Among Gram‐negative biodefence pathogens, Yersinia pestis (plague), Francisella tularensis (tularemia) and, to a lesser extent, Brucella species (brucellosis) are most widely studied. In contrast, Burkholderia mallei (glanders) and B. pseudomallei (melioidosis) have garnered less attention. While the underlying reasons are multifaceted, for example, perceived importance of an organism being listed as a Category A versus B pathogen, B. pseudomallei poses formidable and unique challenges pertaining to development of therapeutic countermeasures. It is fair to say that, in general, Y. pestis, F. tularensis and Brucella species are susceptible to most classes of antibiotics and that the main challenge with these organisms is rapid and accurate diagnosis to enable initiation of proper therapeutic interventions. In contrast, therapeutic countermeasures for B. pseudomallei are limited because of intrinsic resistance (Wuthiekanun and Peacock, 2006; Estes et al., 2010). At present, the recommended acute phase treatment for melioidosis includes β‐lactam antibiotics such as ceftazidime, amoxicillin‐clavulanic acid or carbapenems (e.g. meropenem and imipenem; Peacock et al., 2008). Other efficacious therapeutics such as trimethoprim‐sulfamethoxazole are reserved for eradication phase treatment or potential prophylaxis (Peacock et al., 2008). To complicate matters, Burkholderia species are intrinsically resistant to polymyxins and therefore there is no drug of last resort such as colistin that is being used to treat infections by panresistant so‐called superbugs.
Fundamentally, B. pseudomallei is not unique from other bacteria and intrinsic resistance is achieved using multiple mechanisms documented for other bacteria (Walsh, 2003): (i) exclusion from the cell; (ii) enzymatic inactivation; (iii) target alterations or deletion; and (iv) active efflux from the cell. A fifth mechanism, namely metabolic bypass of the effected enzyme by complementation with an insensitive equivalent, has not yet been reported in B. pseudomallei. Resistance mechanisms can act in synergy to achieve significant levels of resistance. For example, drug efflux is most effective in bacteria with reduced outer membrane permeability (Nikaido, 2001), for example Acinetobacter baumanii, Burkholderia cepacia, Pseudomonas aeruginosa and Stenotrophomonas maltophilia. The outer membrane permeability in these bacteria is between 1% and 11% of that observed in Escherichia coli (Hancock, 1998). Reduced outer membrane permeability is primarily due to the exclusionary properties of porins (Pages et al., 2008) and lipopolysaccharide (LPS) (Raetz et al., 2007). LPS contributes to high‐level polymyxin resistance in species such as Burkholderia (Novem et al., 2009) or mutant strains of P. aeruginosa and S. enterica serovar Typhimurium where the lipid A portion is modified, e.g. by modification with 4‐amino‐4‐deoxyarabinose (Raetz et al., 2007). In summary, the cell envelope of Gram‐negative bacteria, especially the outer membrane, is a major barrier for antibiotics and its contributions to antimicrobial susceptibility are complex (Fig. 1).
Figure 1.
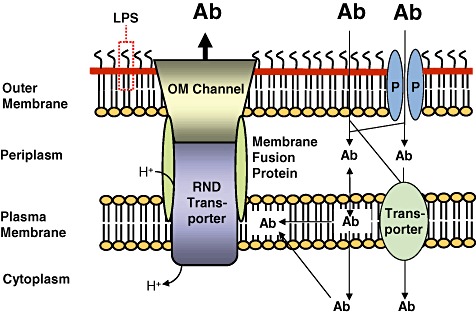
The cell envelope of Gram‐negative bacteria is a major barrier for antibiotics. The cell envelope of Gram‐negative bacteria consists of the plasma membrane, the periplasm and the outer membrane. The outer membrane is the major barrier for antibiotics (Ab). Some antibiotics penetrate this membrane either through porins (P) or by passive diffusion through the outer membrane phospholipid (inner leaflet)‐lipid A (outer leaflet) bilayer. The lipopolysaccharide (LPS) forms another barrier for many antibiotics but polycationic compounds such as gentamicin and colistin are being transported through the outer membrane via interaction with LPS in a process called self‐promoted uptake. Antibiotic molecules then enter the cell from the periplasm either via partition into and passive diffusion through the plasma membrane or are actively transported via transporters into the cytoplasm. Efflux pumps of the resistance nodulation cell division (RND) superfamily are major players in antibiotic resistance of Gram‐negative bacteria. These tripartite systems span the entire cell envelope and are composed of an RND transporter, a membrane fusion protein and an outer membrane (OM) channel. It is generally accepted that RND transporters acquire substrates from the plasma membrane. Efflux via RND pumps is driven by the proton gradient. The setup illustrated in this figure explains why synergy between exclusionary outer membrane and/or cell envelope properties is a powerful mechanism leading to high‐level antibiotic resistance in non‐enteric Gram‐negative bacteria. Although antibiotics may be present outside the bacterial cell in high concentration (illustrated by large bold letters), passive influx through the various compartments of the cell coupled to active efflux via a cell envelope‐spanning efflux system results in low intracellular concentrations of antibiotics (illustrated by smaller letters).
