Abstract
Systems biology provides a framework for assembling models of biological systems from systematic measurements. Since the field was first introduced a decade ago, considerable progress has been made in technologies for global cell measurement and in computational analyses of these data to map and model cell function. It has also greatly expanded into the translational sciences, with approaches pioneered in yeast now being applied to elucidate human development and disease. Here, we review the state of the field with a focus on four emerging applications of systems biology that are likely to be of particular importance during the decade to follow: (a) pathway-based biomarkers, (b) global genetic interaction maps, (c) systems approaches to identify disease genes, and (d) stem cell systems biology. We also cover recent advances in software tools that allow biologists to explore system-wide models and to formulate new hypotheses. The applications and methods covered in this review provide a set of prime exemplars useful to cell and developmental biologists wishing to apply systems approaches to areas of interest.
Keywords: networks, models, high-throughput technology, genome-wide measurements, developmental biology, cell fate, biomarkers, epistasis, genetic interaction, genome-wide association study (GWAS)
INTRODUCTION
Nearly a decade has passed since systems biology was introduced into the language of modern biology (Ideker et al. 2001, Kitano 2002). In that time it has expanded greatly in breadth; it now embraces much of the life sciences and is used to address many research problems across humans and diverse model species (Figure 1). Systems biology has also deepened considerably; many more systematic technologies and methods, both experimental and computational, are in use now than were available a decade ago.
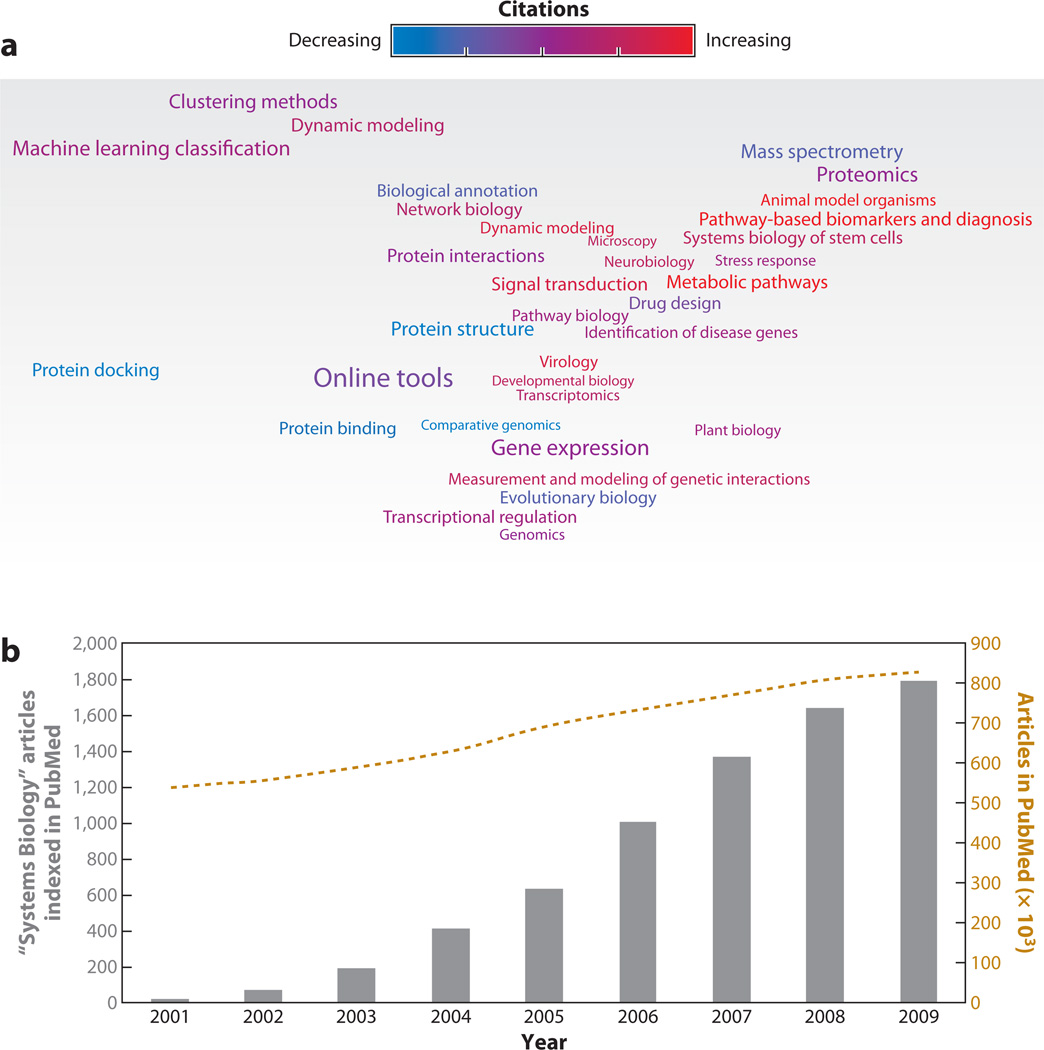
Figure 1.
Meta-analysis of systems biology publications over the past decade. (a) A map of the 34 leading topics in systems biology during the years 2000–2009. The map represents a 2D scaling of the mutual information score between topics, i.e., closely associated topics in the map represent similar themes. The size of the text is roughly proportional to the number of papers. The color gradient indicates a change in rate of citations (from blue to purple to red). Blue indicates topics that were more common prior to 2007; red indicates topics that have been more common since 2007 (see the Supplemental Methods Section for more details on the method and topic word lists). (b) Gray bars show the number of articles indexed in PubMed per year that are labeled with the Medical Subject Heading (MeSH) “Systems Biology.” As a reference, the gold dashed line shows the number of total articles in thousands indexed in PubMed per year.
Yet the field in many ways remains in its infancy. The available genome-scale experimental tools are still in an exponential development phase; new technologies turn the field on its head every few years. Even using current assays, bioinformatic methodology lags significantly behind, such that many more data are generated than possibly can be analyzed or interpreted. Moreover, and perhaps most humbling, the field still has not reached consensus on the definition of systems biology. Part of the reason is that systems biology is in vogue, and some have found it easier to change its definition than to change their research habits. However, it is evident that interesting changes are afoot in biology, and given the newness of some these changes, building consensus may take time.
Systems Biology: A Framework for Modeling Biological Systems from Systematic Measurements
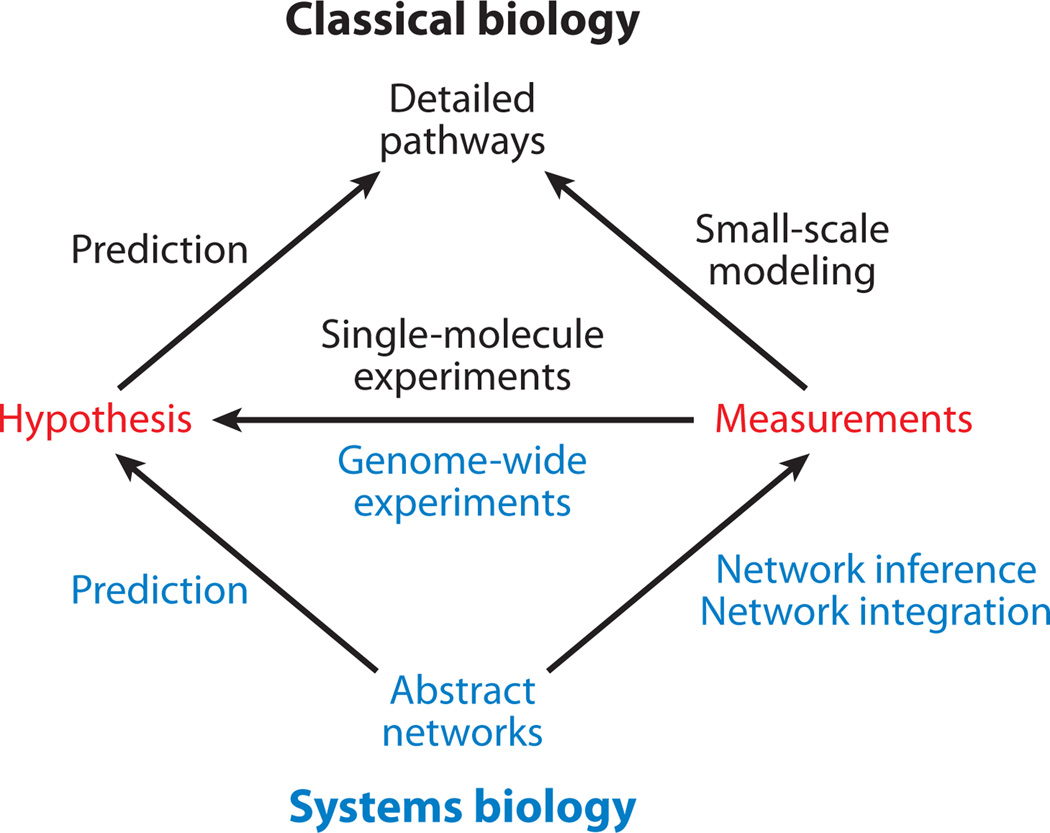
Systems approaches, by necessity, involve systematic data. It is impossible to study a biological system as a whole without them. On one hand, the ability to make genome-wide (or proteome-wide or transcriptome-wide) measurements on a system is arguably the single greatest force driving the rise of systems biology. On the other hand, systems biology is not only about genome-scale measurements; it is about a philosophy and a hypothesis-driven approach for experimental design and analysis (Ideker et al. 2001). Therefore, systems biology does not apply to genome-scale studies that are focused solely on discovery. Rather, it is a framework for using genome-scale experiments to perform predictive, hypothesis-driven science (Figure 2). Using genome-scale data to test hypotheses is nontrivial because it requires that the hypotheses themselves be genome-scale. This, in turn, only becomes possible with a genome-scale model of the system. Of course, systematic technologies are not the only means of measuring biological systems. It is critical that systems-level models are consistent with, and validated by, detailed single-molecule measurements and literature.
