SUMMARY
Background
In nematodes, plants and fungi, RNAi is remarkably potent and persistent due to the amplification of initial silencing signals by RNA-dependent RNA polymerases (RdRPs). In Caenorhabditis elegans (C. elegans), the interaction between the RNA-induced silencing complex (RISC) loaded with primary siRNAs and the target mRNA leads to the recruitment of RdRPs and synthesis of secondary siRNAs using the target mRNA as the template. The mechanism and genetic requirements for secondary siRNA accumulation are not well understood.
Results
From a forward genetic screen for C. elegans genes required for RNAi, we identified rde-10 and through proteomic analysis of RDE-10-interacting proteins, we identified a protein complex containing the new RNAi factor RDE-11, the known RNAi factors RSD-2 and ERGO-1, as well as other candidate RNAi factors. The RNAi defective genes rde-10 and rde-11 encode a novel protein and a RING-type zinc finger domain protein, respectively. Mutations in rde-10 and rde-11 genes cause dosage-sensitive RNAi deficiencies: these mutants are resistant to low dosage, but sensitive to high dosage of double-stranded RNAs (dsRNAs). We assessed the roles of rde-10, rde-11, and other dosage-sensitive RNAi-defective genes rsd-2, rsd-6 and haf-6 in both exogenous and endogenous small RNA pathways using high-throughput sequencing and qRT-PCR. These genes are required for the accumulation of secondary siRNAs in both exogenous and endogenous RNAi pathways.
Conclusions
The RDE-10/RDE-11 complex is essential for the amplification of RNAi in C. elegans by promoting secondary siRNA accumulation.
Keywords: rde-10, rde-11, RNAi, endogenous siRNAs, RNA silencing protein co-factors
INTRODUCTION
RNA interference (RNAi) and related small RNA pathways regulate a broad range of processes including antiviral defense, heterochromatin formation, genome surveillance and gene expression in animals, plants and fungi [1]. In C. elegans, RNAi is initiated by the introduction of long dsRNAs that associate with the dsRNA-binding protein RDE-4 and are processed into ~21–24 nt primary small interfering RNAs (siRNAs) by the RNase III-related enzyme Dicer (DCR-1) [2–4]. Primary siRNAs are loaded onto the RISC containing the Argonaute protein RDE-1 and trigger sequence-specific degradation of mRNAs complementary to the loaded siRNAs [5, 6]. The initial RNAi response is amplified by the recruitment of RdRPs (e.g., RRF-1 and EGO-1) onto target mRNAs [6–11]. RdRPs dramatically increase the effectiveness of RNAi by catalyzing unprimed synthesis of secondary siRNAs using target mRNAs as templates [8–11]. Secondary siRNAs are far more abundant than primary siRNAs and correspond to target mRNA sequences both upstream and downstream of the initial dsRNA trigger [8–11]. Secondary siRNAs associate with a family of worm-specific Argonautes (WAGOs) and potentiate the RNAi response by promoting target mRNA decay and co-transcriptional silencing [6, 12, 13].
Deep sequencing of small RNAs has revealed an extensive repertoire of endogenous siRNAs in many organisms. These endogenous siRNAs play essential roles in maintaining genome integrity at both transcriptional and post-transcriptional levels. In C. elegans, the endogenous siRNAs can be classified into ERGO-1 class 26G, ALG-3/4 class 26G, CSR-1 class 22G and WAGO class 22G siRNAs, based on their length, 5′ nucleotide composition and Argonaute binding partners. 26G siRNAs are primary siRNAs and their biogenesis requires DCR-1, the 3′-5′ exonuclease ERI-1 and the RdRP RRF-3 [14–17]. ERGO-1 class 26G siRNAs function in oocytes and embryos [14, 16], whereas ALG-3/4 class 26G siRNAs are required for normal sperm development [14, 17]. ERGO-1 class 26G siRNAs silence recently acquired duplicated genes, and their accumulation requires the helicase ERI-6/7 and several mutator proteins (proteins first identified as being required for the silencing of transposons) [18, 19]. Biogenesis of 22G siRNAs requires the DCR-related helicase DRH-3, as well as the RdRPs EGO-1 and RRF-1 [12]. CSR-1 class 22G siRNAs are derived from germline-expressed genes and promote chromosome segregation [20]. WAGO class 22G siRNAs silence transposons, pseudogenes, cryptic loci and coding genes in germline and soma [6, 12]. A subset of WAGO class 22G siRNAs are derived from 26G siRNA targets and are dependent on 26G siRNA pathway components for their formation [14–18]. In addition to post-transcriptional down-regulation of target mRNAs in the cytoplasm, certain WAGO class 22G siRNAs which function downstream of ERGO-1 class 26G siRNA pathway also direct co-transcriptional silencing of target genes through association with the Argonaute NRDE-3 in the nuclei of somatic cells [13].
We identified components of a novel protein complex from a forward genetic screen and by mass spectrometry analysis. The key components of this complex are encoded by two new RNAi defective (rde) genes, rde-10 and rde-11. rde-10 encodes a protein only conserved within the nematodes that lacks known functional domains, and rde-11 encodes a RING-type zinc finger domain protein. The RDE-10/RDE-11 complex also contains known RNAi pathway components RSD-2 and ERGO-1. Mutations in rde-10 and rde-11 genes cause a dosage-sensitive RNAi-defective phenotype in that they fail to respond to dsRNAs at relatively low concentrations but are sensitive to dsRNAs at relatively high concentrations. High-throughput sequencing data indicates that the RDE-10/RDE-11 complex is essential for the accumulation of secondary siRNAs that potentiate the silencing effect of primary siRNAs. In addition, by high-throughput sequencing and qRT-PCR, we show that rde-10, rde-11, as well as several other dosage-sensitive rde genes (e.g., rsd-2, rsd-6 and haf-6) regulate the biogenesis/stability of an overlapping subset of endogenous WAGO class 22G siRNAs. A substantial fraction of these 22G siRNAs are secondary to ERGO-1 class 26G siRNAs and target recently acquired duplicated genes. Taken together, our results demonstrate that the dosage-sensitive rde genes are essential for secondary siRNA accumulation to promote efficient exogenous and endogenous RNAi in C. elegans.
