Abstract
Using a combination of X-ray scattering, fluorescence correlation spectroscopy, coarse-grained molecular dynamics (MD) simulations and potential of mean force calculations, we have explored the membrane remodeling effects of monomeric α-synuclein (αS). Our initial findings from multiple approaches are that αS (1) causes a significant thinning of the bilayer and (2) stabilizes positive mean curvature, such that the maximum principle curvature matches that of synaptic vesicles, αS-induced tubules, and the synthetic lipid vesicles to which the protein binds most tightly. This suggests that αS binding to synaptic vesicles likely stabilizes their intrinsic curvature. We then show that αS induces local negative Gaussian curvature, an effect that occurs in regions of αS shown previously via NMR and corroborated by MD simulation to have significant conformational flexibility. The induction of negative Gaussian curvature, which has implications for all curvature-sensing and curvature-generating amphipathic α-helices, supports a hypothesis that connects helix insertion to fusion and fission of vesicles, processes that have recently been linked to αS function. Then, in an effort to explain these biophysical properties of αS, we promote an intrinsic curvature-field model that recasts long-range protein–protein interactions in terms of the interactions between the local curvature fields generated by lipid–protein complexes.
Introduction
Proteins that induce and stabilize membrane curvature facilitate fusion and fission of phospholipid vesicles. Two modes of curvature that are associated with the formation of fusion and fission intermediates are positive mean curvature and negative Gaussian (or saddle) curvature. For example, positive mean curvature is associated with the strongly curved membrane bulge present in early fusion intermediates.1 While mean curvature is more easily characterized than Gaussian curvature, the two can be intimately linked. In both fusion and fission, the presence of positive mean curvature requires a rim of negative Gaussian curvature (i.e., Helfrich's hat model2) to allow for the transition from the source vesicle to the bulging fusion/fission intermediate. Membrane fusion is rendered relatively more favorable by a decrease in the magnitude of the negative Gaussian curvature modulus (proportional to the energy of Gaussian curvature), whereas fission is rendered relatively more favorable by an increase in the magnitude of the negative Gaussian curvature modulus.3
Some amphipathic, α-helical proteins are known to sense and generate curvature in lipid bilayers.4 In certain cases these proteins enhance vesicle fusion through inducing a membrane bulge (e.g., the membrane proximal region of gp41 from HIV-1),3 while in other cases they can enhance vesicle fission (e.g., N-BAR protein).5,6 Positive mean curvature induction and sensing has been studied extensively, through experiments on model systems,6–9 molecular dynamics simulations,10–14 mesoscopic modeling,15–17 and theory,3,18–20 and a consensus is building as to the physical driving forces that dictate this phenomenon. Only recently has a hypothesis been put forth that connects amphipathic helix insertion to membrane fission, positing that protein-induced positive spontaneous curvature results in significant negative values of the Gaussian curvature modulus.3
A number of amyloid proteins form amphipathic α-helices upon binding to anionic lipid membranes. These proteins and their associated diseases include Aβ in Alzheimer's disease, islet amyloid polypeptide (IAPP) in type II diabetes, and α-synuclein (αS) in Parkinson's disease (PD), which is the focus of this study. αS is a 140 amino acid, natively unfolded protein that is the major proteinaceous component of insoluble fibrillar Lewy bodies, a hallmark of PD.21 The precise roles of both native and pathological forms of αS remain unclear, including the range of physiologically observed oligomeric states.22 However, the interaction of monomeric αS with cellular membranes is now thought to be critical to its native function and potentially to its role in PD as well.21,23 Multiple lines of evidence suggest that native αS is involved in regulating synaptic vesicle trafficking, fusion, and fission.9,21,24-27 Recently, Kamp et al. showed that αS is capable of inhibiting fusion of small unilamellar vesicles (SUVs), leading to their hypothesis that αS inhibits membrane fusion by stabilizing the highly curved, stressed bilayer.23 αS has also very recently been shown to fragment mitochondrial membranes, with the suggestion that this action reflects the protein's ability to induce membrane fission.28 Significantly, the physical mechanisms through which monomeric αS exerts its influence on vesicle fusion and fission have not been described.
We have previously shown that monomeric αS binds with substantially greater affinity to lipid vesicles of high curvature.9 Specifically, αS displays only a weak curvature dependence for binding to large unilamellar vesicles (LUVs) ranging in diameter from 75 to 180 nm; however, the binding affinity increases by more than an order of magnitude for ∼45 nm diameter SUVs, which most closely mimic the size of synaptic vesicles. Additionally, it has been shown that at very high protein:lipid ratios (1:40) αS induces vesiculation and tubulation of giant unilamellar vesicles (GUVs), where the tubules have a diameter of ∼40 nm.7 Vesicle curvature has therefore emerged as a central theme in understanding the native function of αS.
Several biophysical studies have suggested that binding of monomeric αS to small vesicles stabilizes their inherent curvature, thereby reducing their propensity to fuse.23,27,29 It is therefore of interest to characterize the structural and mechanical changes induced in a membrane by αS. Here, we have used a combination of X-ray scattering and molecular dynamics (MD) simulation to probe αS-induced thickness changes and curvature induction in membranes built (both in vitro and in silico) to mimic GUVs. We report four major remodeling effects. Namely, αS (1) thins the membrane, (2) stabilizes regions of positive mean curvature, (3) induces negative Gaussian curvature, and (4) slows membrane dynamics.
