 and 2954, respectively. The scan parameters were TR/TE/α = 3.90/1.94/60°, FOV = 240×240×240 mm3, spatial resolution = 1.4×1.4×1.4 mm3. The acquired 3D radial data were reconstructed using the iterative CS reconstruction method and the conventional 3D gridding algorithm with density compensation, and the reconstructed images were compared. We used both the identity transform and Daubechies 4 (42) discrete wavelet transform (DWT) for the sparsity regularization term of the CS reconstruction. We varied the regularization parameter, λ, from 0.01||A*y||∞ to 0.1||A*y||∞ as in (39,43) and manually selected it to get the best image qualities; λ=0.05||A*y||∞ gave satisfactory results for most of the cases with both sparsity regularizations. The DCF for the gridding algorithm was calculated using the iterative procedure proposed in (34). For both the CS reconstruction and the gridding algorithm, a Kaiser-Bessel function with window size 4.0 was used for the convolution kernel (44).
For the second experiment on convergence properties of the reconstruction algorithm, iterative CS reconstructions with both image and wavelet domain regularizations were performed on the phantom data set with 7.5% sampling density and the intermediate images were stored for different numbers of iterations.
Whole-Heart Coronary MRI
Whole-heart coronary MR images were acquired on 9 healthy volunteers (2 male, 26±11 years). 3D free-breathing ECG-triggered SSFP sequences were used for imaging the heart with 3D radial trajectories. A respiratory navigator with 7 mm gating window was used for gating and tracking the respiratory motion (45). The k-space data acquired within the gating window were accepted, and the k-space data acquired outside the gating window were rejected and re-acquired until acquired within the gating window. Within the 7 mm gating window, the position of the imaging volume was adaptively adjusted using a tracking factor of 0.6. The data sets were acquired with Ns = 392 and Ni = 10 for various sampling densities: two data sets with 6.8%, 12.1%, 24.2% and 36.3%, seven data sets with 10%, 20%, 30% and 40%. The scan parameters were as follows: TR/TE/α = 3.9/1.9/60°, FOV = 256×256×256 mm3, spatial resolution = 1.3×1.3×1.3 mm3. The nominal scan time for the data set with sampling density of 40% was reported to be 5 minutes 13 seconds assuming 100% navigator efficiency. For one volunteer, an additional scan with spatial resolution of 1.0×1.0×1.0 mm3 and sampling density of 40% was acquired. The acquired 3D radial data were reconstructed by the three reconstruction methods (i.e. gridding, CS with image domain regularization and CS with wavelet domain regularization), and the reconstructed image quality was compared. We used λ=0.05||A*y||∞ as the regularization parameter for both sparsity regularizations. The DCF for the gridding algorithm was calculated by the same method used in the phantom study and the Kaiser-Bessel function with window size 4.0 was used for the convolution kernel.
The empirical convergence properties of the CS reconstructions were also observed similar to the phantom study. The vessel sharpness and the vessel length of the right coronary artery (RCA) were measured using Soap-Bubble software (46) for quantitative assessment of the quality of the CS reconstruction method. The vessel sharpness is measured using a Deriche algorithm (47) as previously described (48), where vessel sharpness of 1.0 refers to a maximum signal intensity change at the vessel border. The sharpness and the length of the vessels with CS reconstruction were compared with the gridding algorithm using a paired t-test. A P value less than 0.05 was considered to be statistically significant.
RESULTS
GPU Implementation of the CS Reconstruction
Table 1 shows the average time required for the completion of one iteration of the iterative CS reconstruction with CUDA and C++ implementations. The reconstruction was performed on the in vivo data for 4 different sampling densities (10%, 20%, 30% and 40%), which correspond to the sampling parameters (Ns, Np, Ni) = (392, 396, 10), (392, 768, 10), (392, 1152, 10), and (392, 1536, 10), respectively. The measured time is averaged over 100 iterations. The most time-consuming parts of the C++ implementation are the gridding and re-gridding operations, amounting to 67.1%, 79.5%, 85.3% and 88.5% of the total reconstruction time for 10%, 20%, 30% and 40% sampling densities, respectively. The speed-up gains of the GPU implementation over the C++ implementation are also the largest for the gridding and re-gridding operations: 56.5 ~ 58.8 times speed-up for the gridding operation and 111.5 ~ 111.8 speed-up for the re-gridding operation. As the proportion of the gridding and re-gridding operations in the total reconstruction time of the C++ implementation increases, the total speed-up gain of the GPU implementation increases as well. Overall, the speed-up of the CUDA implementation of the CS reconstruction with image domain regularization was 34.3, 43.7, 50.2 and 53.9 for 10%, 20%, 30% and 40% sampling densities, respectively. The speed-up of the CS reconstruction with the wavelet domain regularization was 35.4, 42.7, 48.4 and 51.9 for 10%, 20%, 30% and 40% sampling densities, respectively. The execution time of the gridding operation was about twice as long as the execution time of the re-gridding operation in CUDA implementation for a given sampling density, while the execution time of the gridding and re-gridding operations were nearly the same for C++ implementation. The gridding operation in CUDA is hampered by the cumulative memory writes, which is not present in the re-gridding operation; this results in an increased execution time even if gridding and re-gridding operations have the same thread configuration. The execution time of FFT/IFFT was kept almost constant over different sampling densities for both CUDA and C++ implementations, as the size of reconstruction matrix was the same for all data sets (392×392×392)
Table 1.