RESULTS
Identification of the rde-10 gene from a forward genetic screen
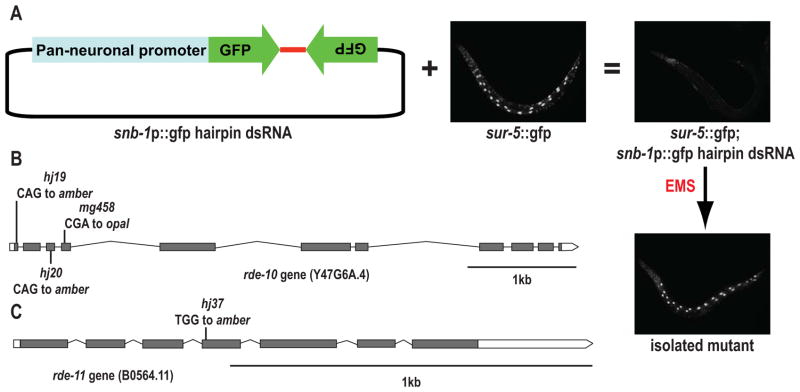
To identify factors required for RNAi or the spreading of silencing signals between tissues, we designed a transgene that initiates RNAi in the nervous system targeting a gfp fusion gene that is more broadly expressed. The RNAi-inducing transgene expresses a gfp hairpin dsRNA under the control of the pan-neuronal snb-1 promoter (Figure 1A). We introduced the neuronal RNAi-inducing transgene into sur-5::gfp transgenic C. elegans, which express nuclear-localized GFP in most somatic cells, and most prominently in intestinal cells. Although the in vivo expressed gfp dsRNA does not affect GFP expression in the neurons, consistent with previous observations showing that neurons are refractory to RNAi, gfp dsRNA spreads to other tissues and potently silences GFP signal in the intestine, muscle and hypodermis. Next we mutagenized the strain using ethyl methanesulfonate (EMS) and screened for mutations that reanimated GFP expression in somatic tissues.
Figure 1.
Identification of rde-10 and rde-11. (A) A schematic diagram of the EMS mutagenesis screen. (B and C) Schematic diagrams of the rde-10 and rde-11 genes and identified alleles. Black boxes, exons; white boxes, 5′ and 3′ UTRs. See also Figure S1.
Sixty-three mutants with a heritable GFP desilencing phenotype were isolated. The first gene identified was rde-10 encoded by Y47G6A.4 (Figure 1B). Three presumptive null alleles, mg458(Arg96Stop), hj19(Gln12Stop) and hj20(Gln73Stop), were identified from this screen and from an independent genetic screen for suppressors of transgene silencing (Yang and Mak, personal communication). The RDE-10 protein contains 627 amino acids (aa) and lacks obvious conservation outside of Caenorhabditis species but is highly conserved within this clade. rde-10 mutants were resistant to many different ingested dsRNAs that target germline-expressed or somatically-expressed genes, including pos-1, elt-2, lin-29, nhr-23, unc-15 and unc-54 dsRNAs (Figure S1A). A transgene comprised of a genomic DNA fragment containing the rde-10 coding sequence plus 770 bp upstream and 200 bp downstream including the probable 3′ UTR rescues an rde-10 null mutant to normal RNAi response to ingested lin-29 dsRNA, confirming that defects in the rde-10 mutants were directly caused by loss of rde-10 activity (Figures S1B–E).
Unlike certain RNAi pathway mutants that are sterile at elevated temperatures, rde-10 mutants produce a normal brood size compared to wild type at either 20°C or 25°C, indicating that rde-10 is not essential for germline development (Figure S1F) [21, 22].
RDE-10 and RDE-11 form a complex that mediates exogenous RNAi
To identify other RNAi pathway components that function together with RDE-10, we performed immunoprecipitation of epitope-tagged RDE-10 followed by tandem mass spectrometry analysis. Transgenic strains were generated expressing a 3XFLAG::GFP::rde-10 fusion construct under the control of rde-10 5′ and 3′ regulatory sequences. The epitope-tagged rde-10 transgene can rescue the RNAi defective phenotype of rde-10 mutants. A control strain was generated expressing a 3XFLAG::GFP fusion construct under the control of the same regulatory sequences. The RDE-10 protein complex was purified using the anti-GFP monoclonal antibody 3E6 or anti-FLAG monoclonal antibody M2 and components of the isolated protein complex were identified by tandem mass spectrometry analysis. Besides the RDE-10 protein, we identified ~84 proteins that coimmunoprecipitated with GFP and/or FLAG antibodies from 3XFLAG::GFP::rde-10-transgenic C. elegans, but were absent in control coimmunoprecipitations from 3XFLAG::GFP-transgenic and wild type C. elegans (Table S1). The most significant interactor of RDE-10, RDE-11, is encoded by B0564.11 (Table 1 and Figure 1C). The RDE-11 protein was present in all RDE-10 coimmunoprecipitation experiments and showed an abundance close to RDE-10 itself, implying that RDE-10 and RDE-11 form a tight complex (Table S1). The RING-type zinc-finger domain containing RDE-11 protein is 316 aa long and conserved in other nematodes at about the same level of conservation as RDE-10.
Table 1.
A major RDE-10 protein interactor is encoded by rde-11(B0564.11). See also Table S1.
| Gene | Sample | % coverage | Number of unique peptides | Number of total peptides | Total % coverage |
|---|---|---|---|---|---|
| rde-10 | rde-10 OE*, GFP IP | 56.1 | 35 | 53 | 72.2 |
| rde-10 SCI+, GFP IP | 26.6 | 14 | 21 | ||
| rde-10 OE, FLAG IP | 38.9 | 20 | 20 | ||
| rde-11 | rde-10 OE, GFP IP | 33.2 | 12 | 17 | 58.5 |
| rde-10 SCI, GFP IP | 21.2 | 6 | 6 | ||
| rde-10 OE, FLAG IP | 25.9 | 7 | 7 | ||
Overexpression,
Single-copy insertion
A mutant allele of rde-11, hj37(Trp117Stop), was isolated in the genetic screen for transgene desilencing (Figure 1C). Similar to rde-10 mutants, the rde-11 mutant is resistant to feeding RNAi but not temperature-sensitive sterile (Figures S1A and S1F). A mutant of a close paralog of rde-11, Y75B8A.10, does not show defects in RNAi (Figures S1G and S1H).