Results
Low-angle X-ray scattering (LAXS) experiments on oriented bilayer stacks ± αS were performed to interrogate the effect of the protein on the thickness of the bilayer. The bilayers were a mixture of 1-palmitoyl-2-oleoyl-sn-phosphatidyl-L-serine (POPS)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (PS/PC) at a 1:3 mole ratio. The αS sample was prepared at a 1:200 protein:lipid ratio. As in previous studies, we used a truncated αS (residues 1–100), which encompasses the complete extended helix lipid binding domain.29,30 The protein concentration used here is an order of magnitude lower than the αS binding saturation level (protein:lipid ratio of 1:20) and was chosen to minimize potential αS–αS interactions while providing high enough signal-to-noise ratio for LAXS experiments.7,9,31 At this concentration we did not observe any vesiculation or tubulation, and there were no observable specific αS–αS interactions or aggregation, as measured by fluorescence correlation spectroscopy (see Figure S1 in the Supporting Information); however, at this protein:lipid ratio nonspecific interactions are likely present. The partition coefficient of truncated αS was found to be 2.55 × 105 ± 4.22 × 104, which suggests almost complete binding of the protein under our conditions.
Figure 1A presents the experimentally derived bilayer form factors (Fexpt(qz)) for both pure and αS-bound bilayers and clearly indicates that αS binding thins the membrane (the curve shifts to higher q). Extracting detailed structural information from Fexpt(qz), e.g., the extent of the thinning and the location of the protein, is nontrivial.32–35 We have taken two approaches to do so. First, we have adapted a recently developed modeling approach (the scattering-length density profile, SDP, model) which relies on the conservation of volume probabilities to determine the overall electron density profile (from which bilayer thickness is extracted as the head-to-head spacing, DHH).36 Specifically, the SDP model fits experimental scattering data (either X-ray or neutron) by jointly minimizing χ2 (evaluating the goodness of fit of FSDP(qz) to Fexpt(qz)) and a penalty term (based on expected ranges for bilayer structural parameters previously determined from atomistic MD simulations). The resulting component volume probabilities can be scaled by the electron content of each group and summed to determine the electron density profile for the system. One solution to the SDP model, FSDP(qz), is given in the overlay in Figure 1A and suggests that αS induces a ∼1 Å decrease in DHH (see the inset in Figure 1A). While variable solutions of the SDP model consistently showed this extent of αS-induced membrane thinning, such an approach is limited in its capacity to elucidate details of protein-induced structural changes to the membrane, including curvature effects. We therefore utilized coarse-grained molecular dynamics (CGMD) simulations (specifically the MARTINI force field37–39) as a second, more robust approach to resolve large-scale changes to membrane structure and dynamics. In the past several years, CGMD has been used extensively to study membranes and the effects of proteins on their structure.11–13,40,41
Figure 1.
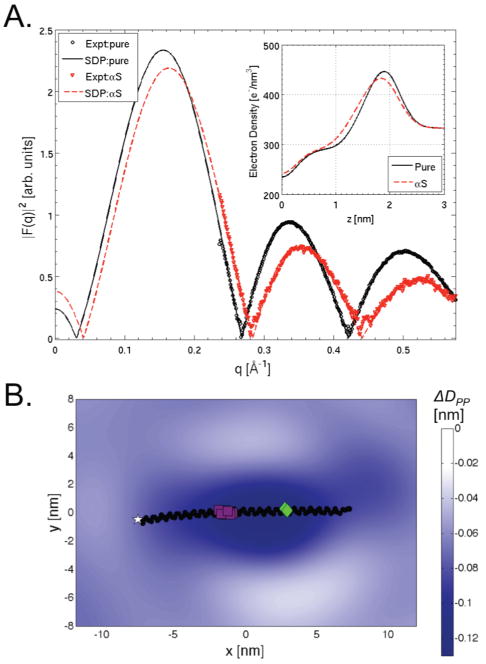
Exploring bilayer thickness. (A) LAXS (expt) and model-fit (SDP) bilayer form factors for pure lipid (black) and αS (red) samples. (B) ΔDPP surfaces plotted versus local (relative to αS) x and y axes for asymmetric CGMD simulations with the average reoriented protein position in black (the white star indicates the N-terminus, purple squares indicate the linker region, and green tilted squares indicate GLY67-GLY68). Color map units are nanometers.
In the sample preparation for the X-ray experiments, the protein is reconstituted into the bilayers through the organic phase and hence is bound to both leaflets. In contrast, in the more physiological scenario αS is bound only to the outer leaflet of a vesicle. Thus, to study both scenarios, we simulated four different system configurations: one pure PS/PC control system and three αS/PS/PC systems (αS on both leaflets, mimicking the X-ray experiments, referred to as the symmetric αS system; αS only on the outer leaflet, mimicking the presumed scenario of αS bound to a vesicle, referred to as the asymmetric αS system; and a single αS on the outer leaflet, referred to as the single-αS system, used to determine protein-linked curvature effects in the absence of protein–protein interactions).