Figure 2.
Overview of the experimental process in classical biology (top) versus systems biology (bottom).
Enabled by advances in genome-scale technology, the available molecular data are growing exponentially. A property of exponential growth is that the amount of data describing a pathway that will be collected in the next year is on par with the amount of data that has ever been collected about that pathway in the history of science. In light of this fact, clearly the main challenge confronting the field is not to look back (incorporating previous findings is critical but will be comparatively easy) but to look forward to how one might plan and interpret the mountains of new data that soon will be generated.
Another principle emerging from systems biology research is that it is not enough to map out the physical components and interactions of a system—one must also map how information propagates through this system in response to perturbations. Similarly, it has proven extremely difficult to infer physical or structural interactions in the system from functional data alone (e.g., expression profiles). Thus, systems approaches must necessarily investigate both the physical and functional aspects of the system. For this reason, many approaches seek to integrate multiple data sets, each of which contains a different slice of information about system structure or state.
Finally, as previous authors have done, we distinguish between systems biology and synthetic biology. Systems biology attempts to understand the workings of natural biological systems; synthetic biology uses this understanding to construct new genetic and biochemical systems in vivo or in vitro. Several good reviews of recent progress in synthetic biology are available elsewhere (Andrianantoandro et al. 2006, Benner & Sismour 2005).
A Systems Approach to the Systems Biology Literature
To obtain a systems-level map of the current status of the field, we performed a meta-analysis of all systems biology publications recorded in PubMed over the past decade (Figure 1). The field has grown from a handful of publications published in 2001 to nearly 2,000 published in 2009. Next, we mined the abstracts of systems biology articles published from 2000 to 2009 to extract popular research topics (Figure 1a). We estimated the trends in publication over the decade and compared the topics prominent in systems biology publications prior to 2007 to those in the latter part of the decade (see the Supplemental Methods Section for details; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). Certain topics, such as gene expression analyses and evolutionary biology, have maintained their places as mainstays of systems biology (Figure 1a). Others are on the rise, such as stem cells and network biology. A few topics, such as protein structure and comparative genomics, show a decline in publication rates. Nonetheless, the increase in breadth and versatility of research carried out under the banner of systems biology sends a clear message.
In the remainder of this review, we describe progress in systems approaches for mapping biological pathways and for using these maps in biomedical research. Guided by topics in the systems biology meta-map (Figure 1a), we focus on four areas in particular. All of these are strongly emerging topics in systems biology over the past few years: pathway-based biomarkers and diagnosis, systematic measurement and modeling of genetic interactions, systems biology of stem cells, and identification of disease genes. Each of these topics has recently been the focal point of significant research progress brought about because of innovative use of systems-wide measurement methods and computational approaches. In addition, we review the software tools available for network visualization and interactive exploration of systems biology data, which can be used to formulate hypotheses for further investigation and discovery.
SYSTEMS APPROACHES TO MOLECULAR DIAGNOSTICS
A first area in which systems approaches have gained recent traction is molecular diagnostics. For complex diseases such as cancer, gene and protein expression profiling have become the methods of choice for identifying diagnostic biomarkers able to diagnose the severity of disease and predict future disease outcomes (reviewed by Asyali et al. 2006, Quackenbush 2006, Cheang et al. 2008). Markers are selected by scoring each individual gene or protein on how well its expression pattern can discriminate between different classes of disease or between cases and controls. The disease status of new patients is predicted using classifiers tuned to the expression levels of the markers.
Despite their promise, expression-based diagnostics continue to face serious challenges owing to their questionable accuracy when predicting patient outcomes in some diseases (Ein-Dor et al. 2005, Sotiriou & Piccart 2007). Problems are thought to arise as the result of at least two factors: cellular heterogeneity within tissues and genetic heterogeneity across patients. The impact of cellular heterogeneity depends on the nature of the disease. For some diseases, such as B cell lymphoma, the diseased cell population is well defined such that it is possible to harvest a relatively pure cell population yielding a distinct expression signature, or to subdivide a mixed B cell population on the basis of expression. In other diseases, such as breast cancer, it has been difficult to cleanly separate tumor from normal cells, such that the resulting expression profile represents an average signal diluted over a mixed cell population.
In contrast, genetic heterogeneity refers to the fact that the same genes may not be dysregulated in each patient. For instance, patient A may have protein A dysregulated, patient B may have protein B dysregulated, patient C may have protein C dysregulated, and so on. Given this disparity across patients who nevertheless may have the same clinical outcomes (e.g., aggressive cancer), classification algorithms have trouble because no single marker is indicative of the status of all (or even most) patients.
To address these problems and improve on expression-based diagnostics, several groups have begun to integrate patient expression profiles with system-wide maps of the pathways in a cell (Anastassiou 2007, Calvano et al. 2005, Doniger et al. 2003, Draghici et al. 2003, Nibbe et al. 2009, Pavlidis et al. 2004, Tian et al. 2005, Ulitsky et al. 2008a,Wei & Li 2007). Depending on the scenario, such pathway maps can involve signaling cascades, transcriptional regulation, or metabolic reactions. They can be as detailed as a series of discrete actions among proteins that lead to a defined end point or functional outcome, or as abstract as a functional annotation on a set of genes. Pathway information provides an overarching layer of organization that can tie seemingly disparate expression responses together into a common pattern. For instance, although any protein A, B, or C may indicate an aggressive form of disease, the knowledge that A, B, and C form a coherent module—e.g., they are subunits of a common protein complex, successive enzymes in a metabolic pathway, or successive steps in a signal transduction cascade—allows us to formulate new biomarker functions that take all of these proteins into account. Some approaches draw this knowledge from known pathways curated from the literature (Subramanian et al. 2005, Vert & Kanehisa 2003); others incorporate pathway knowledge from unbiased networks of physical protein-protein interactions (Chuang et al. 2007, Ma et al. 2007, Taylor et al. 2009, Tuck et al. 2006). In either case, the goal is to identify biomarkers not as lists of individual genes or proteins but as functionally related groups of genes or proteins whose aggregate expression accounts for the phenotypic differences between the different populations of patients. Unlike conventional expression diagnostics based on individual genes, these diagnostic pathway markers provide a strong biological interpretation for the association of an expression profile with a particular type of disease. As a result, the pathway-based approach can be inherently more reliable—which isn’t to say, however, that knowing the pathway relationships assures the success of a diagnostic profile.
In addition to explaining gene expression differences between phenotypes, diagnostic pathways can be used to predict the expression profiles of unknown disease states. Some of these approaches represent pathway activity with a function summarizing the expression values of member genes (Breslin et al. 2005, Guo et al. 2005, Lee et al. 2008); other approaches estimate pathway activation probabilities based on the consistency of changes in gene expression (Efroni et al. 2007, Svensson et al. 2006). Others have engineered normal cells to activate preselected oncogenic pathways to determine gene signatures that can distinguish tumor characteristics (Bild et al. 2006, Glinsky et al. 2005). For example, Bild et al. (2006) overexpressed a panel of oncogenes, one at a time, in primary cultures of human mammary epithelial cells. The goal was to link each oncogene with a distinct set of dysregulated genes. Given these links, they showed that the expression profile of a new tumor sample could be analyzed to identify which oncogenes had been activated.