Average time (s) required for performing main operations in one iteration of the CS reconstruction for each coil with CUDA and C++ implementations for a 3D radial data of size (Ns = # sample, Np = # projection, Ni = # interleaves) and associated speed-up (SU). (DWT: Discrete Wavelet Transform, IDWT: inverse Discrete Wavelet Transform.)
| (Ns, Np, Ni) | (392, 396, 10) | (392, 768, 10) | (392, 1152, 10) | (392, 1536, 10) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CUDA | C++ | SU | CUDA | C++ | SU | CUDA | C++ | SU | CUDA | C++ | SU | |
| FFT | 0.27s | 5.00s | 18.5 | 0.26s | 5.01s | 18.7 | 0.27s | 4.99s | 18.5 | 0.27s | 4.99s | 18.2 |
| IFFT | 0.27s | 5.04s | 18.6 | 0.26s | 5.06s | 18.8 | 0.27s | 5.04s | 18.7 | 0.27s | 5.06s | 18.6 |
| Gridding | 0.31s | 17.59s | 56.5 | 0.58s | 34.04s | 58.1 | 0.86s | 51.00s | 58.7 | 1.15s | 67.84s | 58.8 |
| Re-gridding | 0.15s | 17.64s | 111.5 | 0.30s | 34.12s | 111.8 | 0.45s | 51.11s | 111.8 | 0.60s | 67.99s | 111.6 |
| Thresholding | 0.01s | 1.10s | 69.1 | 0.01s | 1.10s | 68.6 | 0.01s | 1.10s | 68.7 | 0.01s | 1.09s | 67.1 |
| Etc. | 0.50s | 6.07s | 0.51s | 6.39s | 0.50s | 6.41s | 0.52s | 6.45s | ||||
| Total (cs-image) | 1.52s | 52.48s | 34.3 | 1.96s | 85.74s | 43.7 | 2.38s | 119.68s | 50.2 | 2.84s | 153.44s | 53.9 |
| DWT | 0.24s | 8.51s | 35.5 | 0.24s | 8.51s | 35.5 | 0.24s | 8.53s | 35.5 | 0.24s | 8.51s | 35.5 |
| IDWT | 0.21s | 8.72s | 41.5 | 0.21s | 8.74s | 41.6 | 0.21s | 8.74s | 41.6 | 0.21s | 8.74s | 41.6 |
| Total (cs-wavelet) | 1.97s | 69.71s | 35.4 | 2.41s | 102.99s | 42.7 | 2.83s | 136.95s | 48.4 | 3.29s | 170.69s | 51.9 |
The total execution time of the CS reconstruction with image domain regularization for 20% sampled data is 85.74 seconds in C++ implementation and 1.96 seconds in CUDA implementation, yielding 43.7 times speed-up. With a 5 channel phased-array coil and 1000 iterations, the reconstruction of a 3D radial acquisition will take around 5 days in C++ implementation, while it takes around two and a half hours in CUDA implementation. The images reconstructed with the CUDA implementation were visually identical to those reconstructed with the C++ implementation and the normalized mean-squared errors between the two reconstructions were kept less than 10−5 for tested 3D radial datasets.
Phantom Experiment
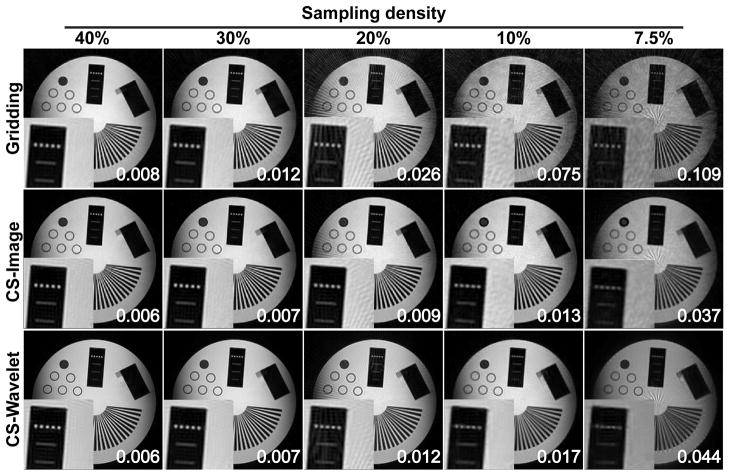
Figure 4 shows the reconstruction results of an example slice of the 3D radial acquisition using the aforementioned algorithms with different sampling densities of 7.5%, 10%, 20%, 30%, and 40%. At the bottom left of each image, a selected region of the phantom is shown at a larger scale. The normalized mean-squared error (MSE) from the reference image with 100% sampling density is also included at the bottom right of each image, calculated as , where xref denotes the reference image from 100% sampled k-space data and xunder denotes the reconstructed image from the undersampled k-space data. Both of the CS reconstructions show improved image quality compared with the conventional gridding reconstruction, and the improvement is more distinct with lower sampling densities. The streaking artifacts degrade the image quality of the conventional gridding reconstructions for lower sampling densities (20, 10, and 7.5%), while most of the streaking artifacts are removed on the CS reconstructed images for the same sampling densities. Overall, the CS reconstructions have less visible artifacts and improved image homogeneity compared with the gridding reconstructions. In particular, the image domain regularization provides better image quality at sharp edges, while the wavelet domain regularization is generally better at removing streaking artifacts. The CS reconstruction with image domain regularization provides the least normalized MSE values at all sampling densities.