The pure and symmetric αS systems both contained 3200 lipids (1:3 PS/PC), with the protein system having a 1:200 protein:lipid ratio (8 proteins per leaflet). The asymmetric αS system contained 3016 lipids and a 1:377 protein:lipid ratio, with 8 proteins bound only to the outer leaflet, and the single-αS system had a 1:3176 protein:lipid ratio. The overall protein concentration was necessarily reduced in the asymmetric αS system to maintain a controlled degree of αS–αS interaction in the leaflet (relative to the symmetric simulation and to the experiment). Because insertion of αS into only one leaflet would inevitably lead to a lateral area mismatch between the two leaflets (and therefore a nonspecific induction of curvature), an extensive series of test simulations were run to determine how many lipids to remove from the outer leaflet such that the lateral area of both leaflets would be the same. Comparison of these test simulations provided a measure of sensitivity of the curvature fields to area mismatch due to an incorrect number of lipids. The effective lateral area per αS was equivalent to ∼23 lipids, with a sensitivity of approximately ±3 lipids having no discernible effect on the local curvature field (discussed below). This is consistent with the experimental value of 14–16 Å2 per residue of an amphipathic helix bound to a lipid/water interface42 (assuming an average surface area of 65 Å2 per phospholipid in a PC–lipid bilayer43). This calculated result also agrees very well with our previous experimental finding.9 We therefore removed a total of 184 lipids from the asymmetric αS monolayer (a total of 24 lipids were removed in the single-αS system). Each system was simulated for a total of 40 μs, with analysis done with 4 ns resolution across the last 24 μs.
Regarding the protein itself, αS has been shown to adopt multiple distinct conformations, either an extended or a horseshoe helix, depending on the substrate curvature and composition. While the horseshoe helix was first proposed for detergent micelles and may also exist on lipid vesicles containing unphysiologically high concentrations of negatively charged lipids, electron spin resonance (ESR) and single-molecule fluorescence resonance energy transfer (smFRET) suggest that the dominant form of αS on more physiologically sized vesicles is the extended helix; we therefore chose our initial αS starting configuration in this conformation.30,44 It is important to point out that this initial choice of secondary structure is constrained in the MARTINI CGMD force field, precluding any unfolding.38 We have therefore run two sets of simulations for each system configuration (seven simulations in total) to minimize any associated artifacts. In the first, we modeled αS secondary structure as a continuous α-helix. While this conformation is unable to unfold into the horseshoe conformations, we observed significant flexibility (bending) of the protein in all directions throughout the simulations. In the second set of simulations, we simulated a broken helix where residues 38–44 were modeled as a random coil (referred to as the linker region). This model is consistent with the sodium dodecyl sulfate (SDS) micelle-bound NMR structure45 and our previous MD study.46 Throughout the broken-helix simulations (results for which are presented in the Supporting Information), the protein sampled both extended and horseshoe conformations, though consistent with experiment, the extended conformation was considerably more probable (see Figure S2, Supporting Information).43 For simplicity, we focus on the continuous, extended helix set of simulations, as the major findings reported here are relatively insensitive to this choice (see Figures S3–S8 and Table S3, Supporting Information).
Regarding bilayer thickness, the simulation results are consistent with the X-ray data and the SDP fit, showing that αS induces a global bilayer thinning in both the symmetric and asymmetric αS systems. As we have noted previously, quantitative determination of DHH from CGMD is complicated by the low resolution of the model.47 A more accessible measure is made via a second common definition of bilayer thickness, DPP, which measures the distance between the component number probabilities for the headgroups in the two leaflets (see Figure S3, Supporting Information). The simulated change in DPP is ∼0.9 and 0.6 Å in the symmetric and asymmetric simulations, respectively.
The true value of molecular simulations is that they provide access to local, detailed information inaccessible to experiments. Figure 1B does just that, illustrating the local changes in bilayer thickness induced by αS, measured as ΔDPP = DPP,αS − DPP,Pure, for the asymmetric system (ΔDPP values for all systems are presented in Figure S4, Supporting Information). For all αS systems, the thinning effect is slightly magnified near the protein, with the asymmetric and single-protein systems coming close to reaching the thickness of the pure system. This suggests that at higher protein density the magnitude of the global thinning effect may be considerably greater. Because these simulations are of large bilayers, all calculations from the simulated lipid coordinates required the proper handling of long-wavelength undulations, specifically defining the undulating reference surface and correcting for the broadening artifact imparted by undulations, for which we have recently developed the necessary computational methods.48,49