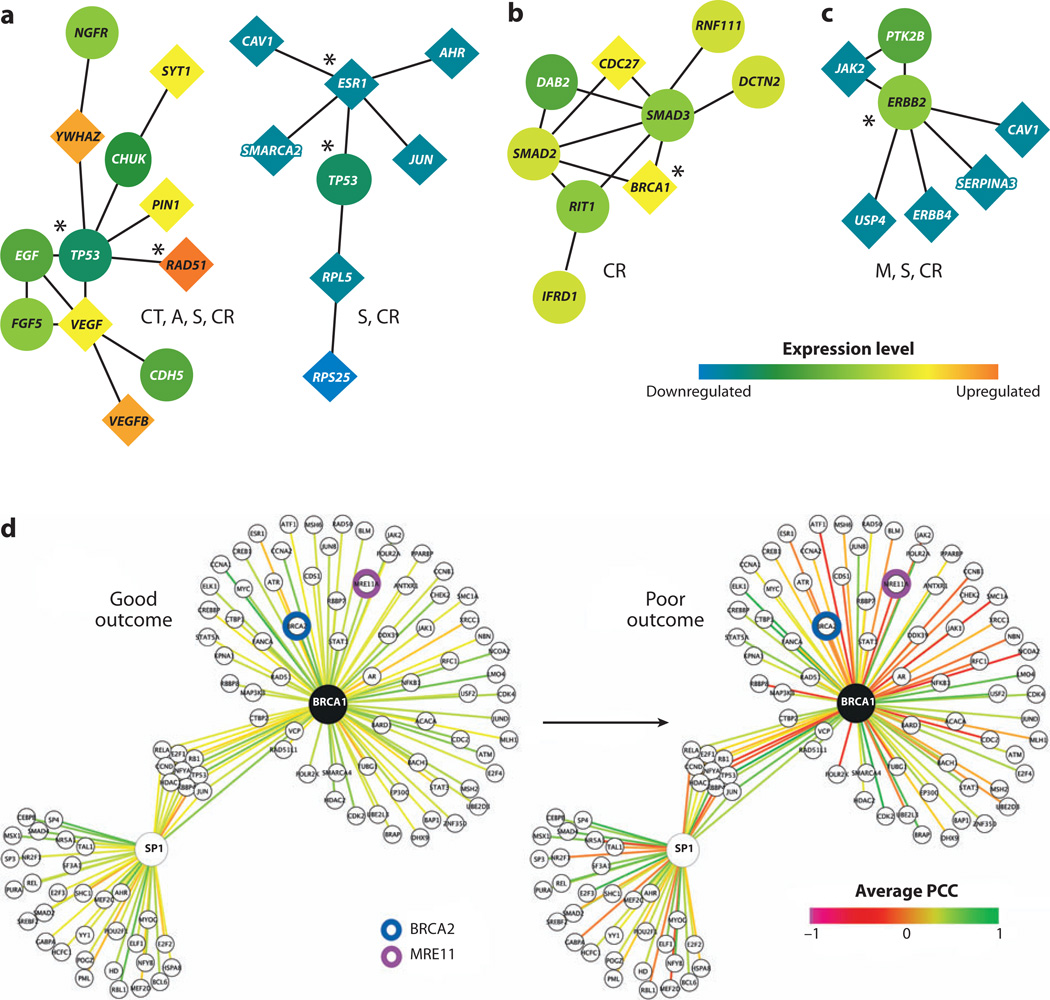
Chuang et al. (2007) demonstrated an approach that mines pathway biomarkers directly from protein-protein interaction networks. Gene expression profiles of breast cancer patients were superimposed on a human protein-protein interaction network to identify protein subnetworks able to predict cancers likely to metastasize within five years (Figure 3a–c). The activity of a subnetwork was inferred by averaging the normalized expression values of its member genes. The dysregulation of a subnetwork was quantified in terms of the mutual information between subnetwork activity and patient phenotype (metastatic or nonmetastatic). Chuang et al. (2007) also showed that subnetwork markers overlap much more extensively between patient cohorts than individual marker genes and are more informative regarding cancer susceptibility.
Figure 3.
Predictive subnetwork markers for breast cancer metastasis. (a–c) Subnetworks identified by Chuang et al. (2007) involving the key susceptibility regulators (a) TP53, (b) BRCA1, or (c) ERRB2. Nodes and links represent human proteins and protein interactions, respectively. The color of each node scales with the change in expression of the corresponding gene for metastatic versus nonmetastatic cancer. The shape of each node indicates whether its gene is significantly differentially expressed (diamond) or not (circle). The predominant cellular functions are listed next to each module: M, metabolism; CT, cell and tissue remodeling; A, apoptosis; S, signaling of cell growth and survival; CR, cell proliferation and replication. Known breast cancer susceptibility genes are marked by asterisks. (d) BRCA1 and its interactors (e.g., BRCA2 and MRE11, as indicated) are highly ordered (green edges indicate correlated expression between protein pairs) in surviving patients, whereas this organization is lost in patients with aggressive cancer. In contrast, interactions involving SP1 are not significantly altered. PCC denotes the Pearson’s correlation coefficient between the expression patterns of two interacting partners. Panels (a–c) are adapted with permission from Chuang et al. (2007). Panel (d) is adapted with permission from Taylor et al. (2009).
Rather than summarizing member gene expression into subnetwork activity, Taylor et al. (2009) proposed to measure changes in interaction coherence between member genes in a subnetwork under different phenotypes (Figure 3d). The interaction coherence in a sample was defined using the difference in expression of the central hub gene in a subnetwork with each of its interacting partners. Although Taylor et al. (2009) and Chuang et al. (2007) differ in the way that they detect pathway dysregulation, both capture a common set of contributions to breast cancer (for example, BRCA1 in Figure 3b versus 3d). Moreover, both studies find that subnetwork markers are more accurate in the classification of breast cancer metastasis than previous predictors based on collections of noninterconnected genes.
In summary, projection of gene expression profiles onto pathway databases or interaction networks is proving to be a powerful approach for understanding disease. On one hand, diagnostic pathways are more reproducible than single genes and can improve the prediction accuracy of disease states. On the other hand, the studies to date are preliminary, and much work is needed before the approach can be translated into advanced diagnostics. One useful direction will be to complement expression and pathway connectivity with other large-scale data sets that include information on genetic perturbations, epigenetic regulation, signal transduction, metabolism, and other factors. Finally, many real and functionally relevant interactions are missing in current protein-protein interaction data sets. Further insights can be expected from reanalysis of the same diseases as the data increase in coverage and quality. Nonetheless, it is clear that constructing functionally coherent, pathway-aggregated biomarkers has great inherent value versus choosing sets of independently selected genes.
GENETIC INTERACTION MAPS: A TOOLBOX WITH IMPLICATIONS FOR CANCER AND DISEASE
Cell function is governed by a large and complex network of combinatorial interactions among genes, collectively referred to as genetic interactions. Recently, several systems biology studies in yeast, fly, worm, and mammalian cell lines have made important strides in our ability to map this genetic interaction network and its impact on function. Classically, a genetic interaction is defined as the phenomenon whereby combined mutations at several genes produce a phenotype that is unexpected from any of the single mutants (Avery & Wasserman 1992).Genetic interactions are often quantified under the assumption that combining two unrelated (independent) mutations should result in a multiplicative effect on phenotype, such that any deviation is considered an indication of a genetic interaction (Bandyopadhyay et al. 2008, Collins et al. 2006, Costanzo et al. 2010, Dixon et al. 2009, Mani et al. 2008b, St Onge et al. 2007). A phenotypic score that is less than expected is a negative or “aggravating” interaction, whereas a score that is greater than expected is a positive or “alleviating” interaction. An extremely negative genetic interaction that is often studied is the “synthetic lethal” in which the combined gene mutations result in cell death.