Figure 4.
Comparison of conventional 3D gridding reconstruction vs. 3D iterative CS reconstruction with different sparsity regularization (image domain and wavelet domain) for a 3D radial acquisition using four different sampling densities (40%, 30%, 20%, 10%, and 7.5%). The number of iterations were 3000 and 500 for CS with image domain sparsity and wavelet domain sparsity, respectively. For high sampling densities all three reconstruction methods yield comparable image qualities. For lower densities, both CS reconstructions provide superior image qualities compared with the gridding algorithm, while CS with image domain sparsity shows better results at sharp edges and CS with wavelet domain sparsity is better at smooth surfaces. The normalized mean-squared errors are also included at the right bottom of the images.
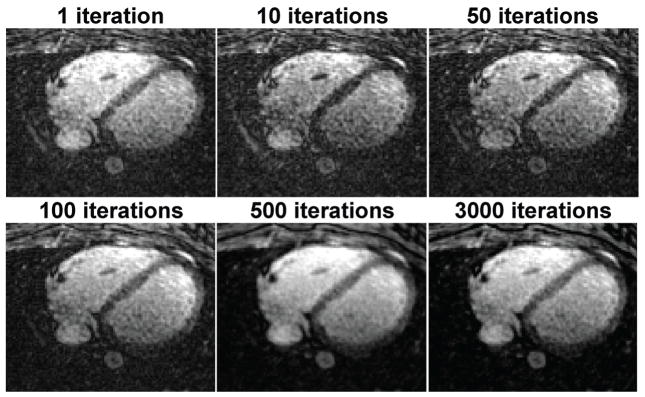
Figure 5 depicts the resulting images generated by the CS reconstruction with image domain regularization for different numbers of iterations. The streaking artifacts in the earlier iterations are gradually removed as the number of iterations increases, while the image loses the sharpness at the edges of the phantom object and becomes slightly more blurry up to 500 iterations. After additional 2500 iterations, the sharpness of the object is improved and the image looks more refined with preserved edges. A similar trend in the convergence of the CS algorithm is observed for the reconstructions with the wavelet transform as the regularization term. However, no improvement in the image quality was observed after 500 iterations in this case.
Figure 5.
CS reconstruction with image domain regularization for a phantom imaged with 3D radial with sampling density of 7.5% at different numbers of iterations, initiated with the conventional gridding reconstruction. The streaking artifacts are gradually removed with some blurring up to 500 iterations, however, with additional iterations the streaking artifacts are suppressed with improved sharpness.
In Vivo Experiment
Figure 6 and 7 show example slices of axial and reformatted sagittal views from 3D whole-heart acquisitions with isotropic 1.3 mm spatial resolution reconstructed with the gridding reconstruction, as well as iterative CS reconstruction with image and wavelet domain regularizations for four different sampling densities (6.8, 12.1, 24.2 and 36.3%). The images reconstructed with gridding present streaking artifacts and high-frequency noise-like artifacts, especially at lower sampling densities. Both CS reconstructions were able to substantially suppress these artifacts at lower densities. While the wavelet domain regularization provides cleaner and more homogeneous results in the blood pool, the image domain regularization provides more detailed and sharper edges. The wavelet domain regularization results in checkerboard-like artifacts in the reconstructed image with 6.8% sampling density.
Figure 6.
Example slices of axial views from 3D whole-heart images reconstructed with the conventional 3D gridding reconstruction and iterative CS reconstruction (with 1000 iterations for image domain regularization and 500 iterations for wavelet domain regularization) for different sampling densities. For all sampling densities, CS reconstructions have less high-frequency streaking artifacts, and the improvement in the image quality is more distinct at lower sampling densities.
Figure 7.
Example slices of sagittal views from 3D whole-heart images reconstructed by conventional 3D gridding reconstruction and iterative CS reconstruction (with 1000 iterations for image domain regularization and 500 iterations for wavelet domain regularization) for different sampling densities. For all the sampling densities, CS reconstructions have less high-frequency streaking artifacts, and the improvement in the image quality is more distinct at lower sampling densities.
Figure 8 illustrates the resulting images of the CS reconstruction with image domain regularization for different numbers of iterations. The artifacts associated with undersampling are gradually removed and the image quality improves as the number of iterations increases. The blurring of the image during the iterations shown in the phantom (Figure 5) was not observed in the in-vivo result. Between 500 and 3000 iterations, there is a slight improvement in the image quality but it was less prominent than the phantom case. Similar trends were observed for the wavelet domain regularized CS reconstruction but no visual improvement was observed after 500 iterations.