Each of the simulations consistently position αS (ZαS) between the phosphate and the carbonyl-glycerol distribution, ZαS ≈ 17.5 Å from the bilayer center (∼3.5 Å below the peak in the phosphate distribution). Because these simulations were all initiated with αS in solution, there remained the possibility of a kinetic barrier to sampling deeper, potentially stable, protein positions. Thus, to determine the equilibrium location of αS in the membrane more carefully, we followed two distinct computational strategies. First, we calculated a potential of mean force (PMF), which reports on the thermodynamic energy as a function of the protein depth in the membrane, and thus can be used to predict the equilibrium value of ZαS on the basis of the lowest calculated energy.50,51 PMF calculations have been used recently to explore both protein–protein and lipid–protein interactions (e.g., transmembrane α-helix packing and orientation as well as the partitioning characteristic of amphipathic α-helices into the bilayer).50,52–55 To calculate the PMF, we relied upon the weighted-histogram analysis method, which applies a biasing potential to increase sampling of conformational space along a specific reaction coordinate.51,55–57 As shown in Figure S9A (Supporting Information), the PMF predicts a strong energetic dependence on ZαS, with a minimum at 18.1 Å (∼3 Å below the phosphates). Second, we ran a series of unbiased simulations that were initiated with the protein at various depths in the bilayer (see Figure S9B), each of which converged to a similar ZαS. Collectively, our computational results agree well with previously published EPR data which suggested that αS partitions between 1 and 4 Å below the headgroup phosphates.58 The result is also roughly consistent with our previous all-atom simulation in a different bilayer.46 In that case, we simulated the horseshoe helix conformation in a 100% PS bilayer and found that the protein embedded 3–5 Å below the headgroup phosphates.46
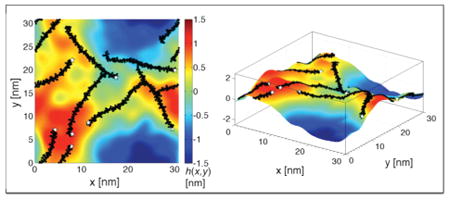
Having established reasonable consistency with the X-ray experiments as regards overall bilayer structure, we turned our attention to curvature. Figure 2A shows snapshots from the asymmetric αS simulation, represented as a surface whose color map indicates the height deviations from the average (flat) phosphate surface. These surfaces are quite distinct from local thickness profiles (Figure 1B; Figure S4, Supporting Information). Instead, they represent local monolayer surfaces and are descriptive of the curvature induction present in the protein-containing leaflet. (We see similar effects in the symmetric αS system, but focus on the more physiologically relevant asymmetric system from here on. A complete detailing of all curvature field surfaces for both rigid and flexible αS systems in the symmetric, asymmetric, and single-αS configurations can be found in the Supporting Information.) Throughout the simulation, it was visually clear that the lateral locations of αS were strongly correlated with regions of positive height (warm colors in the figure indicating local, positive monolayer curvature). This can be seen in Figure 2A (top), which shows the outer leaflet with the protein at two distinct time points, as well as in Figure S6A (Supporting Information), which shows the average height fluctuations at the location of each residue in the protein. Figure 2A (bottom) shows the inner leaflet at the same two time points and demonstrates a coupled remodeling of the bilayer between leaflets: proteins in the outer leaflet induce a correlated curvature in the protein-free inner leaflet.
Figure 2.
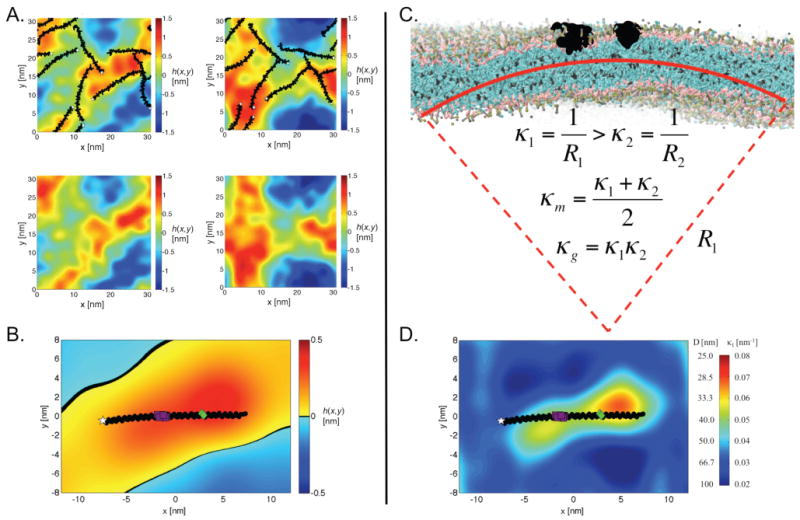
αS stabilizes positive mean curvature. (A) Representative height surfaces, h(x,y) = z, for the outer leaflet of the asymmetric system at two time points (18.2 and 32.8 μs scaled simulation times, left and right columns, respectively) showing both the outer leaflet (top row) and the inner leaflet (bottom row). αS (black), with the N-terminus (white star) highlighting the protein orientation, is correlated with regions of positive surface fluctuation (hot color map). (B) Time- and protein-averaged height surface for the asymmetric system. Panels A and B color map units are nanometers. (C) Snapshot of the asymmetric system showing positive mean curvature with two αS proteins (black) positioned at the apex. Mean (κm) and Gaussian (κg) curvatures are determined from the principle curvatures (κ1 and κ2), where κ1 is the maximum curvature and κ2 is the minimum curvature, with κ1 ⊥ κ2. (D) Local κ1 and corresponding vesicle diameter, D = 2/〈κ1〉, for the asymmetric system (the white star indicates the N-terminus, purple squares indicate the linker region, and green tilted squares indicate GLY67–GLY68).