Several validated small RNA pathway components were identified in our proteomic analysis of RDE-10 interacting proteins (Table S1), including ERGO-1, a primary Argonaute associated with endogenous 26G siRNAs, RSD-2 and the RISC component VIG-1[6, 23, 24].
Mutations in rde-10 and rde-11 cause dosage-sensitive RNAi-defective phenotype
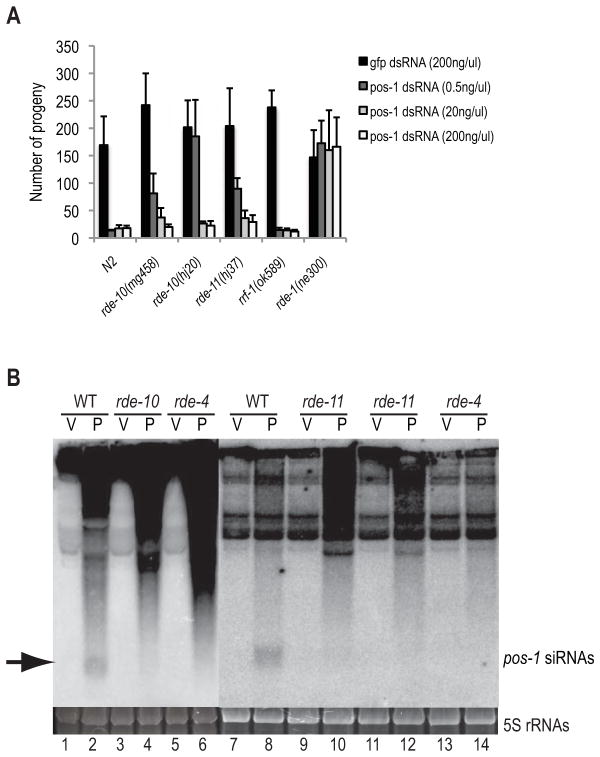
To dissect the roles of rde-10 and rde-11 in exogenous RNAi, we analyzed the response of rde-10 and rde-11 mutants to different concentrations of pos-1 dsRNA targeting a germline-essential gene. Feeding the worms with E. coli that produce dsRNAs delivers very low concentrations of dsRNAs, while injection can introduce dsRNAs at a wide range of concentrations. Wild type C. elegans were sensitive to feeding and injected pos-1 dsRNA and produced dead embryos (Figures 2A and S1A). rde-10 and rde-11 mutants were resistant to feeding pos-1 dsRNA and produced a full brood of viable progeny (Figure S1A). rde-10 and rde-11 mutants were partially resistant to pos-1 dsRNA injected at low concentrations (e.g., 0.5ng/μl), producing a brood of ~80 to 180 progeny, but were sensitive to pos-1 dsRNA injected at higher concentrations (e.g., 20ng/μl and 200ng/μl), causing embryonic lethality, similar to wild type (Figure 2A). The rrf-1 mutants were sensitive to pos-1 dsRNA even at low concentrations presumably because rrf-1 and ego-1 function redundantly in the germline for the biogenesis of secondary siRNAs (Figure 2A) [7, 8]. The rde-1 mutants were resistant to high concentrations of pos-1 dsRNA because these mutants do not produce functional primary or secondary siRNAs (Figure 2A). The dosage-sensitive phenotype of rde-10 and rde-11 mutants suggests that the RDE-10/RDE-11 complex regulates the efficacy of the exogenous RNAi pathway.
Figure 2.
rde-10 and rde-11 are dosage-sensitive RNAi-defective mutants. (A) Synthesized pos-1 dsRNA was injected into both gonads of WT, rde-10, rde-11, rrf-1 and rde-1 day one adult worms, and the total number of viable progeny was counted. gfp dsRNA was introduced by gonadal injection and served as controls for brood size. (B) Northern blot assays of pos-1 siRNAs (as indicated by the arrow). V and P: total RNAs extracted from worms fed with vector control and pos-1 dsRNA, respectively. rde-4 mutant was used as a control for lack of siRNA accumulation. 5S rRNAs stained with ethidium bromide are shown as a loading control.
To determine whether rde-10 and rde-11 affect siRNA accumulation in response to exogenous RNAi, Northern blot assays were done with RNA isolated from wild type, rde-10 and rde-11 mutant animals fed with pos-1 dsRNA. pos-1 siRNAs at about 21–24 nt were readily detectable in wild type (Figure 2B, lanes 1, 2, 7 and 8), but were absent in rde-10 and rde-11 mutant animals (Figure 2B, lanes 3, 4, and 9–12), as well as in rde-4 mutants that are defective at an early step in RNAi (Figure 2B, lanes 5, 6, 13 and 14) [2]. These results indicate that rde-10 and rde-11 are required for the formation or stability of siRNAs.
rde-11 is required for the biogenesis of secondary siRNAs in response to exogenous RNAi
To determine whether rde-10 and rde-11 regulate the biogenesis of either primary or secondary siRNAs, we analyzed small RNAs from WT and rde-11 mutant animals exposed to exogenous pos-1 dsRNA by high-throughput sequencing. Primary siRNAs bear 5′ monophosphates and can be captured by 5′ ligation using T4 RNA ligase during small RNA library generation [9, 10]. Secondary siRNAs contain 5′ triphosphates, which are resistant to ligation, and thus require treatment with tobacco acid pyrophosphatase (TAP) to reduce polyphosphates to monophosphates prior to ligation [9, 10]. Since secondary siRNAs are depleted in libraries in which the small RNAs are not first subjected to TAP treatment, we developed small RNA libraries from both non-treated and TAP treated RNA to distinguish primary and secondary siRNAs. In TAP treated samples, which contain both primary and secondary siRNAs, pos-1 siRNAs were reduced by ~93% in rde-11 mutants compared to WT (Figure 3A). In contrast, in non-treated samples, which are enriched for primary siRNAs, the levels of pos-1 siRNAs were similar in WT and rde-11 mutant animals (Figure 3B).