Figure 8.
An example slice from 3D data-set (sampling density = 6.8%) of the coronary arteries reconstructed using CS with image domain regularization at different iterations. The high-frequency artifacts are gradually removed throughout the iterations up to 500 iterations. Slight improvement was observed after 500 iterations, but it was less prominent that the phantom case (Figure. 5).
Figure 9 depicts reformatted RCA images from 3D whole-heart data with a spatial resolution of 1.0×1.0×1.0 mm3 and sampling density of 40%, reconstructed by the iterative CS reconstruction with the image domain regularization. The data set is retrospectively undersampled to get 10% and 20% sampling densities, and the reconstructed images are shown. Due to the isotropic resolution of the 3D radial acquisition in all three dimensions, the image can be reformatted retrospectively in an arbitrary angle to obtain a desirable imaging plane for visualizing the vessels. Table 2 summarizes the quantitative results of the 3D whole-heart images from 6 complete data sets with sampling densities of 10%, 20%, 30% and 40%. The measured vessel lengths increase as the sampling density increases for all the reconstruction methods, but the vessel lengths are not significantly different among the three reconstruction methods. The CS reconstruction with image domain regularization provides higher vessel sharpness for all sampling densities, and the improvements are statistically significant for sampling densities of 10%, 20% and 30% compared with the gridding reconstruction. The CS reconstruction with wavelet domain regularization, however, does not show significant improvement in the vessel sharpness over the gridding reconstruction for any of the sampling densities.
Figure 9.
Reformatted images of the RCA with isotropic resolution of (1.0 mm)3 from wholeheart 3D radial data with three sampling densities (40%, 20% and 10%) by the iterative CS reconstruction with image domain regularization and 1000 iterations on GPU. The actual scan time with sampling density of 40% was 7 minutes 28 seconds with the navigator gating efficiency of 54%. The RCA is clearly visualized with the CS reconstruction for all sampling densities, while slight blurring of the image and residual artifacts are observed at low sampling density (10%).
Table 2.
Mean ± standard deviation of normalized vessel sharpness and vessel length (cm) measured for conventional gridding reconstruction and iterative CS reconstructions. CS reconstruction with image domain regularization improves the vessel sharpness for sampling densities 10%, 20% and 30% compared with the gridding reconstruction.
| Sampling density | Reconstruction method | RCA sharpness | RCA length (cm) |
|---|---|---|---|
| 10% | CS-Image | 0.65±0.05*,# | 7.29±3.02 |
| CS-Wavelet | 0.52±0.06 | 7.35±2.95 | |
| Gridding | 0.53±0.03 | 6.99±2.82 | |
| 20% | CS-Image | 0.61±0.03*,# | 7.32±4.10 |
| CS-Wavelet | 0.54±0.04 | 7.08±3.91 | |
| Gridding | 0.51±0.04 | 6.89±3.61 | |
| 30% | CS-Image | 0.60±0.05* | 8.41±2.82 |
| CS-Wavelet | 0.54±0.03 | 8.50±2.76 | |
| Gridding | 0.54±0.02 | 8.52±2.86 | |
| 40% | CS-Image | 0.64±0.04 | 9.07±3.28 |
| CS-Wavelet | 0.59±0.06 | 9.13±3.26 | |
| Gridding | 0.58±0.05 | 8.78±3.40 |
: P<0.05 compared with the gridding reconstruction,
P<0.05 compared with the CS reconstruction with wavelet domain regularization
DISCUSSION
In this study, we have evaluated the implementation of a GPU-accelerated CS reconstruction for 3D radial imaging. GPU allows substantial reduction of the reconstruction time for 3D radial imaging. Phantom and in vivo whole-heart coronary MRI studies demonstrated the efficacy of CS reconstruction in removing the streaking artifacts, especially at high acceleration rates.
For CUDA implementation of the CS reconstruction, the computations in gridding/re-gridding operations can be assigned to the device code either by dividing the 3D radial data points among threads (radial point driven) or by dividing the Cartesian grid points among threads (Cartesian point driven). The radial point driven assignment is a simple and intuitive approach and has a minimum number of memory reads (writes) in gridding (re-gridding), but results in a large amount of data sharing among threads and cumulative memory writes in gridding as illustrated in Figure 3(a). Each CUDA thread assigned to the radial data point will read the memory to get the measured k-space value for the sample point and distribute the value to the neighboring Cartesian grid points inside the convolution window. The Cartesian grid point will have different contributions from different radial sample points, resulting in cumulative memory access among different CUDA threads. In our experiment, the execution time of the gridding operation was only twice as long as the re-gridding operation despite the massive cumulative memory access and atomic operations. On the other hand, the Cartesian point driven assignment has minimum number of memory writes (reads) in gridding (re-gridding). However, one must compute the list of the radial points associated with the Cartesian grid point within the convolution window for every thread, which requires additional computations and/or additional memory usage. The Cartesian point driven assignment is illustrated in Figure 3(b). Each CUDA thread assigned to the Cartesian grid point will read the memory to get the measured k-space values from neighboring radial sample points inside the convolution window, combine the values and write on the memory for the Cartesian point only once. The Cartesian point driven assignment has an uneven workload distribution over different threads and causes poor compute to global memory access ratio for outer k-space points, especially at low sampling densities. Each thread assignment strategy has its advantages and disadvantages, and it is not simple to determine which one is superior to the other. In this paper, we used radial point driven assignment; more study and optimization on the thread allocation and memory management can be done in the future for further speed-up of the parallel implementation.