In general, the time-averaged curvature effects are best interrogated by averaging over all proteins and recasting the bilayer as a whole in terms of the flexible midplane surface. Figure 2B does just this, presenting the time- and protein-averaged bilayer topology map. Consistent with the individual snapshots shown in Figure 2A, αS sits atop a region of positive height (on a hill). The shape of the hill is quite interesting, with four main features: (1) it is ellipsoidal along the long axis of the protein, making it more of a ridge than a hill, (2) it is slightly skewed from the long axis of the protein, (3) the height of the hill is not constant along the protein, with a peak toward the C-terminal region, and (4) the hill gives way to a valley. Regarding the skew of the hill, we considered if this were due to protein–protein interactions, but see the same characteristic in the single-αS system (Figure S5B, Supporting Information). Interestingly, these anisotropies are reduced in the broken helix system (Figure S5C). Thus, the nuances of protein-induced changes in bilayer topology are likely dictated by sequence—perhaps due to regions of increased flexibility as we will discuss below—and suggest more complexity than is typically considered in the modeling of amphipathic α-helices as characterless rigid rods.
Figure 2C presents a zoomed-in cross-section from the asymmetric system. We have chosen a snapshot where the curvature effect is visually obvious for illustrative purposes only and do not intend to suggest that two proteins in proximity (as pictured) are a requirement for curvature induction (they are not). In Figure 2C, the red arc defines the principle curvature κ1.20 That is, the main curvature effect is in the direction orthogonal to the long axis of the protein (as described above, the protein sits on a ridge), a characteristic that has been recognized in other studies of amphipathic helix-induced curvature.7,18,19 κ2 is orthogonal to κ1, spans the longitudinal axis of the proteins, and is used in the calculation of the mean curvature.
To quantify the curvature effects, and to confirm that the effect is propagated across the two leaflets (as is apparent in Figure 2C), we have calculated the time- and protein-averaged mean curvature field, 〈κm〉. These curvature fields describe the curvature along the membrane midplane and provide a description of the average local environment near the protein, where each of the individual protein's curvature fields are aligned relative to the protein's orientation and averaged over all proteins and time frames. For the symmetric αS systems, only proteins on a single leaflet were included in the averaging. Analysis of the inner leaflet produced consistent results relative to the leaflet orientation (i.e., an opposite sign for 〈κm〉). We find that, consistent with our expectation, αS colocalizes to regions of positive bilayer curvature. This, taken with the demonstration in Figure 2A, shows that αS curvature induction is communicated across the bilayer to the inner leaflet, consistent with the recent finding regarding αS-induced tubulation.7
Figure 2D casts these findings in terms of the more familiar lipid vesicle diameter. Specifically, the plot illustrates how the local αS-induced curvature corresponds to the global curvature of a vesicle (i.e., its diameter) to which αS is bound. This diameter is calculated from the time- and protein-averaged principle curvature 〈κ1〉 as D = 2/〈κ1〉. Remarkably, the extrapolated diameter correlates almost perfectly with that observed in our experimental measurements of binding affinity,9 the equilibrium diameters of αS-induced tubules, and the size of synaptic vesicles59 (D ≈ 40 nm in each case).We note that, in the case of synaptic vesicles, possible interactions between αS and other synaptic proteins60–62 may contribute to curvature stabilization/sensing. In the single-protein system we see a similar pattern of mean curvature induction (see Figures S6 and S8, Supporting Information), suggesting that the overall curvature effects of a high density of αS on the bilayer can most likely be attributed to the sum of local effects induced by single proteins.
In addition to the mean and principle curvatures, we have calculated the effect of αS on Gaussian curvature (κg), a property that can be associated with both fusion- and fission-favorable intermediates.3 κg can be used to distinguish between a locally convex surface (κ1 and κ2 are both positive or both negative) and a local saddle (κ1 and κ2 have opposite signs). Negative values of κg are associated with fusion or fission intermediate states. By definition, a flat periodic bilayer must have a globally averaged 〈κg〉 of zero; however, stabilized spontaneous curvature would induce a nontrivial locally averaged 〈κg〉. For our protein-free bilayer, all locally defined 〈κg〉 ≈ 0. Figure 3 presents the time- and protein-averaged local 〈κg〉 for the asymmetric system, showing that there is an induction of negative 〈κg〉 at the center of the helix, spanning almost one-third of the protein's longitudinal axis. Here, where the protein is positioned at the apex of a region of positive mean curvature (Figure 2), there is a Gaussian curvature field with regions of positive Gaussian curvature colocalized at the maxima and minima of the mean curvature field with a pronounced region of negative Gaussian curvature stabilized at the center of the protein. Additionally, each extreme in the mean curvature field is completely surrounded by a ring of negative Gaussian curvature. This is consistent with the Helfrich hat model.2 To our knowledge, this is the first such demonstration of amphipathic α-helix-induced Gaussian curvature.
Figure 3.
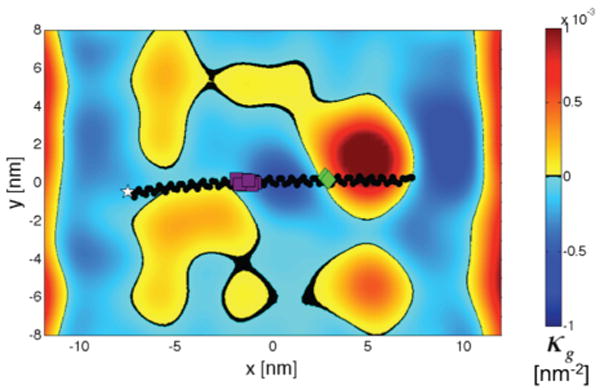
αS induces negative Gaussian curvature. Time-averaged, reoriented 〈κg〉 determined from the continuous helix asymmetric system. Two regions of known increased flexibility, the linker region for the horseshoe helix conformation (purple) and GLY67–GLY68 (green), border a region of stabilized negative 〈κg〉. Color map scale units are inverse square nanometers.