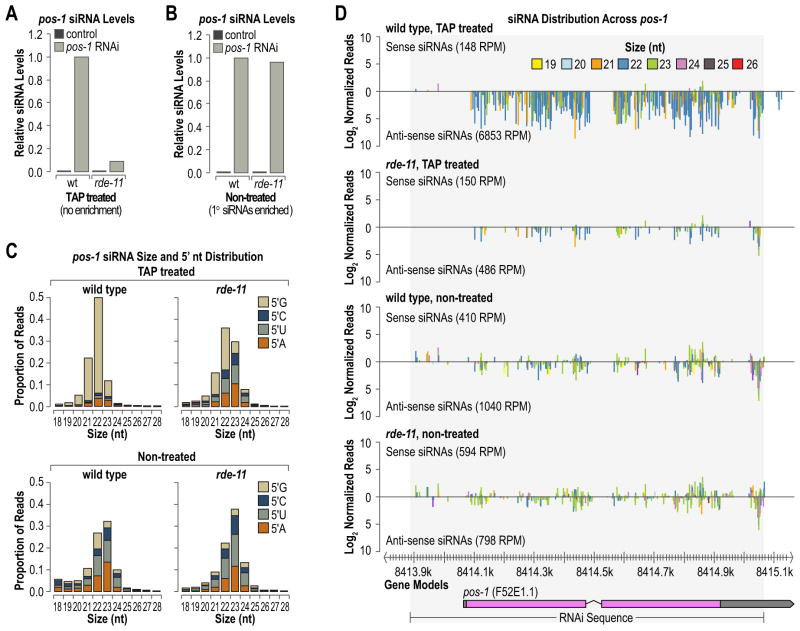
Figure 3.
rde-11 mutants fail to accumulate secondary siRNAs in response to exogenous pos-1 RNAi. (A and B) Ratio of pos-1 siRNA reads in rde-11 mutants to WT (WT = 1.0) from total non-treated (A) or TAP treated (B) RNA. (C) pos-1 siRNA size and 5′ nucleotide distribution in WT and rde-11 mutant worms. (D) siRNA distribution across the pos-1 genomic region in WT and rde-11 mutants. The shaded region indicates the sequence corresponding to the pos-1 dsRNA used for RNAi.
In WT TAP treated samples, pos-1 siRNAs were predominantly 22 nt long and contained 5′G, characteristics of secondary siRNAs (Figure 3C). siRNAs from the non-treated WT samples were predominantly 23 nt and contained a 5′U or 5′A (Figure 3C). This indicates that primary siRNAs are typically 23 nt, consistent with C. elegans Dicer products [3], and biased against a 5′G. In rde-11 TAP treated samples, the 22 nt 5′G-containing siRNAs were depleted and 23 nt 5′A, 5′T and 5′C-containing siRNAs were enriched (Figure 3C). When plotted along the pos-1 region of genomic DNA sequence, siRNAs from the TAP treated samples were predominantly 22 nt, in the antisense orientation and aligned to the pos-1 gene sequence (Figure 3D). In non-treated samples, siRNAs were predominantly 23 nt, of both sense and antisense orientation and derived from both the pos-1 gene sequence as well as upstream sequence corresponding to the exogenous dsRNA used for RNAi treatment (Figure 3D). In rde-11 mutants, siRNAs were uniformly depleted across pos-1 in TAP treated samples, but were more or less unaffected in non-treated samples (Figure 3D). Taken together, these data indicate that rde-11 is essential for the accumulation of secondary siRNAs, but dispensable for the biogenesis of primary siRNAs. Additionally, the results suggest that primary siRNAs can be distinguished from secondary siRNAs by their length and 5′ nt.
Mutations in rde-10 and rde-11 affect the levels of a subset of WAGO class endogenous 22G siRNAs downstream of ERGO-1 class 26G siRNAs
To determine if rde-10 and rde-11 act in the pathways that produce primary or secondary endogenous siRNAs, we used Taqman qRT-PCR to measure the levels of ERGO-1 class O1 26G siRNA (C40A11.10), ALG-3/4 class S5 26G siRNA (ssp-16) and WAGO class X-cluster 22G siRNA [14]. In rde-10 and rde-11 mutants, the levels of O1 26G siRNA were similar to wild type (Figure 4A). In contrast, mut-16 mutants displayed an ~98% reduction in O1 26G siRNA. The levels of S5 26G siRNA in rde-10 and rde-11 mutants were also indistinguishable from wild type, but greatly reduced in eri-1 mutants (Figures 4B). In contrast to 26G siRNAs, the X-cluster 22G siRNA was strongly depleted in rde-10 and rde-11 mutants (Figures 4C and 4D). However, a germline-expressed 22G siRNA, derived from eri-6 (T01A4.3), was unaffected in rde-10 mutants as determined by Northern blot (Figure 4D) [25]. These results indicate that rde-10 and rde-11 are essential for the formation or stability of a subset of endogenous siRNAs.
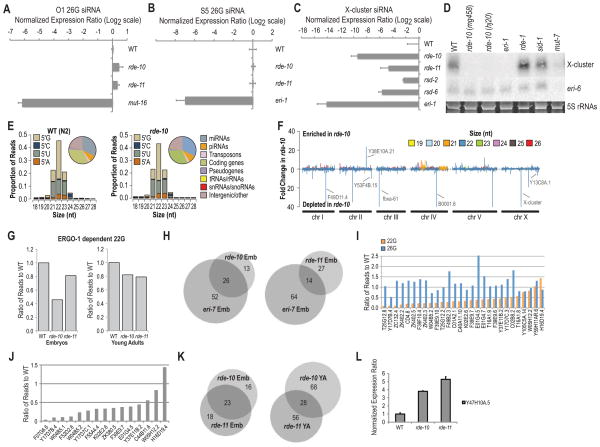
Figure 4.