When viewed globally over a genome, the network of genetic interactions becomes quite large. To appreciate the magnitude of such a network, consider that among the approximately 30,000 human genes there are on the order of a billion (30,0002 = 900,000,000) potential pair-wise genetic interactions. In a recent near-comprehensive screen of genetic interactions in the yeast Saccharomyces cerevisiae, more than 3% of gene pairs showed signs of genetic interaction in rich media conditions (Costanzo et al. 2010). Moreover, genetic interactions need not involve only pairs of genes; rather, they can involve much larger combinations.
Detection of Genetic Interactions in Model Species and Humans
The development of rapid screening techniques for genetic interactions, such as synthetic genetic arrays (SGAs) (Tong et al. 2001, 2004), diploid synthetic lethality analysis by microarray (dSLAM) (Ooi et al. 2003), and epistatic miniarray profiles (E-MAP) (Schuldiner et al. 2005), have allowed the quantification of genetic interaction profiles for the majority of genes in S. cerevisiae. Whereas studies of this scope have yet to be implemented in higher organisms, limited genetic interaction screens in human cell lines and model organisms such as Caenorhabditis elegans and Drosophila melanogaster, as well as screens in Schizosaccharomyces pombe, have already been conducted (Bakal et al. 2008, Bommi-Reddy et al. 2008, Dixon et al. 2008, Lehner et al. 2006, Roguev et al. 2007; for a comprehensive review of epistasis and genetic interaction data sources, see Dixon et al. 2009).
Explicit construction of double gene knockouts in mammals remains a laborious process. Viable alternatives, such as testing combinations of RNA interference (RNAi) knockdowns (Bommi-Reddy et al. 2008, Yang & Stockwell 2008, Zender et al. 2008), are emerging but will naturally take time to mature into genome-scale research tools. In the meantime, a potential role for genetic interaction networks in humans comes from the unlikely direction of statistical genetics, and in particular genome-wide association studies (GWAS). GWAS involves rapidly scanning genetic markers along the genome [such as single nucleotide polymorphisms (SNPs) or copy number variations (CNVs)] to find genetic variations associated with a particular phenotype, such as a heritable trait or disease (Hirschhorn & Daly 2005). However, in many cases GWAS has thus far failed to explain more than a few percent of the genetic contribution to a particular disease, especially for common diseases such as type II diabetes, hypertension, or bipolar disorder (Donnelly 2008, Maher 2008). Evidence is emerging, however, that some of the missing heritability is attributable to combinatorial genetic interactions within and across pathways (Peng et al. 2009, Torkamani et al. 2008, Wang et al. 2007). The need for inclusion of combinatorial genetic interactions also showcases the importance of developing new approaches to systems-level analysis of genetic interactions (Benfey & Mitchell-Olds 2008).
Characteristics of Genetic Networks
Studies in yeast have shown the relative robustness of the cell to systematic deletions, as only a small subset (~20%) of genes are essential in rich media conditions (Dixon et al. 2009). Hillenmeyer et al. (2008), however, showed that under a variety of stress conditions this list is in fact expanded to include most protein-coding genes (~97%). As for the effect seen on single deletions, St Onge et al. (2007) reported a ~24-fold increase in the number of genetic interactions observed after exposure to methyl-methane sulfonate (MMS), a known DNA-damaging agent. Comparisons of normal versus stress conditions suggest that although the genetic network contains some degree of redundancy, it is a highly optimized response mechanism (Costanzo et al. 2010).These experiments illustrate that one should beware of confusing redundancy with robustness.
Topological analysis of the yeast genetic network showed a negative correlation between a gene’s number of genetic interactions and the fitness of its deletion mutant, i.e., hubs in the genetic network tend to have a higher impact on fitness (St Onge et al. 2007). Furthermore, hubs exhibit higher pleiotropy, as estimated by the variety of functional annotations of genetic interactions connected with the hub. A gene’s number of genetic interactions was also found to be correlated with its conservation across yeast species, suggesting that genetic interactions have substantial evolutionary effects (Costanzo et al. 2010). Comparison of genetic interaction networks across different yeasts or between yeast and metazoans suggests that evolutionary conservation is greater at the network level, where the topological characteristics are similar, than at the level of individual interactions, which are not always shared (Dixon et al. 2009, Roguev et al. 2008).
Integration with Physical Interactions
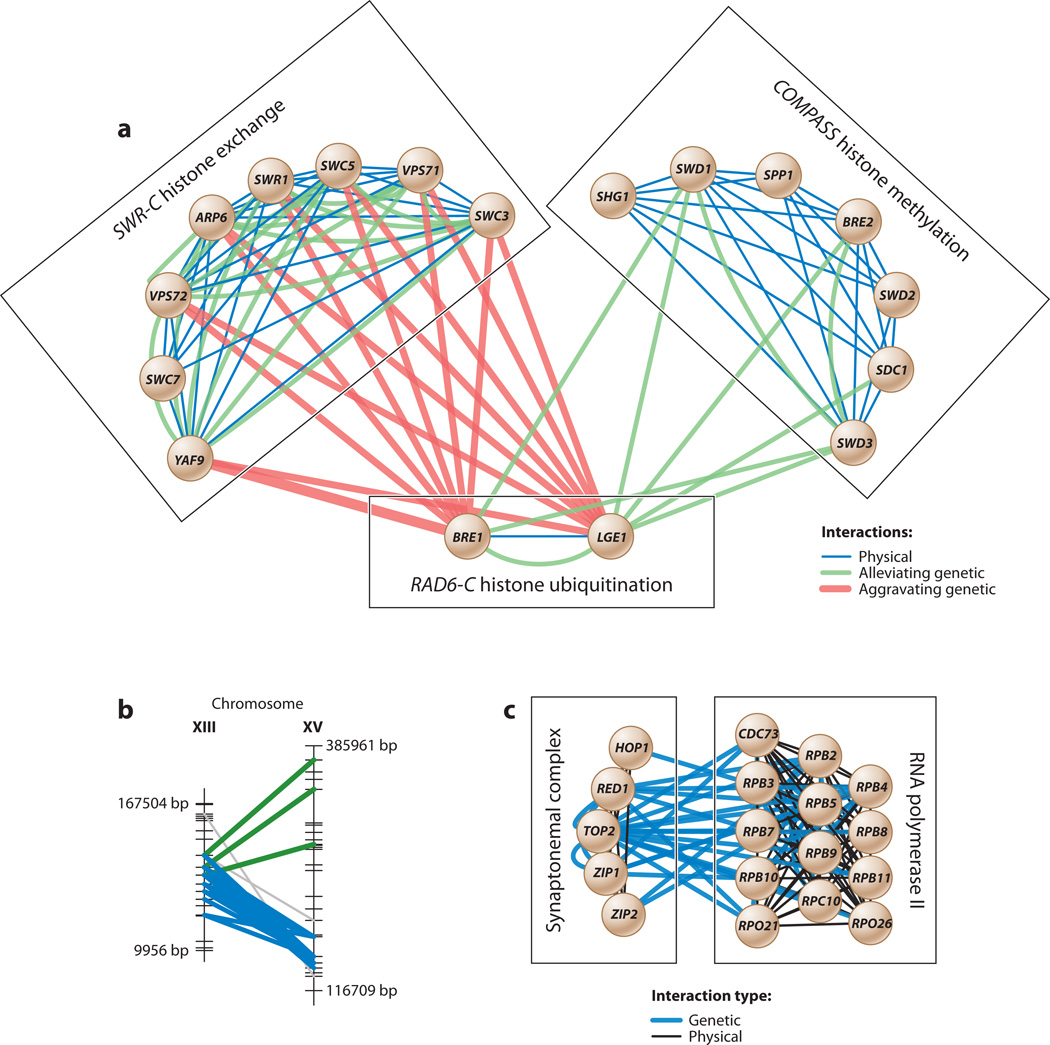
Several studies (Bandyopadhyay et al. 2008, Pu et al. 2008, Ulitsky et al. 2008b) have attempted to integrate genetic interaction networks with networks of physical interactions between proteins. As an example, Bandyopadhyay et al. (2008) scored the likelihoods of a protein pair operating either within the same protein complex or between functionally related complexes on the basis of the strength of its genetic and physical interactions. They first learned the appropriate pattern of physical and genetic interactions from known protein complexes curated in databases. Protein pairs with a strong genetic but weak physical interaction typically were found to operate between two functionally related complexes. An agglomerative clustering procedure was then used to merge the protein pairs into increasingly larger complexes and to identify pairs of complexes interconnected by bundles of many strong genetic interactions. Figure 4a shows three example complexes enriched for aggravating genetic interactions (i.e., synthetic lethality).