Most interestingly, the region of greatest negative Gaussian curvature is flanked by the linker region of the horseshoe helix conformation (experimentally shown to have increased disorder on highly curved substrates) and Gly67–Gly68, both of which are regions of αS that have been shown, under varying conditions, to have increased flexibility and disorder.45,46,63 It has been hypothesized that conversion from the vesicle-bound extended helix to the horseshoe conformation may regulate the stability and fusion of docked synaptic vesicles.27,29,64 The CGMD simulations in this study do not address this putative conformational switch. However, our observation of αS-induced, localized negative 〈κg〉 suggests the possibility that these regions of the protein may be important in future investigations of how—and under which circumstances—αS may facilitate, rather than inhibit, fusion, as well as participate in fission.
Lastly, because fusion and fission are intimately linked to the rigidity of the membrane, and the formation of fusion/fission intermediates is a dynamic process, we have used the simulations to investigate the effects of αS on bilayer dynamics. Additionally, results from Figures 1 and 2 suggest competing influences: decreased thickness is often presumed to correlate with decreased rigidity, while increased static curvature may be presumed to correlate with increased rigidity. We therefore calculated a relaxation time, τ, for the membrane from the time correlation of the height surface (the monolayer representation as shown in Figure 2A). This relaxation time provides a measure of the dynamics of the bilayer, as it characterizes the propagation of the thermally driven undulation waves. We have found that αS causes a 2 orders of magnitude increase in the undulation relaxation time (τpure ≈ 5 ns and ταS ≈ 500 ns for the pure and symmetric continuous helix systems, respectively). Whether and how this change in relaxation time relates to curvature and bilayer rigidity, and by extension fusion and fission processes, is an important and open question.
Discussion
The processes of membrane fusion and fission involve rearrangement of the phospholipid bilayer into nonlamellar structures. Access to these structural states requires the bilayer to cross high-energy barriers, a process that can be facilitated, or inhibited, by proteins that change the membrane's structure and dynamics. These alterations can include, for example, reductions in thickness, curvature strain, and rigidity. The main goal of this study has been to understand how proteins such as αS can have such profound effects, both biophysically and biologically. The biophysical effects include enhanced binding affinity to highly curved bilayers,9 inhibition of the fusion of small, synaptic-like vesicles,23 and tubulation and vesiculation of large, flat membranes.7,28 Biologically, these effects include the stalling of vesicles in exocytic transport pathways26 and fragmentation of mitochondrial membranes.28 That the membrane binding region of αS, a relatively simple 100 amino acid α-helix, appears to have these multiple roles raises several important and fundamental questions about the biophysical interactions between amphipathic α-helices and membranes. In particular, what is the relationship between a protein's ability to sense and/or generate curvature? Are all amphipathic α-helices the same, or are there sequence requirements that endow specific curvature effects? That is, is it sufficient to view amphipathic helices as simple, rigid rods that insert into the membrane and cause it to bend?
The equilibrium diameters of curvature in our simulated systems correspond remarkably with those of the SUVs to which αS binds with greatest affinity and to the diameters of tubules induced in GUVs. This result supports the assertion that αS–lipid complexes possess an intrinsic curvature field (Figure 2D), which results from the balance of intermolecular interactions between a single protein, several shells of surrounding and affected lipids, water, and ions. We say that the curvature field is intrinsic because it represents the lowest energy state of the local membrane complex and is dictated by the specific protein and lipid type. In this more continuum-like view, protein–protein interactions within the membrane are recast as interactions between individual, intrinsic curvature fields. At high protein density, these interactions between fields reinforce curvature induction and can lead to global rearrangements of the bilayer, namely, tubulation and vesiculation. More specifically, when αS binds to a flat membrane (e.g., a mitochondrial membrane or a GUV), the process of curvature induction alleviates the nonequilibrium curvature stress as the system remodels to adopt its intrinsic curvature (this is the process we have simulated). Conversely, if αS interacts with a small lipid vesicle, one whose own intrinsic curvature field roughly matches that of the protein–lipid system, there is no net molecular driving force to cause the membrane to remodel. Instead, the protein stabilizes the highly curved vesicle through a reinforcement of the matched curvature fields. This notion should be used to explain the increased binding affinity of αS to highly curved vesicles (curvature sensing). Thus, curvature generation and sensing arise from the same set of thermodynamic driving forces; from a thermodynamics point-of-view, they are the same thing.
The results have important implications for physical and thermodynamic theories that address curvature induction via amphipathic α-helices. Current models treat the protein as a rigid rod that wedges into the lipid headgroup region of the bilayer.3,18,19 This wedge increases the effective lateral area of the headgroups but, depending upon the depth of insertion, can have less of an effect on the lipid chains. This results in an area mismatch within the monolayer that must be reconciled by the induction of curvature. The specific character of the protein-induced curvature field, for example, the magnitude of curvature and the overall shape of the field (isotropic or anisotropic), has to this point been investigated in terms of amphipathic α-helical structure (namely, its length) and depth of insertion.18,19 It is in this context that we view αS as a particularly important molecule for understanding the generation of curvature that plays a central role in fusion (or its inhibition) and fission. Our finding that the transitions between phases of stabilized Gaussian curvature are precisely flanked by two highly flexible regions within αS (Figure 3) strongly suggests that flexibility within helices, dictated by the specific amino acid sequence, is a highly important feature. That is, regions of increased protein dynamics correlate with the stabilization of negative Gaussian curvature, which is linked to the formation of fusion/fission-favorable intermediates.