rde-10 and rde-11 genes regulate the levels of a subset of WAGO class 22G siRNAs. (A) Ratio of O1 26G siRNA in rde-10 and rde-11 mutant embryos to WT after log2 transformation as determined by Taqman qRT-PCR (WT = 0; i.e., Log2 1.0). Total RNA extracted from mut-16 was used as a negative control. miR-35 was used for normalization. (B) Ratio of S5 26G siRNA in rde-10 and rde-11 mutant L4 larvae/young adults to WT after log2 transformation as determined by Taqman qRT-PCR (WT = 0; i.e., Log2 1.0). Total RNA extracted from eri-1 was used as a negative control. let-7 was used for normalization. (C) Ratio of X-cluster siRNA in various mutant strains at the adult stage to WT after log2 transformation as determined by Taqman qRT-PCR (WT = 0; i.e., Log2 1.0). Total RNA extracted from eri-1 was used as a negative control. miR-1 was used for normalization. All error bars are standard deviations calculated from three technical replicates. (D) Northern blot assays of X-cluster and eri-6 (T01A4.3) siRNAs. 5S rRNAs stained with ethidium bromide are shown as a loading control. Total RNAs were extracted from day one gravid adult worms. eri-1 mutant was used as a negative control for X-cluster siRNA expression, while mut-7 mutant was used as a negative control for X-cluster and eri-6 (T01A4.3) siRNA expression. (E) Small RNA size and 5′ nucleotide distribution in WT and rde-10 mutant day one gravid adult worms. (Insets) Pie charts display the proportion of reads corresponding to the indicated features. (F) Enrichment or depletion of small RNAs in rde-10 mutant worms relative to WT worms. Total small RNA reads were calculated for 5 kb bins in 1 kb increments across each chromosome. (G) Ratio of 22G siRNA reads derived from ERGO-1 class 26G siRNA target mRNAs in rde-10 and rde-11 mutants to WT (WT = 1.0) at embryo and L4 larvae/young adult stages. (H) Venn diagrams of rde-10, rde-11 and eri-7 dependent siRNAs. (I) Ratio of NRDE-3 class 22G and upstream ERGO-1 class 26G siRNA reads in rde-10 embryos to WT embryos (WT = 1.0) (J) Ratio of RRF-3 dependent 22G siRNA reads from somatic target mRNAs in rde-10 and rde-11 mutant embryos to WT embryos (WT = 1.0). (K) Venn diagrams of rde-10 and rde-11 dependent siRNAs at embryo and L4 larvae/young adult stages. (L) Ratio of Y47H10A.5 target mRNA levels in rde-10 and rde-11 mutants to WT (WT = 1.0) as determined by qRT-PCR. All error bars are standard deviations calculated from three technical replicates. See also Figure S2 and Tables S2-S7.
To more broadly assess the roles of rde-10 and rde-11 in endogenous small RNA pathways, we performed Illumina deep sequencing of small RNA cDNA libraries derived from wild type, rde-10 and rde-11 mutants at embryo and young adult stages, as well as wild type and rde-10 mutants at the day one gravid adult stage (Figures S2A–C). Small RNA libraries from adult wild type and rde-10 mutant animals showed similar size and 5′ nt distribution, with the most abundant species being 22 nt long and containing a 5′G (Figure 4E). miRNAs and siRNAs derived from predicted coding genes were the predominant classes of small RNAs (Figure 4E, insets). None of the known classes of small RNAs were broadly depleted in rde-10 mutants, although several clusters of 22G siRNAs were strongly reduced (Figure 4F). Similar results were observed in rde-10 and rde-11 embryo and young adult libraries (Figures S2D–G). Consistent with our qRT-PCR results (Figures 4A and 4B), both the ERGO-1 and ALG-3/4 classes of 26G siRNAs were unaffected in rde-10 and rde-11 mutants (Figures S2D–E, the 26 nt peaks in the bar plots are either ERGO-1 class 26G siRNAs in the embryo libraries (Figure S2D) or ALG-3/4 class 26G siRNAs in the young adult libraries (Figure S2E)).
To determine if rde-10 and rde-11 function in a specific endogenous RNAi pathway, we assessed the siRNAs depleted in rde-10 and rde-11 mutants for their dependence on other endogenous siRNA factors using small RNA deep sequencing datasets from mut-16, eri-7 and rrf-3 mutants and NRDE-3 and ERGO-1 coimmunoprecipitation assays [13, 15, 16, 18, 19]. mut-16 is essential for the accumulation of WAGO class but not CSR-1 class 22G siRNAs derived from thousands of genes [12, 19]. eri-7 and rrf-3 are required for the accumulation of ERGO-1 class 26G siRNAs and their downstream WAGO class 22G siRNAs that interact with the Argonaute NRDE-3 [15, 18]. In rde-10 mutant gravid adults, 37 features, which includes coding genes, pseudogenes and transposons, that yielded ≥10 siRNA reads per million total small RNA reads (RPM) in at least one of the libraries analyzed, were depleted of siRNAs by 67% or more, relative to wild type (Table S2). Of these, 34 were also reduced in mut-16 mutant gravid adults (Table S2) [19]. Thirty-nine features were depleted of siRNAs in rde-10 mutant embryos, of which 33 were also reduced in mut-16 mutant embryos (Tables S3 and S4). These results indicate that rde-10 is required for the accumulation of a subset of WAGO class 22G siRNAs. The majority of rde-10 target genes have multiple paralogs in the genome, which is characteristic of target genes regulated by the ERGO-1 class 26G siRNAs and their downstream WAGO class 22G siRNAs [16, 18]. 22G siRNA reads derived from ERGO-1 targets were reduced by 60% and 20% in rde-10 mutant embryo and young adult libraries, respectively (Figure 4G and Tables S3 and S5). There was no reduction in the corresponding upstream 26G siRNAs from ERGO-1 targets. Of the 39 features depleted of siRNAs in rde-10 mutant embryos, 26 were also depleted of siRNAs in eri-7 mutant embryos (Figure 4H) [18]. Of the 116 NRDE-3 targets analyzed [13], 28 yielded ≥ 10 RPM in either rde-10 mutant or wild type embryo libraries of which 23 were depleted of 22G siRNAs by ≥50% in rde-10 mutants (Figure 4I). In contrast, 26G siRNA reads from each of the NRDE-3 targets analyzed were unchanged or slightly elevated in rde-10 mutants relative to wild type (Figure 4I). Of 23 previously identified RRF-3 target genes [15], 15 yielded ≥10 RPM 22G siRNAs in the wild type embryo library and of these, 12 had ≥50% or more decrease in 22G siRNA reads in the rde-10 mutant embryos relative to wild type (Figure 4J).