Figure 4.
(a) Complexes associated with RAD6-C histone ubiquitination. Protein-protein interactions are enriched among the proteins within each of the three complexes; in contrast, genetic interactions are enriched both within and between complexes. Adapted with permission from Bandyopadhyay et al. (2008). COMPASS, complex of proteins associated with SET1; SWR-C, SWR1 complex; RAD6-C, RAD6 complex. (b) Interacting genomic loci (green and blue) that represent significantly dense groups of marker-marker interactions in a genome-wide association study. (c) Interacting complexes spanned by dense bundles of genetic interactions recovered from the same study. Adapted with permission from Hannum et al. (2009).
Hannum et al. (2009) used a similar integrative approach to analyze and reinforce genetic interactions extracted from GWAS (Figures 4b,c). They first identified pairs of SNP markers whose combined state was associated with the expression phenotypes of one or more genes. A biclustering method was then used to discover consecutive intervals of these SNP pairs on two distinct chromosomes and define a genetic interaction network. Similar to Bandyopadhyay et al. (2008), genetic interactions were shown to be strongly enriched within and between known protein interaction complexes. The key difference, however, was that these genetic interactions had been inferred from GWAS rather than generated using directed mutations.
Genetic Interaction-Based Approaches to Cancer Therapy
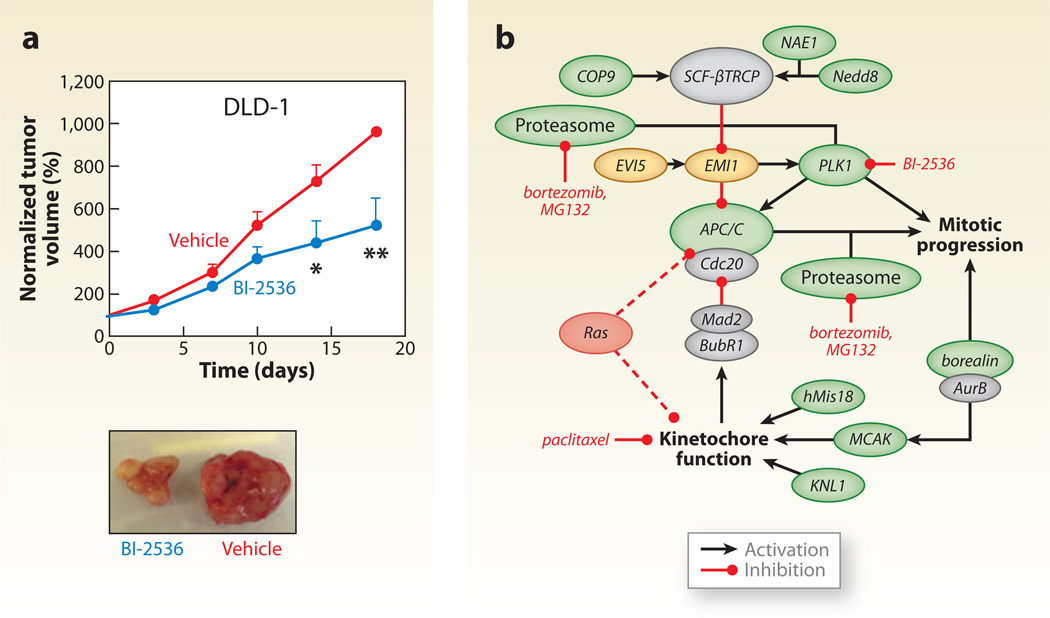
A prominent treatment for cancer is to kill proliferating cancer cells through DNA damage. Because DNA-damaging agents are also toxic to normal tissue, there has been a great deal of interest in developing DNA-damage sensitizers that act specifically on cancer cells via synthetic lethal interactions (Michod & Widmann 2007). In effect, the goal of these studies is to identify and target proteins encoded by genes that are synthetic lethal with cancer-causing mutations. In pioneering work, two groups (Luo et al. 2009, Scholl et al. 2009) have reported promising results from screens focused on finding synthetic lethal relationships with the KRAS oncogene (Figure 5).
Figure 5.
A model of mitotic regulation by Ras. (a) BI-2536, a PLK1 inhibitor, attenuates tumor growth in colorectal cancer cells (DLD-1 cell line) in vivo. Representative images of tumors after treatment are shown. (b) A model in which oncogenic Ras introduces mitotic stress that can be exacerbated to produce lethality by interfering with kinetochore and APC/C (anaphase-promoting complex) function. Genes shaded green are RSL (Regulators of Sex-Limitation) genes, whereas yellow genes cause Ras-specific lethality when overproduced. Red dashed lines illustrate genetic connections between Ras and aspects of mitotic regulation that lead to mitotic stress. Adapted with permission from Luo et al. (2009).
Many sensitizers have been or are currently being investigated. Most notably, much attention has been given to a new class of sensitizers known as PARP inhibitors (Farmer et al. 2005). These drugs target an enzyme involved in the base excision repair pathway, which is synthetic lethal with the homologous recombination pathway genes BRCA1 and BRCA2 that are commonly mutated in breast cancer. In addition, farnesyltransferase inhibitors have reached phase III clinical trials, an inhibitor to the cell cycle checkpoint kinaseChk1 is in phase II, and diverse other compounds, such as ataxia telangiectasia mutated (ATM) kinase inhibitors, are under preclinical development. Significant opportunity remains to identify many other potential molecular targets for tumor sensitization, and to date, DNA damage response pathways appear to be a hotbed of such targets. Thus, in addition to the long-term goals of comprehensively mapping the genetic interaction network in different cells under various conditions, the systematic discovery of genetic interactions has the potential to profoundly change the treatment of cancer (Mendes-Pereira et al. 2009, Morgan et al. 2010).
SYSTEMS APPROACHES TO IDENTIFY DISEASE GENES
The search for disease-causing genes is a long-standing goal of human genetics. Despite several success stories [e.g., identification of the genetic basis of cystic fibrosis (Rommens et al. 1989), Tay-Sachs (Harding 1983), and Huntington’s disease (Myers 2004)], many diseases with quantifiably substantial genetic components continue to elude detailed genetic explanations (Culverhouse et al. 2002, Moore 2003). For this reason, systems approaches are playing an increasing role in this area through the computational integration of multiple types of genome-wide measurements (Adler et al. 2006, Ergün et al. 2007, Franke et al. 2006, Lage et al. 2007, Mani et al. 2008b, Mullighan et al. 2007, Oti et al. 2006, Tomlins et al. 2005, Yao et al. 2006). Several groups have promoted the idea that similar diseases are caused by mutations in different genes that are part of the same functional module (Goh et al. 2007, Oti & Brunner 2007). The approaches differ in the underlying data sets used, but most of them involve superimposing a set of candidate genes alongside a set of known disease genes on a physical or functional network (Franke et al. 2006, Lage et al. 2007, Oti et al. 2006).