The proposed implementation of the 3D radial acquisition still takes a long time to be clinically feasible. For example, we have used 3000 iterations for the CS reconstruction of the phantom data (3443 voxels) and 1000 iterations for the in-vivo data (3923 voxels), and the final reconstruction times for the data with 20% sampling density were around 5 hours and 2.5 hours, while the conventional gridding algorithm takes a few minutes with the C++ implementation for any case. However, CS reconstruction with fewer iterations (e.g. 100 iterations) still provides improved image quality compared to the gridding algorithm, and the reconstruction time in this case is around 10–16 minutes with 20% sampling density with the GPU implementation.
Multiple GPUs can be used for further speed-up as the reconstruction of the individual coil images can also be parallelized among the GPUs. In this paper, the proposed CS algorithm only utilizes the sparsity property of the image for the reconstruction of the undersampled k-space data and does not exploit the coil sensitivity information from multiple coils which may potentially enable further undersampling. Techniques aiming to combine parallel imaging and CS for even higher acceleration rates have been proposed (10,49–51), and GPU implementations were also proposed for some of the approaches (52,53). The 3D radial CS reconstruction may also be combined with such techniques for further acceleration. The main issue of combining parallel imaging for the reconstruction of the 3D radial acquisition is the huge amount of the data and the limited size of the GPU’s global memory. The data from multiple coils cannot be stored on the GPU’s global memory at the same time and the reconstruction process needs to be divided into smaller jobs to fit on the GPU’s memory. This will result in frequent memory access between the main system memory and the GPU’s memory, serial execution of the divided processes, and additional handling of the shared data between the divided processes. As GPU hardware is fast developing for general purpose computing, it is expected that these limitations will be resolved, enabling efficient implementation of more advanced reconstruction methods without complicated designing and optimization for GPU programming.
We have utilized the identity transform and the Daubechies 4 wavelets as the sparsifying transforms for the CS reconstruction. The baseline assumption for successful CS reconstruction is that the MR images are sparse in these transform domains. Wavelets have been applied in many MR reconstruction studies (50,54) but the use of image domain sparsity has been limited to applications such as MR angiography (55,56). The 3D radial trajectories are generally oversampled in the read-out direction and this results in an increased FOV larger than the prescribed FOV. The 3D image then contains redundant areas where there is not much signal, making the image sparse in the image domain itself. Both image domain and wavelet domain regularizations have provided improved image quality compared with the conventional gridding algorithm, but exhibited some issues that need to be improved. The CS reconstruction with image domain regularization has a slow convergence speed with the iterative algorithm described in this paper. The CS reconstruction with wavelet domain regularization provides a better convergence speed than the image domain regularization, but shows checkerboard-like and blocky artifacts at low sampling densities. The two step iterative CS reconstruction algorithm used in this paper enables simple and efficient coefficient-wise thresholding for the thresholding step in Eq. [5] only when the sparsifying transform is given by a unitary matrix. The image domain (identity transform) and Daubechies wavelet transform satisfy this condition, while the well-known and commonly used TV regularization does not. Use of other sparsifying transforms or more advanced techniques that can adaptively capture object-specific sparsity nature (57,58) can improve the CS reconstruction, which requires further investigation.
CONCLUSION
We have implemented a GPU-accelerated iterative CS reconstruction method for 3D radial acquisitions and evaluated its performance in 3D whole-heart coronary MRI. The CS reconstruction method improved the image quality of highly undersampled 3D radial data sets compared to the conventional gridding reconstruction and the GPU implementation was able to substantially reduce the reconstruction time.
Acknowledgments
The authors would like to thank Jaime Shaw for help with proofreading.
The project described was supported by NIH R01EB008743-01A2, AHA SDG-0730339N and NIH UL1 RR025758-01, Harvard Clinical and Translational Science Center, from the National Center for Research Resources.