In rde-11 mutants, a subset of WAGO class 22G siRNAs was also depleted (Figures S2D–G). Twenty-one out of 41 features depleted of siRNAs by 67% or more in rde-11 mutant embryos were also depleted of siRNAs in mut-16 embryos (Tables S4 and S6). 22G siRNA reads derived from ERGO-1 target genes were reduced by 20% in both rde-11 mutant embryos and young adults relative to wild type (Figure 4G). This modest reduction was likely caused by substantial decreases in 22G siRNA reads derived from a small subset of ERGO-1 target genes (Figure 4H and Tables S6–7). There was partial overlap in the features depleted of siRNAs in rde-10 and rde-11 mutant embryos and young adults (Figure 4K and Tables S3 and S5–7).
WAGO class 22G siRNAs downregulate their targets by triggering both mRNA degradation and co-transcriptional silencing [12, 13]. The mRNA levels of most of the rde-10 and rde-11 regulated genes were unchanged in the mutants compared to wild type as measured by qRT-PCR, presumably because of moderate decrease of siRNA levels. However, the mRNA levels of Y47H10A.5 gene, which yielded high number of siRNA reads in the wild type and the ~80% fewer reads in the mutants, were upregulated by 4- to 5-fold in rde-10 and rde-11 mutants compared to wild type (Figure 4L). The accumulation of Y47H10A.5 siRNAs requires RDE-1 [26], consistent with a similar role of the RDE-10/RDE-11 complex downstream of RDE-1 in endogenous RNAi.
Taken together, these results indicate that rde-10 and rde-11 are essential for the accumulation of a subset of WAGO class 22G siRNAs that are primarily derived from ERGO-1 targets.
rsd-2, rsd-6 and haf-6 mutants display siRNA defects that overlap with rde-10 and rde-11 mutants
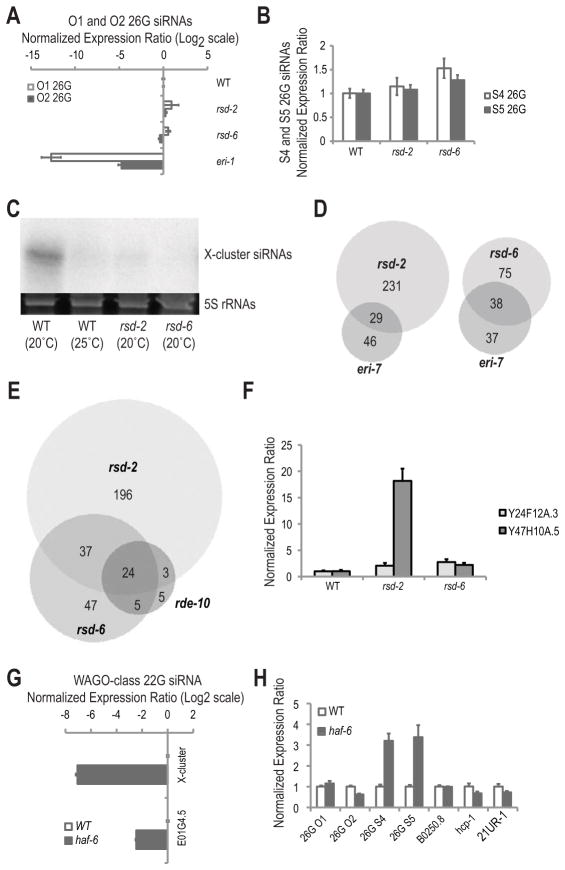
RSD-2 interacts with RDE-10 and the TUDOR domain protein RSD-6 (Table S1) [23]. Given that rsd-2 and rsd-6 mutants show similar dosage-sensitive RNAi-defective phenotype to rde-10 and rde-11 mutants, we investigated the specific roles of these genes in endogenous small RNA pathways. We measured the levels of ERGO-1 class O1 (C40A11.10) and O2 (E01G4.7) 26G siRNAs, as well as ALG-3/4 class S4 (deps-1) and S5 (ssp-16) 26G siRNAs by Taqman qRT-PCR assays (Figures 5A and 5B) [14]. The levels of these 26G siRNAs were similar to wild type in rsd-2 and rsd-6 mutants (Figures 5A and 5B). However, the levels of the WAGO class X-cluster 22G siRNA were strongly depleted in rsd-2 and rsd-6 mutants relative to wild type by both Northern blot assay and Taqman qRT-PCR (Figures 4C and 5C).
Figure 5.
rsd-2, rsd-6 and haf-6 are required for the accumulation of a subset of WAGO class 22G siRNAs. (A) Ratio of O1 and O2 26G siRNAs in rsd-2 and rsd-6 mutant embryos to WT embryos after log2 transformation as determined by Taqman qRT-PCR (WT = 0; i.e., Log2 1.0). Total RNA extracted from eri-1 was used as a negative control. miR-35 was used for normalization. (B) Ratio of S4 and S5 26G siRNAs in rsd-2 and rsd-6 mutant strains to WT at L4 larval/young adult stage as determined by Taqman qRT-PCR (WT = 1.0). miR-1 was used for normalization. (C) Northern blot assays of X-cluster siRNA. 5S rRNAs stained with ethidium bromide are shown as a loading control. Total RNA extracted from wild type N2 strain raised at the elevated temperature 25°C was used as a negative control of X-cluster siRNA expression [37]. (D) Venn diagrams of rsd-2, rsd-6 and eri-7 dependent siRNAs. (E) Venn diagram of rsd-2, rsd-6 and rde-10 dependent siRNAs. (F) Ratio of Y24F12A.3 and Y47H10A.5 target mRNA levels in rsd-2 and rsd-6 adults to WT adults (WT = 1.0) as determined by qRT-PCR. (G) Ratio of X-cluster and E01G4.5 22G siRNAs in haf-6 mutant embryos to WT embryos after log2 transformation as determined by Taqman qRT-PCR (WT = 0; i.e., Log21.0). miR-35 was used for normalization. (H) Ratio of a panel of representative small RNAs in haf-6 mutant to WT as determined by Taqman qRT-PCR (WT = 1.0). For O1, O2, B0250.8 and hcp-1 siRNA assays, total RNAs were extracted from embryos and miR-35 was used for normalization. For S4 and S5 siRNA assays and the 21UR-1 piRNA assay, total RNAs were extracted from L4 larvae/young adult worms and miR-1 was used for normalization. All error bars are standard deviations calculated from three technical replicates. See also Figure S3 and Tables S8-S9.