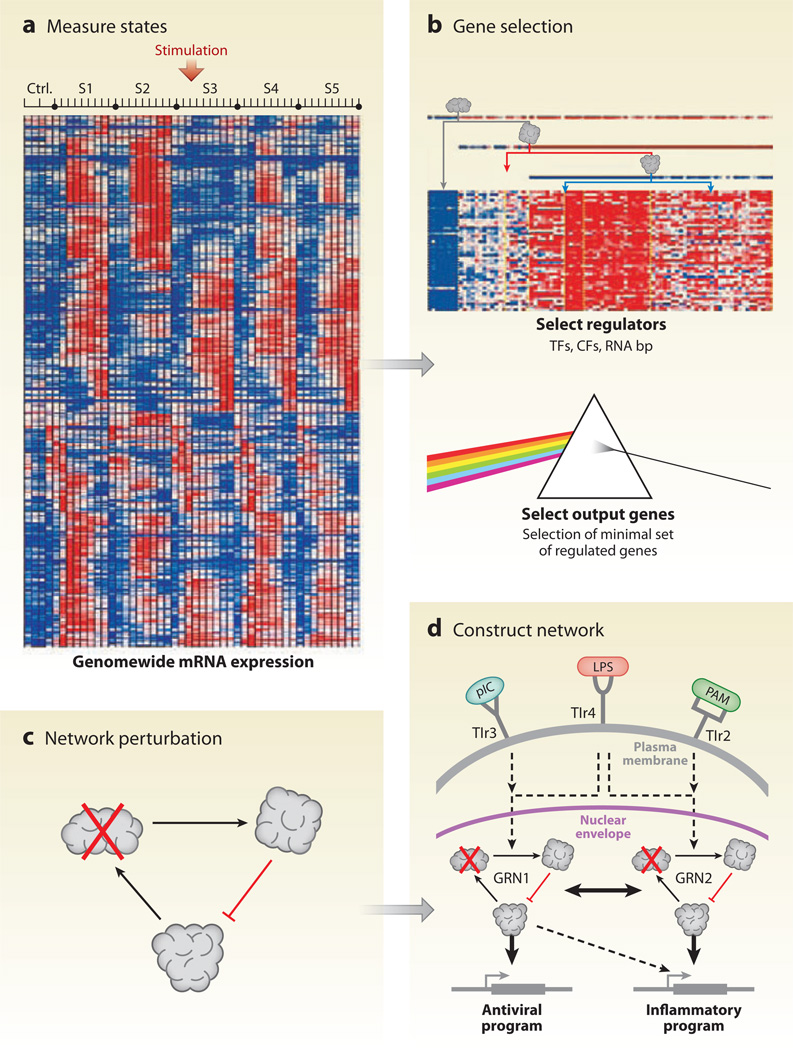
Other methods do not depend on prior knowledge of disease genes but instead infer molecular interaction networks to locate susceptibility genes. For example, Amit et al. (2009) used an RNAi perturbation strategy in mouse dendritic cells to reconstruct the transcriptional network downstream of the Toll-like receptors (TLRs), an important protein family in initiation of pathogen-specific immune responses (Figure 6). Candidate regulators were chosen on the basis of a time course of mRNA expression measured after stimulation with pathogen-derived components. The regulators serve as a gene signature of the immune response in the presence of pathogens. In particular, they identified 144 candidate regulators whose expression changed in response to at least one stimulus. Next, each of the candidate regulators was perturbed (knocked down) by a library of validated lentiviral short hairpin RNAs. Gene expression profiles were gathered under each perturbation and used to infer the regulatory network. The final network included 24 core regulators, affecting the expression patterns of multiple targets, four of which were validated experimentally. They further identified 76 fine-tuners with fewer targets. Together these networks shed light on the regulatory dynamics of the immune response in mammals.
Figure 6.
A systematic strategy for network reconstruction. (a) Cell state is measured using array-based mRNA expression profiles. (b) From these data, a set of putative regulators is selected. TF, transcription factor; CF, chromatin modifier factor; RNA bp, RNA-binding protein. (c) The network is perturbed with lentiviral short hairpin RNA (shRNA) against each regulator, followed by measurement of signature genes. (d) These shRNA profiling measurements are used to inform network reconstruction. Adapted with permission from Amit et al. (2009).
Another approach for de novo identification of disease genes was developed by Wang et al. (2009),who dissected gene expression profiles to infer posttranslational modulators of the MYC transcription factor. Modulators affect a transcription factor at the level of phosphorylation, acetylation, and ubiquitination and are difficult to detect systematically using largescale methods (Linding et al. 2007). However, key modulators were efficiently identified by computing an information-theoretic measure of correlation between the expression profile of MYCand its direct transcriptional targets, given the expression of a “third party” candidate modulator. Candidate modulators were selected for which the expression level was found to significantly influence the correlation between MYC and its targets. Using a similar information-theoretic measure, Mani et al. (2008a) constructed a network of B cell transcriptional interactions and interrogated it for cancer genes. A similar information-theoretic measure was used to find pairs of interactions in the network that gain or lose correlation when comparing a B cell lymphoma tumor with a reference B cell. They demonstrated that pathways enriched for such high and low correlations may be implicated in pro-oncogenic processes. As a specific example, their method recovered BCL2 and SMAD1 in follicular B cell lymphoma. Both of these are oncogenes known to cause cancer but that are not detected through a standard analysis of differential gene expression. Although this approach used expression measurements and is thus unable to capture the effects of posttranslational regulation, the framework can be easily extended to include measurements of protein level as such high throughput data become more commonly available.
Ergün et al. (2007) discovered key mediators in metastatic and nonrecurrent prostate cancers through the use of a regulatory interaction network constructed from a reference set of 1,144 microarray expression profiles spanning seven different cancer types. The known prostate cancer metastasis genes, androgenic receptor (AR) and other genes from the AR pathway, were recovered among the top modulators in metastatic samples but not in non-metastatic ones.
STEM CELL SYSTEMS BIOLOGY AND COMPUTATION OF CELL FATE
Cell fate decisions involve coordinated dynamic expression and regulatory control of hundreds of genes in response to both internal and external stimuli. To dissect the complex interplay among these regulatory pathways, recent studies in stem cell biology have begun to combine classical experimental techniques with emerging high-throughput experimental techniques such as screens for RNAi, genome-wide mRNA expression profiling, large-scale chromatin immunoprecipitation (ChIP), and mass spectrometry–based proteomics (Chen et al. 2008, Kidder et al. 2008, Spooncer et al. 2008). How these vast amounts of data can be used to build a quantitative and predictive model of cell fate control is one of the key challenges in systems biology and stem cell research.
Numerous efforts have been devoted to characterizing the molecular components involved in self-renewal of embryonic stem (ES) cells and differentiation of stem cells along specific lineages. Owing to dramatic advances in genome-wide ChIP technology, the target genes of 20 key ES cell transcription factors, including NANOG (Boyer et al. 2005, Loh et al. 2006, Mathur et al. 2008), OCT4 (Boyer et al. 2005, Loh et al. 2006, Mathur et al. 2008), SOX2 (Boyer et al. 2005), and other factors (Boyer et al. 2005, 2006; Cole et al. 2008; Jiang et al. 2008; Johnson et al. 2008; Kidder et al. 2008; Kim et al. 2008; Liu et al. 2008; Loh et al. 2006; Mathur et al. 2008), have now been identified. An ES cell transcriptional circuit has been assembled through integration of these separate ChIP studies, which cover approximately 50,250 putative protein-DNA interactions that have been identified specifically in ES cells (MacArthur et al. 2009). Moreover, several studies have reported that epigenetic regulation of the key transcription factors by way of chromatin structure (Bernstein et al. 2006, Guenther et al. 2007, Mikkelsen et al. 2007) or DNA methylation (Fouse et al. 2008, Lagarkova et al. 2006, Shen et al. 2006, Yeo et al. 2007) also contributes to the maintenance of pluripotence (reviewed by Bibikova et al. 2008). In addition to epigenetic marks, microRNAs (miRNAs) (Marson et al. 2008) and signaling pathways (Chen et al. 2008) have also been connected to the dynamic balance of ES transcriptional control.
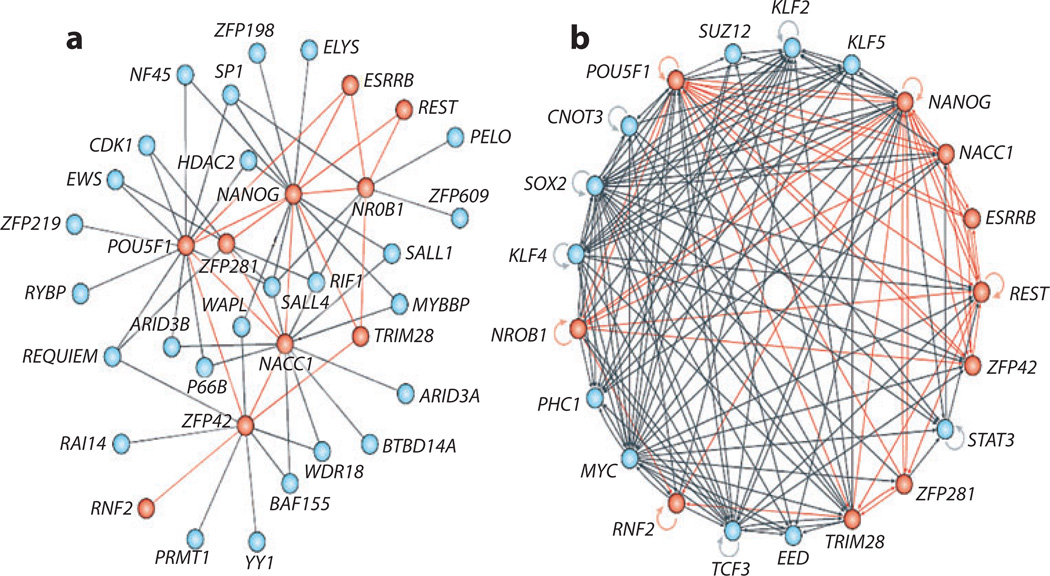
Wang et al. (2006) reported a different take on stem cell systems biology; they assembled a high-quality protein-protein interaction network centered on the NANOG transcription factor in mouse ES cells. They used iterative immunoprecipitation experiments to pull down proteins that physically associate withNANOG, after which mass spectrometry was used to identify the components of the NANOG interactome. Interestingly, the NANOG interactome is highly enriched in the transcription factors of the core ES cell transcriptional circuit, and many of these factors also regulate the expression of other members of the NANOG protein-protein interaction network. This indicates that stem cell fate control is highly dynamic and involves combinatorial interactions between key transcription factors and the genes that encode them. Figure 7 shows the current model of this intrinsically complex but coordinated protein-protein and protein-DNA interaction network.