Our findings indicate two defining features of the monomeric form of the αS amphipathic helix: (1) it is quite long (∼150 Å), and (2) it is very flexible. These features distinguish the protein from, for example, the well-studied N-BAR protein, which also generates curvature. N-BAR contains an amphipathic α-helix (H0), but unlike αS its curvature-inducing mechanism relies heavily upon a large scaffolding domain, as the H0 helix alone is insufficient to induce enough curvature to tubulate or vesiculate membranes.11 H0 is quite short compared to the membrane binding domain of αS (3-fold fewer residues). Being significantly shorter, one expects the intrinsic curvature field for H0 to be more isotropic than that of αS, which due to its length is highly anisotropic (Figure 2D). Anisotropy in both the mean and Gaussian curvature fields allows for a directional alignment and reinforcement of neighboring proteins' fields when protein–protein interactions are important (as they are in tubulation and vesiculation, and as they would be expected to be in fission intermediates). We expect that this reinforcement effect is diminished in the case of a shorter protein such as H0. Indeed, we see evidence of this in the simulations of the horseshoe configuration of αS (see Figures S5–S8, Supporting Information, for evidence of more isotropic curvature fields). Of course, due to its length, H0 also lacks the same capacity for multiple flexible segments, so parsing curvature effects based on helix length and flexibility requires further investigation. Such research will be critical in building a complete and robust picture of how subtleties in sequence dictate the structure, dynamics, and function of amphipathic α-helices. Clarifying how these proteins alter membrane behavior will be a fundamental part in continuing efforts to understand both native and pathological mechanisms in cellular membrane biophysics, for example, in Parkinson's disease.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health (NIH) Grant GM44976 (S.T.-N.), a postdoctoral fellowship from the Max Kade Foundation (E.S.), and the Ellison Medical Foundation (E.R.). X-ray scattering data were taken at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation (NSF) and the National Institutes of Health/National Institute of General Medical Sciences under National Science Foundation Award DMR-0225180. Parts of this work were carried out in the Characterization Facility, University of Minnesota, which receives partial support from the NSF through the Materials Research Science and Engineering Centers (MRSEC) program. We especially thank Dr. Arthur Woll for obtaining our beam and for general support during our data collection at the G1 station and Dr. John F. Nagle for assisting with data collection. All CGMD simulations and data analysis were performed at the Minnesota Supercomputing Institute (MSI). We thank Dr. Ralf Langen and Dr. Ad Bax for their helpful comments and discussion.
Footnotes
Supporting Information: Details for protein expression, purification, and labeling, fluorescence correlation spectroscopy, X-ray scattering, and molecular dynamics methods, data analysis, and complete refs 26, 28, 59, and 61. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.McMahon HT, Kozlov MM, Martens S. Cell. 2010;140:601. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Helfrich W. Liq Cryst. 1989;5:1647. [Google Scholar]
- 3.Kozlov MM, McMahon HT, Chernomordik LV. Trends Biochem Sci. 2010;35:699. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgart T, Capraro BR, Zhu C, Das SL. Ann Rev Phys Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. Science. 2004;303:495. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 6.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. EMBO J. 2006;25:2898. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varkey J, Isas JM, Mizuno N, Jensen MB, Bhatia VK, Jao CC, Petrlova J, Voss JC, Stamou DG, Steven AC, Langen R. J Biol Chem. 2010;285:32486. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madsen KL, Bhatia VK, Gether U, Stamou D. FEBS Lett. 2010;584:1848. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 9.Middleton ER, Rhoades E. Biophys J. 2010;99:2279. doi: 10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blood PD, Voth GA. Proc Natl Acad Sci USA. 2006;103:15068. doi: 10.1073/pnas.0603917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes F, Loura LM, Chichon FJ, Carrascosa JL, Fedorov A, Prieto M. Biophys J. 2008;94:3065. doi: 10.1529/biophysj.107.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arkhipov A, Yin Y, Schulten K. Biophys J. 2009;97:2727. doi: 10.1016/j.bpj.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campelo F, Fabrikant G, McMahon HT, Kozlov MM. FEBS Lett. 2010;584:1830. doi: 10.1016/j.febslet.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 14.Cui H, Lyman E, Voth GA. Biophys J. 2011;100:1271. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayton GS, Lyman E, Voth GA. Faraday Discuss. 2010;144:347. doi: 10.1039/b901996k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayton GS, Voth GA. Biophys J. 2010;99:2757. doi: 10.1016/j.bpj.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown M, Thurmond R, Dodd S, Otten D, Beyer K. Phys Rev E. 2001;64:010901. doi: 10.1103/PhysRevE.64.010901. [DOI] [PubMed] [Google Scholar]
- 18.Zemel A, Ben-Shaul A, May S. J Phys Chem B. 2008;112:6988. doi: 10.1021/jp711107y. [DOI] [PubMed] [Google Scholar]