To more comprehensively examine the roles of rsd-2 and rsd-6 in endogenous small RNA pathways, we performed Illumina deep sequencing of small RNA cDNA libraries derived from wild type, rsd-2 and rsd-6 mutant day one gravid adults (Figure S2A). The levels of 22G siRNAs derived from several clusters across the genome were strongly reduced in rsd-2 and rsd-6 mutants relative to wild type (Figures S3A–B). Two hundred and sixty features were depleted of siRNAs by ≥67% in rsd-2 mutants, of which 223 were also depleted in mut-16 mutants (Figure S3C and Table S8) [19]. One hundred and thirteen features were depleted of siRNAs in rsd-6 mutants and 106 of these also had reduced siRNA levels in mut-16 mutants (Figure S3D and Table S9) [19]. Of the features depleted of siRNAs in rsd-2 and rsd-6 mutant day one gravid adults, 29 and 28 were also depleted of siRNAs in eri-7 mutants, respectively (Figure 5D) [18]. There was also considerable overlap between features that were depleted of siRNAs in rsd-2, rsd-6 and rde-10 mutants (Figure 5E). In addition to their roles in the accumulation of ERGO-1 dependent 22G siRNAs which express in the soma, rsd-2 and rsd-6 also function in the germline. Ninety-nine and 44 features that had reduced siRNA levels in rsd-2 and rsd-6 mutants, respectively, were derived from genes that are enriched for siRNAs in the germline (Figures S3E and S3F) [12]. To determine if the reductions in siRNA levels observed in rsd-2 and rsd-6 mutants results in upregulation of the target mRNAs, we measured the levels of Y24F12A.3 and Y47H10A.5 mRNAs by qRT-PCR (Figure 5F). Y24F12A.3 mRNA levels were increased for more than 2-fold in rsd-2 and rsd-6 mutants compared to the wild type. Y47H10A.5 mRNAs were elevated for 18.2- and 2.2-fold in rsd-2 and rsd-6 mutants, respectively.
haf-6 mutants also show dosage-sensitive RNAi-defective phenotype [27]. haf-6 encodes a half-molecule ATP-binding cassette (ABC) transporter protein. In order to assess the specific requirements of haf-6 in endogenous small RNA pathways, we measured the levels of several small RNAs using Taqman qRT-PCR assays. The levels of ERGO-1-dependent WAGO class X-cluster and E01G4.5-derived 22G siRNAs were substantially reduced in haf-6 mutants relative to wild type (Figure 5G). In contrast, a germline-enriched WAGO class 22G siRNA derived from B0250.8, whose biogenesis is independent of ERGO-1 pathway components, was unaffected (Figure 5H). In addition, the levels of ERGO-1 class O1 and O2 26G siRNAs, CSR-1 class hcp-1-derived 22G siRNA, as well as piRNA 21UR-1 were unchanged in haf-6 mutants relative to wild type (Figure 5H) [12, 20, 28]. Interestingly, ALG-3/4 class 26G siRNAs were moderately elevated in haf-6 mutants, indicating that haf-6 may negatively regulate the biogenesis of ALG-3/4 class 26G siRNAs (Figure 5H).
Taken together, our results indicate that rsd-2, rsd-6 and haf-6 regulate the levels of a subset of WAGO class 22G siRNAs, many of which are downstream of ERGO-1 class 26G siRNAs and are also dependent on rde-10 and rde-11.
DISCUSSION
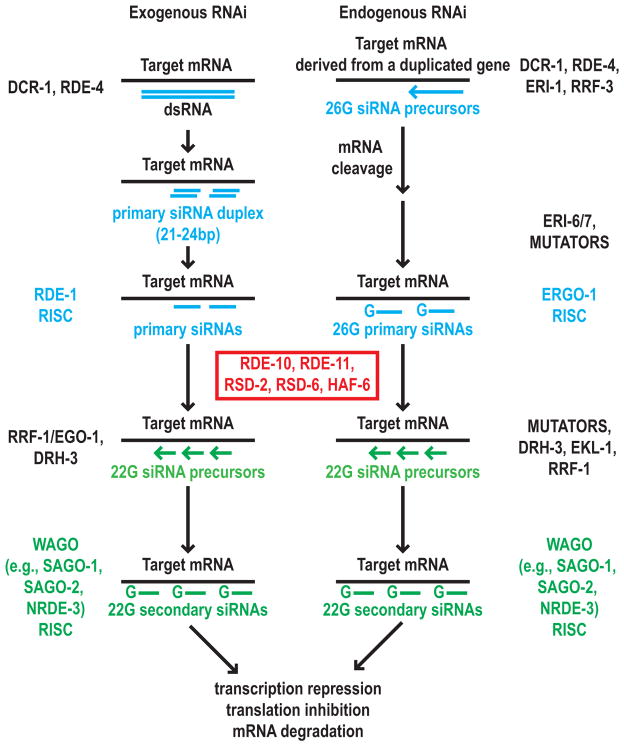
In this study we characterized a new protein complex containing RDE-10, RDE-11, and RSD-2 that promotes the amplification of C. elegans RNAi response. We showed that these dosage-sensitive RNAi-defective genes affect the biogenesis and/or stability of secondary siRNAs in both exogenous and endogenous RNAi pathways (Figure 6). When exogenous dsRNAs are present at high concentrations, enough primary siRNAs can be generated by DCR-1 to trigger an RNAi response, and the function of RDE-10/RDE-11 complex is dispensable. However, at low dsRNA concentrations, secondary siRNA amplification, which requires the RDE-10/RDE-11 complex, is essential to mediate RNAi. Therefore, these genes promote the effectiveness of exogenous RNAi.
Figure 6.
A model of the roles of the dosage-sensitive RNAi-defective genes in both exogenous and endogenous RNAi pathways.