Why is B. pseudomallei unique among Gram‐negative biodefence pathogens with respect to drug discovery efforts? Although outer membrane permeability has not yet been directly assessed in B. pseudomallei, the intrinsic resistance of this bacterium to many antibiotics can most likely be directly attributed to synergy between exclusion and active efflux from the cell. This notion is supported by the finding that antibiotic susceptibilities of efflux pump expressing strains compared with their isogenetic pump mutant counterparts are vastly different and could not simply be explained by expression of efflux pumps alone. For example, aminoglycoside and macrolide susceptibilities of wild‐type and AmrAB–OprA efflux pump mutant strains differ up to 100‐fold and 16‐fold respectively (Moore et al., 1999; Trunck et al., 2009). Similarly, the clindamycin susceptibility of B. pseudomallei is greatly (> 16‐fold) affected by the expression status of the BpeAB–OprB efflux pump (Mima and Schweizer, 2010). Although outer membrane barrier properties may look alike, our experiences indicate that even bacteria like B. pseudomallei and P. aeruginosa with similar outer membrane permeabilities behave quite differently in terms of antibiotic susceptibility profiles. Expression of the BpeEF–OprC efflux pump in B. pseudomallei results in high level resistance [as judged by minimal inhibitory concentrations (MIC)] to chloramphenicol (512 µg ml−1) and trimethoprim (> 32 µg ml−1) (T. Mima and H. Schweizer, unpubl. obs.). In contrast, expression of the same efflux pump in P. aeruginosa only results in modest increases in resistance with MICs of 8 µg ml−1 for both chloramphenicol and trimethoprim (Kumar et al., 2006). This rather dramatic difference is not due to lack of transcription or translation, but likely because the outer membrane properties of B. pseudomallei and P. aeruginosa are quite different despite similar relative outer membrane permeabilities of Pseudomonas and Burkholderia species.
Our experiences have shown us that commonly used Gram‐negatives bacteria such as E. coli and P. aeruginosa, including TolC or pump mutants to assess roles of efflux, are often inappropriate surrogates for drug anti‐B. pseudomallei discovery efforts. To this end, we have generated isogenetic B. pseudomallei efflux pump proficient (expressing) and deficient mutants in either the virulent (and therefore select agent) strain 1026b (DeShazer et al., 1997) or its derivative Bp82 (Propst et al., 2010), which is excluded from select agent listings and can be handled in a BSL2+ laboratory with local Institutional Biosafety Committee jurisdiction. We have employed these strains to test novel compounds for anti‐B. pseudomallei activity. The ketolide cethromycin showed efficacy against clinical and environmental strains but expression of the AmrAB–OprA efflux pump resulted in high‐level resistance (Mima et al., 2011b). In contrast, the activity of the new monosulfactam BAL30072 was not significantly affected by efflux (Mima et al., 2011a).
In our hands, the less pathogenic but closely related BSL2 agent B. thailandensis (Brett et al., 1998; Yu et al., 2006) is an appropriate surrogate for B. pseudomallei. It for example possesses the equivalent cadre of efflux pumps and we have generated the corresponding panel of isogenetic B. thailandensis efflux pump proficient (expressing) and deficient mutants.