References
- 1.Weber OM, Martin AJ, Higgins CB. Whole-heart steady-state free precession coronary artery magnetic resonance angiography. Magn Reson Med. 2003;50(6):1223–1228. doi: 10.1002/mrm.10653. [DOI] [PubMed] [Google Scholar]
- 2.Nehrke K, Bornert P, Mazurkewitz P, Winkelmann R, Grasslin I. Free-breathing whole-heart coronary MR angiography on a clinical scanner in four minutes. J Magn Reson Imaging. 2006;23(5):752–756. doi: 10.1002/jmri.20559. [DOI] [PubMed] [Google Scholar]
- 3.Bi X, Carr JC, Li D. Whole-heart coronary magnetic resonance angiography at 3 Tesla in 5 minutes with slow infusion of Gd-BOPTA, a high-relaxivity clinical contrast agent. Magn Reson Med. 2007;58(1):1–7. doi: 10.1002/mrm.21224. [DOI] [PubMed] [Google Scholar]
- 4.Alley MT, Napel S, Amano Y, Paik DS, Shifrin RY, Shimakawa A, Pelc NJ, Herfkens RJ. Fast 3D cardiac cine MR imaging. J Magn Reson Imaging. 1999;9(5):751–755. doi: 10.1002/(sici)1522-2586(199905)9:5<751::aid-jmri21>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Barger AV, Grist TM, Block WF, Mistretta CA. Single breath-hold 3D contrast-enhanced method for assessment of cardiac function. Magn Reson Med. 2000;44(6):821–824. doi: 10.1002/1522-2594(200012)44:6<821::aid-mrm1>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 6.Kozerke S, Tsao J, Razavi R, Boesiger P. Accelerating cardiac cine 3D imaging using k-t BLAST. Magn Reson Med. 2004;52(1):19–26. doi: 10.1002/mrm.20145. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen TD, Spincemaille P, Weinsaft JW, Ho BY, Cham MD, Prince MR, Wang Y. A fast navigator-gated 3D sequence for delayed enhancement MRI of the myocardium: comparison with breathhold 2D imaging. J Magn Reson Imaging. 2008;27(4):802–808. doi: 10.1002/jmri.21296. [DOI] [PubMed] [Google Scholar]
- 8.Peters DC, Appelbaum EA, Nezafat R, Dokhan B, Han Y, Kissinger KV, Goddu B, Manning WJ. Left ventricular infarct size, peri-infarct zone, and papillary scar measurements: A comparison of high-resolution 3D and conventional 2D late gadolinium enhancement cardiac MR. J Magn Reson Imaging. 2009;30(4):794–800. doi: 10.1002/jmri.21897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin T, Hu HH, Pohost GM, Nayak KS. Three dimensional first-pass myocardial perfusion imaging at 3T: feasibility study. J Cardiovasc Magn Reson. 2008;10:57. doi: 10.1186/1532-429X-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otazo R, Xu J, Axel L, Sodickson D. Combination of compressed sensing and parallel imaging for highly-accelerated 3D first-pass cardiac perfusion MRI. 2010 May; Stockholm. Proceedings of the 18th Scientific Meeting of ISMRM; p. 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters DC, Korosec FR, Grist TM, Block WF, Holden JE, Vigen KK, Mistretta CA. Undersampled projection reconstruction applied to MR angiography. Magnetic Resonance in Medicine. 2000;43(1):91–101. doi: 10.1002/(sici)1522-2594(200001)43:1<91::aid-mrm11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 12.Thedens DR, Irarrazaval P, Sachs TS, Meyer CH, Nishimura DG. Fast magnetic resonance coronary angiography with a three-dimensional stack of spirals trajectory. Magnetic Resonance in Medicine. 1999;41(6):1170–1179. doi: 10.1002/(sici)1522-2594(199906)41:6<1170::aid-mrm13>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.Bhat H, Yang Q, Zuehlsdorff S, Li K, Li D. Contrast-enhanced whole-heart coronary magnetic resonance angiography at 3T with radial EPI. Magnetic Resonance in Medicine. 2011:82–91. doi: 10.1002/mrm.22781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat H, Ge L, Nielles-Vallespin S, Zuehlsdorff S, Li D. 3D radial sampling and 3D affine transform-based respiratory motion correction technique for free-breathing whole-heart coronary MRA with 100% imaging efficiency. Magnetic Resonance in Medicine. 2011;65(5):1269–1277. doi: 10.1002/mrm.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stehning C, Bornert P, Nehrke K, Eggers H, Dossel O. Fast isotropic volumetric coronary MR angiography using free-breathing 3D radial balanced FFE acquisition. Magnetic Resonance in Medicine. 2004;52(1):197–203. doi: 10.1002/mrm.20128. [DOI] [PubMed] [Google Scholar]
- 16.Barger AV, Block WF, Toropov Y, Grist TM, Mistretta CA. Time-resolved contrast-enhanced imaging with isotropic resolution and broad coverage using an undersampled 3D projection trajectory. Magnetic Resonance in Medicine. 2002;48(2):297–305. doi: 10.1002/mrm.10212. [DOI] [PubMed] [Google Scholar]