We showed that rde-10, rde-11, rsd-2, rsd-6 and haf-6 genes are required for the accumulation of a subset of WAGO class 22G siRNAs (Figure 6). Consistent with the identification of ERGO-1 in RDE-10 proteomics, our data indicated that some of these 22G siRNAs act downstream of ERGO-1 class 26G siRNAs and silence target genes expressed in the soma. In our model (Figure 6), primary 26G siRNA biogenesis requires DCR-1 and RRF-3, and the target mRNAs are cleaved by DCR-1 during the biogenesis of primary siRNA duplex [18]. The primary Argonaute ERGO-1 interacts with the primary siRNA duplex and degrades the passenger strand [18]. The association between the ERGO-1-bound primary siRNA and target mRNA leads to recruitment of DRH-3 and RRF-1 for secondary 22G siRNA biogenesis [12]. DRH-3 and RRF-1 are not present in the RDE-10 protein complex. However, we identified ERGO-1 and the RISC component VIG-1 in RDE-10 proteomics. The RDE-10 complex may bridge the step between primary siRNA target recognition and RDRP recruitment to facilitate secondary siRNA accumulation. These somatically expressed secondary siRNAs interact with SAGO-1 and SAGO-2 in the cytoplasm [6, 12]. These secondary Argonautes do not have endonuclease activity and may recruit other cellular mRNA degradation machinery for target mRNA decay. Secondary siRNAs also interact with NRDE-3 in the nucleus and silence target gene expression co-transcriptionally [13]. To date, the only published proteomics studies in the C. elegans RNAi field are the characterization of DCR/ERI and DRH-3 complexes essential for endogenous 26G and 22G siRNA production, respectively [12, 25]. Our data suggest that the RDE-10 complex is distinct from and likely functions subsequent to the DCR/ERI complex and before the DRH-3 complex.
Some of the RDE-10 interacting proteins have been identified as potential regulators of small RNA pathways in previous RNAi-based genome-wide screens, but have not been further characterized (e.g., MATH-33 and PAB-1). The MATH-33 protein has a meprin-associated Traf homolog (MATH) domain and also shares sequence homology with ubiquitin carboxyl-terminal hydrolases. MATH-33 is required for cosuppression in the C. elegans germline, an RNAi-related pathway in which introduction of exogenous repetitive sequences causes repression of the endogenous homologous genes [29]. PAB-1 is a polyadenylate-binding protein that plays a role in at least two RNAi-dependent processes, transposon silencing in the germline as well as transcriptional silencing of a transgene in the soma [30, 31]. PAB-1 forms a complex with ATX-2, the C. elegans ataxin-2 ortholog [32]. We also identified ATX-2 in the RDE-10/RDE-11 protein complex.
We showed that rsd-2, rsd-6, and haf-6 genes regulate the levels of a subset of WAGO class 22G siRNAs. RSD-2 and RSD-6 associate with endoplasmic reticulum, while HAF-6 is a plasma membrane protein [23, 27, 33]. It would be intriguing to identify the physical interactors of these membrane-associated RNAi factors. A subset of germline-enriched siRNAs were depleted in rsd-2 and rsd-6 mutants which may explain the germline-related phenotypes of these mutants: rsd-2 mutants display elevated levels of germline transposon mobilization at high temperatures, while rsd-6 mutants display high incidence of males, as a result of an increase in X chromosome nondisjunction, and sterility at elevated temperatures [33]. Our data suggest that these dosage-sensitive RNAi-defective genes encode amplification phase factors that promote and extend the potency of RNAi-based surveillance.
EXPERIMENTAL PROCEDURES
Bristol N2 was used as control wild-type strain. All strains were incubated at 20°C unless otherwise described. Large-scale immunoprecipitation was performed based on a previously published protocol [34]. dsRNA synthesis and injection were carried out as described [35]. Total RNA isolation, small RNA Northern blot, small RNA deep sequencing, Taqman small RNA qRT-PCR and target mRNA qRT-PCR were performed as described [19, 36]. Detailed experimental procedures are in Supplemental Information. The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSEXXXXX).
Supplementary Material
HIGHLIGHTS.
rde-10 and rde-11 are required for low-dosage RNAi
rde-10 and rde-11 regulate a subset of endogenous siRNAs
rsd-2, rsd-6 and haf-6 act in the same pathway as rde-10 and rde-11
RDE-10 and RDE-11 form a complex specifically required for siRNA amplification
Acknowledgments
We are very grateful to Huan Yang and Ho Yi Mak for rde-10 and rde-11 mutant and transgenic strains, as well as communicating unpublished data. We thank Darryl Conte, Weifeng Gu, Pedro Batista, and Daniel Chaves for providing protocols for immunoprecipitation and generation of small RNA cDNA libraries for deep sequencing; Xiaoyun Wu for protocols for large-scale immunoprecipitation; John Asara for mass spectrometry analysis; Christian Daly, Jiangwen Zhang, Mark Borowsky, Tammy Gillis and Kaleena Shirley for Illumina deep sequencing and computational assistance; the Caenorhabditis Genetics Center and Shohei Mitani for strains. This work was supported by a grant to G.R. from National Institute of Health (GM44619), fellowships to C.Z. and S.E.F. from the MGH ECOR Fund for Medical Discovery, fellowship to T.A.M. from the Damon Runyon Cancer Research Foundation (DRG-2029-09), fellowship to S.M.G. from the American Heart Association (0825938D), and fellowships to C.G.R. from the European Molecular Biology Organization and the Human Frontier Science Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 3.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 6.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 8.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 9.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 10.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 11.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011;7:e1002369. doi: 10.1371/journal.pgen.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 23.Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol. 2004;14:111–116. doi: 10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 24.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 25.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 26.Correa RL, Steiner FA, Berezikov E, Ketting RF. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000903. doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundaram P, Echalier B, Han W, Hull D, Timmons L. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3678–3688. doi: 10.1091/mbc.E06-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vastenhouw NL, Fischer SE, Robert VJ, Thijssen KL, Fraser AG, Kamath RS, Ahringer J, Plasterk RH. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 31.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- 33.Han W, Sundaram P, Kenjale H, Grantham J, Timmons L. The Caenorhabditis elegans rsd-2 and rsd-6 genes are required for chromosome functions during exposure to unfavorable environments. Genetics. 2008;178:1875–1893. doi: 10.1534/genetics.107.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 37.Habig JW, Aruscavage PJ, Bass BL. In C. elegans, high levels of dsRNA allow RNAi in the absence of RDE-4. PLoS One. 2008;3:e4052. doi: 10.1371/journal.pone.0004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.