- 19.Campelo F, McMahon HT, Kozlov MM. Biophys J. 2008;95:2325. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown FL. Annu Rev Phys Chem. 2008;59:685. doi: 10.1146/annurev.physchem.59.032607.093550. [DOI] [PubMed] [Google Scholar]
- 21.Auluck PK, Caraveo G, Lindquist S. Annu Rev Cell Dev Biol. 2010;26:211. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 22.Bartels T, Choi JG, Selkoe DJ. Nature. 2011;477(7362):107–U123. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, Eimer S, Winklhofer KF, Haass C. EMBO J. 2010;29:3571. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, Chaudhry FA, Nicoll RA, Edwards RH. Neuron. 2010;65:66. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang YJ, Hay J. C Mol Biol Cell. 2010;21:1850. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper AA, et al. Science. 2006;313:324. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamp F, Beyer K. J Biol Chem. 2006;281:9251. doi: 10.1074/jbc.M512292200. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, et al. J Biol Chem. 2011;286:20710. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. J Biol Chem. 2010;285:28261. doi: 10.1074/jbc.M110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trexler AJ, Rhoades E. Biochemistry. 2009;48:2304. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J, Tieleman DP, Nagle JF, Kučerka N, Tristram-Nagle S. Biochim Biophys Acta, Biomembr. 2009;1788:1387. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyatskaya Y, Liu YF, Tristram-Nagle S, Katsaras J, Nagle JF. Phys Rev E. 2001;63:0119071. doi: 10.1103/PhysRevE.63.011907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YF, Nagle JF. Phys Rev E. 2004;69:040901. doi: 10.1103/PhysRevE.69.040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kučerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Biophys J. 2005;88:2626. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kučerka N, Perlmutter JD, Pan J, Tristram-Nagle S, Katsaras J, Sachs JN. Biophys J. 2008;95:2792. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kučerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Biophys J. 2008;95:2356. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. J Phys Chem B. 2007;111:7812. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 38.Monticelli L, Kandasamv SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. J Chem Theory Comput. 2008;4 doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 39.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 40.Perlmutter JD, Sachs JN. J Am Chem Soc. 2011;133:6563. doi: 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 41.Baoukina S, Tieleman DP. Biophys J. 2010;99:2134. doi: 10.1016/j.bpj.2010.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Atkinson D, Small DM. J Biol Chem. 2003;278:37480. doi: 10.1074/jbc.M303133200. [DOI] [PubMed] [Google Scholar]
- 43.Nagle JF, Tristram-Nagle S. Biochim Biophys Acta, Biomembr. 2000;1469:159. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. J Am Chem Soc. 2008;130:12856. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulmer TS, Bax A, Cole NB, Nussbaum RL. J Biol Chem. 2005;280:9595. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 46.Perlmutter JD, Braun AR, Sachs JN. J Biol Chem. 2009;284:7177. doi: 10.1074/jbc.M808895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perlmutter JD, Sachs JN. Biochim Biophys Acta, Biomembr. 2009;1788:2284. doi: 10.1016/j.bbamem.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Brandt EG, Braun AR, Sachs JN, Nagle JF, Edholm O. Biophys J. 2011;100:2104. doi: 10.1016/j.bpj.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun AR, Brandt EG, Edholm O, Nagle JF, Sachs JN. Biophys J. 2011;100:2112. doi: 10.1016/j.bpj.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gkeka P, Sarkisov L. J Phys Chem B. 2010;114:826. doi: 10.1021/jp908320b. [DOI] [PubMed] [Google Scholar]
- 51.Hub JS, De Groot BL, Van der Spoel D. J Chem Theory Comput. 2010;6:3713. [Google Scholar]
- 52.Li L, Vorobyov I, Allen TW. J Phys Chem B. 2008;112:9574. doi: 10.1021/jp7114912. [DOI] [PubMed] [Google Scholar]
- 53.Li L, Vorobyov I, Allen TW. Biochim Biophys Acta. 2012;1818(2):135–145. doi: 10.1016/j.bbamem.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 54.MacCallum JL, Bennett WF, Tieleman DP. Biophys J. 2011;101:110. doi: 10.1016/j.bpj.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chetwynd A, Wee CL, Hall BA, Sansom MS. Biophys J. 2010;99:2534. doi: 10.1016/j.bpj.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roux B. Comput Phys Commun. 1995;91:275. [Google Scholar]
- 57.Souaille M, Roux B. Comput Phys Commun. 2001;135:40. [Google Scholar]
- 58.Jao CC, Der-Sarkissian A, Chen J, Langen R. Proc Natl Acad Sci USA. 2004;101:8331. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamori S, et al. Cell. 2006;127:831. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 60.Darios F, Ruiperez V, Lopez I, Villanueva J, Gutierrez LM, Davletov B. EMBO Rep. 2010;11:528. doi: 10.1038/embor.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Reitbock P, et al. Brain. 2010;133:2032. doi: 10.1093/brain/awq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC. Science. 2010;329:1663. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bussell R, Jr, Eliezer D. J Mol Biol. 2003;329:763. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 64.Kjaer L, Giehm L, Heimburg T, Otzen D. Biophys J. 2009;96:2857. doi: 10.1016/j.bpj.2008.12.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