- 17.Lai P, Huang F, Li Y, Nielles-Vallespin S, Bi X, Jerecic R, Li D. Contrast-kinetics-resolved whole-heart coronary MRA using 3DPR. Magnetic Resonance in Medicine. 2010;63(4):970–978. doi: 10.1002/mrm.22246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu T, Korosec FR, Block WF, Fain SB, Turk Q, Lum D, Zhou Y, Grist TM, Haughton V, Mistretta CA. PC VIPR: A High-Speed 3D Phase-Contrast Method for Flow Quantification and High-Resolution Angiography. AJNR Am J Neuroradiol. 2005;26(4):743–749. [PMC free article] [PubMed] [Google Scholar]
- 19.O’Sullivan JD. A Fast Sinc Function Gridding Algorithm for Fourier Inversion in Computer Tomography. Medical Imaging, IEEE Transactions on. 1985;4(4):200–207. doi: 10.1109/TMI.1985.4307723. [DOI] [PubMed] [Google Scholar]
- 20.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magnetic Resonance in Medicine. 2007;57(6):1086–1098. doi: 10.1002/mrm.21236. [DOI] [PubMed] [Google Scholar]
- 21.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magnetic Resonance in Medicine. 1999;42(5):952–962. [PubMed] [Google Scholar]
- 22.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magnetic Resonance in Medicine. 2002;47(6):1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 23.Pruessmann KP, Weiger M, Bornert P, Boesiger P. Advances in sensitivity encoding with arbitrary k-space trajectories. Magnetic Resonance in Medicine. 2001;46(4):638–651. doi: 10.1002/mrm.1241. [DOI] [PubMed] [Google Scholar]
- 24.Seiberlich N, Breuer FA, Blaimer M, Barkauskas K, Jakob PM, Griswold MA. Non-Cartesian data reconstruction using GRAPPA operator gridding (GROG) Magnetic Resonance in Medicine. 2007;58(6):1257–1265. doi: 10.1002/mrm.21435. [DOI] [PubMed] [Google Scholar]
- 25.Chang T, He L, Fang T. MR Image Reconstruction from Sparse Radial Samples using Bregman Iteration. Proceedings of the 13th Scientfic Meeting of ISMRM; Seattle. p. 696. Proceedings of the 13th Scientfic Meeting of ISMRM. [Google Scholar]
- 26.Beck A, Teboulle M. A Fast Iterative Shrinkage-Thresholding Algorithm for Linear Inverse Problems. SIAM Journal on Imaging Sciences. 2009;2(1):183–202. [Google Scholar]
- 27.Doneva M, Eggers H, Rahmer J, Bornert P, Mertins A. Highly Undersampled 3D Golden Ratio Radial Imaging with Iterative Reconstruction. Proceedings of the 16th Scientific Meeting of ISMRM; Toronto, Canada. 2008. p. 336. [Google Scholar]
- 28.Sorensen TS, Schaeffter T, Noe KO, Hansen MS. Accelerating the Nonequispaced Fast Fourier Transform on Commodity Graphics Hardware. Medical Imaging, IEEE Transactions on. 2008;27(4):538–547. doi: 10.1109/TMI.2007.909834. [DOI] [PubMed] [Google Scholar]
- 29.Sorensen TS, Atkinson D, Schaeffter T, Hansen MS. Real-Time Reconstruction of Sensitivity Encoded Radial Magnetic Resonance Imaging Using a Graphics Processing Unit. Medical Imaging, IEEE Transactions on. 2009;28(12):1974–1985. doi: 10.1109/TMI.2009.2027118. [DOI] [PubMed] [Google Scholar]
- 30.Knoll F, Unger M, Diwoky C, Clason C, Pock T, Stollberger R. Fast reduction of undersampling artifacts in radial MR angiography with 3D total variation on graphics hardware. Magnetic Resonance Materials in Physics, Biology and Medicine. 2010;23(2):103–114. doi: 10.1007/s10334-010-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchgraber G, Knoll F, Freiberger M, Clason C, Grabner M, Stollberger R. Fast Regridding using LSQR on Graphics Hardware. 2010 May; Stockholm. Proceedings of the 18th Scientific Meeting of ISMRM; p. 4959. [Google Scholar]
- 32.Stone SS, Haldar JP, Tsao SC, Hwu WmW, Sutton BP, Liang ZP. Accelerating advanced MRI reconstructions on GPUs. Journal of Parallel and Distributed Computing. 2008;68(10):1307–1318. doi: 10.1016/j.jpdc.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong STS, Roos MS. A strategy for sampling on a sphere applied to 3D selective RF pulse design. Magnetic Resonance in Medicine. 1994;32(6):778–784. doi: 10.1002/mrm.1910320614. [DOI] [PubMed] [Google Scholar]
- 34.Pipe JG, Menon P. Sampling density compensation in MRI: Rationale and an iterative numerical solution. Magnetic Resonance in Medicine. 1999;41(1):179–186. doi: 10.1002/(sici)1522-2594(199901)41:1<179::aid-mrm25>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KO, Pipe JG. Convolution kernel design and efficient algorithm for sampling density correction. Magn Reson Med. 2009;61(2):439–447. doi: 10.1002/mrm.21840. [DOI] [PubMed] [Google Scholar]
- 36.Zwart NR, Johnson KO, Pipe JG. Efficient sample density estimation by combining gridding and an optimized kernel. Magn Reson Med. 2011 doi: 10.1002/mrm.23041. in press. [DOI] [PubMed] [Google Scholar]