Figure 7.
Core embryonic regulatory networks for cell fate decisions. (a) High-confidence protein-protein interactions between the transcription factor NANOG and NANOG-associated proteins. An iterative proteomics approach was adapted to identify proteins that physically associate with NANOG and NANOG-associated proteins by using affinity purification in conjunction with mass spectrometry (Wang et al. 2006). (b) Transcription factor binding (protein-DNA) interactions from the data generated by various recent high-throughput chromatin immunoprecipitation (ChIP) experiments. Reproduced with permission from MacArthur et al. (2009).
Müller et al. (2008) reconstructed an extended stem cell regulatory network using a computational approach to integrate publicly available gene expression profiles and protein interaction networks. They first clustered pluripotent, multipotent, and differentiated human cells on the basis of gene expression and identified a set of genes that are specifically upregulated in undifferentiated pluripotent cells (pluripotency-related genes). Next, using a previously compiled network of human protein-protein and protein-DNA interactions including those in the NANOG interactome (Wang et al. 2006), a collection of subnetworks induced by these pluripotency-related genes was identified using a graph-theoretic algorithm (Ulitsky & Shamir 2007). This collection of subnetworks, which the authors name PluriNet, contains mostly novel interactions; few have been well characterized in stem cells. Nonetheless, the collection seems to represent common cellular machineries shared by pluripotent cells (including ES cells, embryonal carcinomas, and induced pluripotent cells).
Another recent study has revealed a large map of transcription factor combinations that may point the way to understanding, and perhaps altering, cell fate decisions. Using the mammalian two-hybrid (M2H) system, Ravasi et al. (2010) generated a database of all pairwise protein-protein interactions among the majority (~1,200) of human transcription factors. From these data, they extracted an interaction network of 15 homeobox transcription factors for which the expression levels were strongly associated with tissue type. The homeobox network was also shown to be capable of stratifying the stem cell expression profiles that had been collected by Muller et al. (2008) into the germ layer from which each was derived (endoderm, mesoderm, ectoderm). It has long been appreciated that combinatorial transcription factor interactions play an important role in cell commitment to different tissue lineages; the work by Ravasi et al. (2010) maps out precisely what some of these combinations are.
All of the studies described above support the idea that pluripotency and self-renewal are under tight control by a dynamic and highly complex regulatory network involving protein-protein interactions, transcription factors, signaling pathways, miRNAs, and other epigenetic modifiers. Meanwhile, follow-up experiments are needed to test these inferred regulatory interactions and their effects on stem cell fate. Integration of large-scale RNAi perturbations with genome-wide ChIP experiments and subsequent gene expression profiling (Ding et al. 2009, Hu et al. 2009) has been shown to be useful in confirming a set of transcriptional interactions and their effects on ES cell fate regulation. A next step is to understand how different internal and external stimuli can affect the dynamics of the regulatory network in ES cells. The thorough understanding of such dynamics will enable human control over cell fate decisions and, ultimately, tissue engineering and regenerative medicine.
SYSTEMS BIOLOGY SOFTWARE
Considerable time and resources have been expended on developing computational tools for answering systems-level research questions. To effectively analyze systems data, a software tool must meet several requirements. First, it must handle genome-scale data sets. Second, the tool must not be restricted to a single data type but be able to integrate multiple measurements of a system. Third, the software should assist with mapping and modeling of networks and pathways from component data sets. Fourth, it should provide an intuitive interface and visual display of both the data and models.
A number of software packages have been developed to address these requirements. Typically, these packages view the landscape of biological data as belonging to either of two categories: (a) data pertaining to molecular components and their states, and (b) data pertaining to molecular interactions. In what follows, we give a sampling of some of these robust integrative software tools available for systems biology research. Some bioinformatics software is intended for those with an in-depth knowledge of computer science; we focus instead on software tools that are geared toward cell biologists.
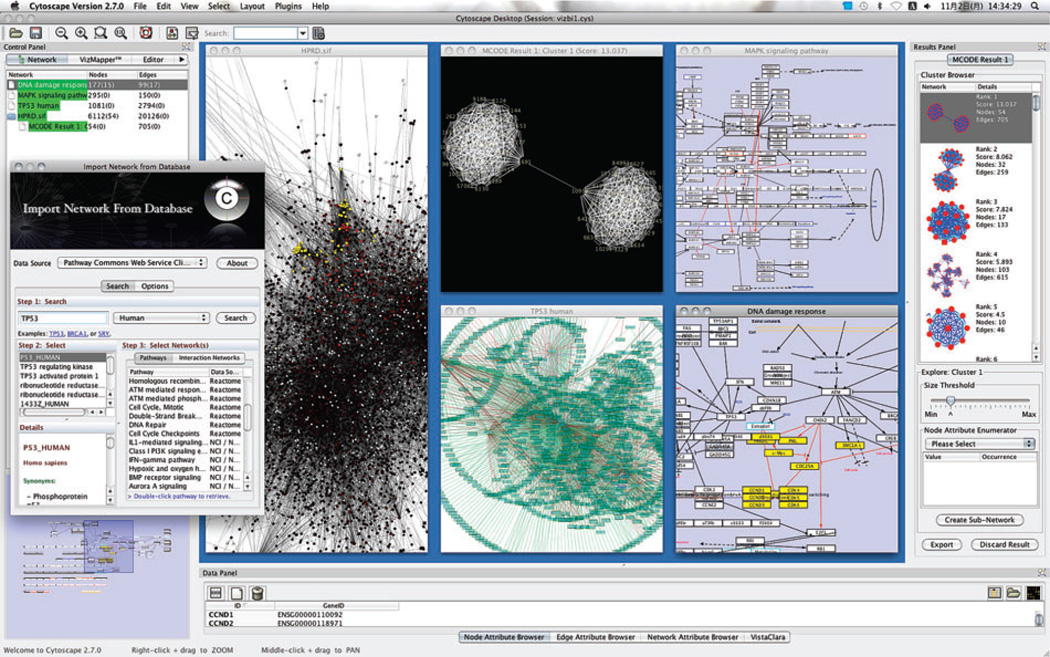
Cytoscape is a free bioinformatics environment for integration, visualization, and query of biological networks (Figure 8). Cytoscape’s core software component provides functionality for data import and export, integration of molecular states with molecular interactions, network and integrated data visualization, and data filtering and query tools. Cytoscape’s VizMapper enables attribute-to-visual mappings, which control visual aspects of nodes and edges (e.g., shape, color, size) based on their molecular states (called node attributes). Such mappings allow overlay of multiple data types in a network context.
Figure 8.
Graphical user interface of Cytoscape. Each window showcases a different analysis or visualization of protein interaction networks and integrated data.
Cytoscape is developed in Java and disseminated under an open source license (the GNU Lesser General Public License, a permissive software license published by the Free Software Foundation). It has been integrated with many other software tools, including stand-alone applications (e.g., geWorkbench, http://wiki.c2b2.columbia.edu/workbench/index.php), Web sites such as a network image generator (e.g., Harvard Gene Functional Annotation Prediction Browser, http://func.med.harvard.edu/site/yeast/), and major network and pathway databases, including the Biomolecular Interaction Network Database (BIND, http://www.bind.ca/), Reactome (http://www.reactome.org/), the Database of Interacting Proteins (http://dip.doe-mbi.ucla.edu/), the Michigan Molecular Interactions database (MiMI http://mimi.ncibi.org), and Pathway Commons (http://pathwaycommons.org). Commercial software companies have also used Cytoscape, including Oracle, Agilent GeneSpring and GeneGO (see below).