Conclusions
Whole‐cell screening is an important step in the drug discovery process. Our findings with B. pseudomallei indicate that it is imperative to choose proper strains for whole‐cell screening. Even seemingly closely related species or species with similar outer membrane permeabilities may possess quite disparate cell envelope properties. One must especially be careful about choice of surrogate strains and recognize that Gram‐negatives are not all created equal. For example, in the context of drug discovery efforts E. coli strains may be perfectly good surrogates for Y. pestis and F. tularensis, but in most instances they are likely inappropriate surrogates for B. pseudomallei. By choosing inappropriate surrogates, properties of antibiotics may be misjudged (e.g. propensity for efflux) or antibiotics with activity against the targeted bacterium may be entirely missed. Modern genetic technologies facilitate construction of suitable screening strains, which may include proper surrogates (e.g. B. thailandensis for B. pseudomallei). Burkholderia mallei is extremely closely related to and widely considered a clone of B. pseudomallei but is generally more susceptible to antibiotics than B. pseudomallei because most strains are lacking or not expressing some of the resistance mechanisms, for example the AmrAB–OprA efflux pump (Nierman et al., 2004). Once can therefore generalize that when a compound shows efficacy against B. pseudomallei it is also efficacious against B. mallei. In a sense, then, B. thailandensis and B. pseudomallei are suitable surrogates for B. mallei.
Acknowledgments
I am grateful to the students and postdocs at Colorado State University that have contributed to various aspects of this work. Funding was provided by NIH NIAID grant AI065357.
References
- Brett P.J., DeShazer D., Woods D.E. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei‐like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- DeShazer D., Brett P., Carlyon R., Woods D. Mutagenesis of Burkholderia pseudomallei with Tn5‐OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J Bacteriol. 1997;179:2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes D.M., Dow S.W., Schweizer H.P., Torres A.G. Present and future therapeutic strategies for melioidosis and glanders. Expert Rev Anti Infect Ther. 2010;8:325–338. doi: 10.1586/eri.10.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E.W. Resistance mechanisms in Pseudomonas aeruginosa and other non‐fermentative bacteria. Clin Infect Dis. 1998;27(1):S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- Kumar A., Chua K.‐L., Schweizer H. Method for regulated expression of single‐copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF‐OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob Agents Chemother. 2006;50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima T., Schweizer H. The BpeAB‐OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad‐spectrum drug efflux system. Antimicrob Agents Chemother. 2010;54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima T., Kvitko B.H., Rholl D.A., Page M.G., Desarbre E., Schweizer H. In vitro activity of BAL30072 against Burkholderia pseudomallei. Int J Antimicrob Agents. 2011a;38:157–159. doi: 10.1016/j.ijantimicag.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima T., Schweizer H., Xu Z.‐Q. In vitro activity of cethromycin against Burkholderia pseudomallei and investigation of mechanism of resistance. J Antimicrob Chemother. 2011b;66:73–78. doi: 10.1093/jac/dkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.A., DeShazer D., Reckseidler S., Weissman A., Woods D.E. Efflux‐mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W.C., DeShazer D., Kim H.S., Tettelin H., Nelson K.E., Feldblyum T. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci USA. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol. 2001;12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- Novem V., Shui G., Wang D., Bendt A.K., Sim S.H., Liu Y. Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin Vaccine Immunol. 2009;16:1420–1428. doi: 10.1128/CVI.00472-08. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages J.‐M., James C.E., Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram‐negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- Peacock S.J., Schweizer H., Dance D.A.B., Smith T.L., Gee J.E., Wuthiekanun V. Consensus guidelines on the management of accidental laboratory exposure to Burkholderia pseudomallei and Burkholderia mallei. Emerg Infect Dis. 2008;14:e2. doi: 10.3201/eid1407.071501. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propst K.L., Mima T., Choi K.H., Dow S.W., Schweizer H. A Burkholderia pseudomallei ΔpurM mutant is avirulent in immune competent and immune deficient animals: candidate strain for exclusion from Select Agent lists. Infect Immun. 2010;78:3136–3143. doi: 10.1128/IAI.01313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C.R., Reynolds C.M., Trent M.S., Bishop R.E. Lipid A modification systems in gram‐negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunck L.A., Propst K.L., Wuthiekanun V., Tuanyok A., Beckstrom‐Sternberg S.M., Beckstrom‐Sternberg J.S. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis. 2009;3:e519. doi: 10.1371/journal.pntd.0000519. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. American Society for Microbiology Press; 2003. [Google Scholar]
- Wuthiekanun V., Peacock S.J. Management of melioidosis. Expert Rev Anti Infect Ther. 2006;4:445–455. doi: 10.1586/14787210.4.3.445. [DOI] [PubMed] [Google Scholar]
- Yu Y., Kim H.S., Chua H.H., Lin C.H., Sim S.H., Lin D. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 2006;6:46. doi: 10.1186/1471-2180-6-46. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