- 37.Rasche V, Proksa R, Sinkus R, Bornert P, Eggers H. Resampling of data between arbitrary grids using convolution interpolation. Medical Imaging, IEEE Transactions on. 1999;18(5):385–392. doi: 10.1109/42.774166. [DOI] [PubMed] [Google Scholar]
- 38.Daubechies I, Defrise M, De Mol C. An iterative thresholding algorithm for linear inverse problems with a sparsity constraint. Communications on Pure and Applied Mathematics. 2004;57(11):1413–1457. [Google Scholar]
- 39.Wright SJ, Nowak RD, Figueiredo MAT. Sparse Reconstruction by Separable Approximation. Signal Processing, IEEE Transactions on. 2009;57(7):2479–2493. [Google Scholar]
- 40.NVIDIA’s Next Generation CUDA Compute Architecture: Fermi. white paper. NVIDIA Corporation; 2009. [Google Scholar]
- 41.Frigo M, Johnson SG. The Design and Implementation of FFTW3. Proceedings of the IEEE. 2005;93(2):216–231. [Google Scholar]
- 42.Daubechies I. Ten lectures on wavelets. Philadelphia, PA: Society for Industrial and Applied Mathematics; 1992. [Google Scholar]
- 43.Kim S, Koh K, Lustig M, Boyd S, Gorinvesky D. An interior-point method for large-scale l1-Regularized Least Squares. IEEE Journal of Selected Topoics in Signal Processing. 2007;1(4):606–617. [Google Scholar]
- 44.Jackson JI, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding [computerised tomography application] Medical Imaging, IEEE Transactions on. 1991;10(3):473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- 45.Scott AD, Keegan J, Firmin DN. Motion in cardiovascular MR imaging. Radiology. 2009;250(2):331–351. doi: 10.1148/radiol.2502071998. [DOI] [PubMed] [Google Scholar]
- 46.Etienne A, Botnar RM, van Muiswinkel AMC, Boesiger P, Manning WJ, Stuber M. “Soap-Bubble” visualization and quantitative analysis of 3D coronary magnetic resonance angiograms. Magnetic Resonance in Medicine. 2002;48(4):658–666. doi: 10.1002/mrm.10253. [DOI] [PubMed] [Google Scholar]
- 47.Deriche R. Fast Algorithms for Low-Level Vision. IEEE Trans Pattern Anal Mach Intell. 1990;12(1):78–87. [Google Scholar]
- 48.Botnar RM, Stuber M, Danias PG, Kissinger KV, Manning WJ. Improved coronary artery definition with T2-weighted, free-breathing, three-dimensional coronary MRA. Circulation. 1999;99(24):3139–3148. doi: 10.1161/01.cir.99.24.3139. [DOI] [PubMed] [Google Scholar]
- 49.Liang D, Liu B, Wang J, Ying L. Accelerating SENSE using compressed sensing. Magn Reson Med. 2009;62(6):1574–1584. doi: 10.1002/mrm.22161. [DOI] [PubMed] [Google Scholar]
- 50.Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med. 2010;64(2):457–471. doi: 10.1002/mrm.22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knoll F, Clason C, Bredies K, Uecker M, Stollberger R. Parallel imaging with nonlinear reconstruction using variational penalties. Magn Reson Med. 2011 doi: 10.1002/mrm.22964. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uecker M, Zhang S, Frahm J. Nonlinear inverse reconstruction for real-time MRI of the human heart using undersampled radial FLASH. Magn Reson Med. 2010;63(6):1456–1462. doi: 10.1002/mrm.22453. [DOI] [PubMed] [Google Scholar]
- 53.Murphy M, Keutzer K, Vasanawala S, Lustig M. Clinically Feasible Reconstruction Time for L1-SPIRiT Parallel Imaging and Compressed Sensing MRI. 2010 May; Stockholm. Proceedings of the 18th Scientific Meeting of ISMRM; p. 4854. [Google Scholar]
- 54.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 55.Cukur T, Lustig M, Nishimura DG. Improving non-contrast-enhanced steady-state free precession angiography with compressed sensing. Magn Reson Med. 2009;61(5):1122–1131. doi: 10.1002/mrm.21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akcakaya M, Hu P, Chuang ML, Hauser TH, Ngo LH, Manning WJ, Tarokh V, Nezafat R. Accelerated noncontrast-enhanced pulmonary vein MRA with distributed compressed sensing. J Magn Reson Imaging. 2011;33(5):1248–1255. doi: 10.1002/jmri.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doneva M, Bornert P, Eggers H, Stehning C, Senegas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med. 2010;64(4):1114–1120. doi: 10.1002/mrm.22483. [DOI] [PubMed] [Google Scholar]
- 58.Akcakaya M, Basha TA, Goddu B, Goepfert LA, Kissinger KV, Tarokh V, Manning WJ, Nezafat R. Low-dimensional-structure self-learning and thresholding: regularization beyond compressed sensing for MRI reconstruction. Magn Reson Med. 2011;66(3):756–767. doi: 10.1002/mrm.22841. [DOI] [PMC free article] [PubMed] [Google Scholar]