The Cytoscape core is extended through a straightforward plugin architecture, which allows rapid prototyping and development of advanced computational analyses and features. The active involvement in Cytoscape plugin development by many third-party programmers attests to the success of Cytoscape as an open source bioinformatics computing environment. Since 2004 (Cytoscape v2.0–v2.6), more than 74 publicly available plugins have been developed, 46 of which have maintained full compatibility with the latest Cytoscape releases (v2.5 or v2.6) (Cline et al. 2007, Shannon et al. 2003).
NAViGaTOR, another open source network visualization package, is an alternative to Cytoscape. Its use of hardware-based graphics accelerators using Open Graphics Library (openGL) allows fast rendering and visualization of extremely large networks. Interesting options include the ability to visualize graphs using both 2D and 3D views and the ability to collapse nodes into a single “meta node.” NAViGaTOR supports an application programming interface (API) for future plug-ins as well as a variety of data formats. It boasts a lasso selection option and a book marking feature to facilitate manual layout and other operations on a network (Brown et al. 2009).
VisANT is a lightweight network visualization tool able to run as a browser-based applet or as a standalone Java program. Of particular interest is its name resolution feature, which attempts to map all nodes in the network to distinct gene names such that two proteins coded by a single gene are always mapped as one entity. This name-mapping feature is one of the most easy to use and streamlined of any software package we review here; it is well designed for the common case with scalability in mind. For large data sets, VisANT has been tested at representing more than 200,000 nodes on a machine with 1 Gb of random-access memory (RAM). Another interesting feature is the representation of metagraphs, whereby a single node can contain a subgraph. VisANT is also integrated with an online database featuring more than 450,000 interactions in dozens of organisms (Hu et al. 2008).
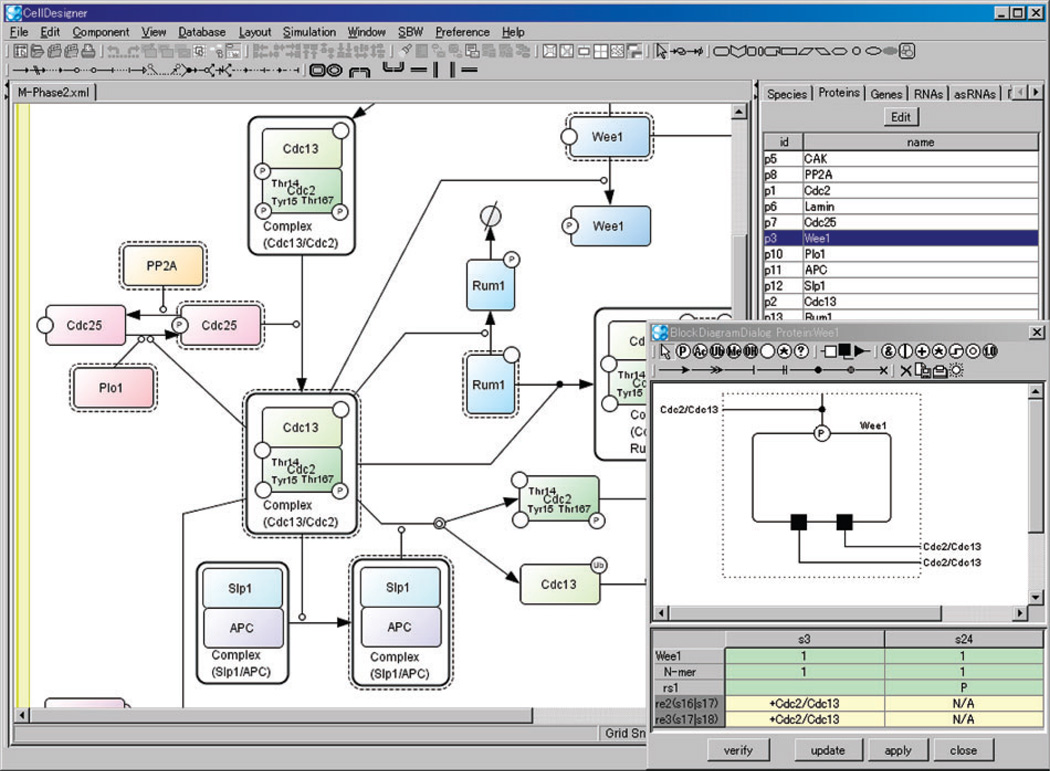
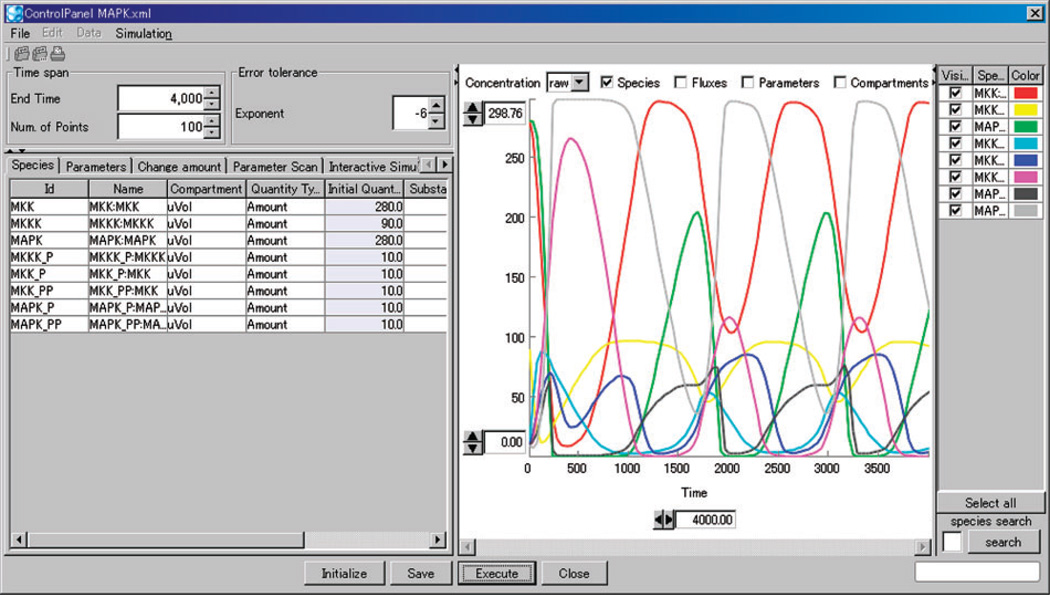
Cell Designer is a structured diagram editor for drawing gene regulatory and biochemical networks (Figures 9 and 10). Users can browse or modify networks as process diagrams (Kitano et al. 2005) and store the networks in systems biology markup language (Hucka et al. 2003), a standard for representing models of biochemical and gene regulatory networks. A unique feature of Cell Designer is that networks are able to link with simulations. Users can view the dynamics of a network under the input parameters through an intuitive graphical interface. Cell Designer is implemented in Java and thus supports various operating systems. The recent releases integrate several simulation/analysis software packages (Funahashi et al. 2008).
Figure 9.
Screenshot of Cell Designer when drawing a network as process diagrams.
Figure 10.
Screenshot of Cell Designer when stimulating a network model given different input parameters.
Another open-source option is Pathway Assist. The focus of this tool is an automated natural language processing-based information extraction system for protein-protein and gene-gene functional interactions. Pathway Assist also provides a native curated database of protein interactions and cellular pathways. Its text-mining tool can extract biological interactions by reading digital text documents (e.g., biomedical journal articles and abstracts). It efficiently scans sentences, searching for co-occurrences of biological terms and connecting verbs (e.g., the keywords “binds,” “inhibits,” “modulates” or “phosphorylates”) between the co-occurring terms. The bundled database contains at present approximately 500,000 biological interactions among more than 50,000 proteins from several organisms extracted from the current literature (Nikitin et al. 2003). It is available for download upon request.
Finally, two commercial packages are also available—GeneGO (Nikolsky et al. 2005) and Ingenuity—both of which offer a comprehensive product aimed at industry and academia.
SUMMARY AND CONCLUSIONS
In this review, we have visited four nascent and emerging areas in the field of systems biology. An overarching principle, and one we have tried to highlight throughout, is that systematic measurement techniques coupled with the use of network models lead to the discovery of novel biology and medicine. Although one can implement this paradigm in several ways, we have attempted to point out some of the exemplars that have led to big wins in the study of development and disease.
One of the main challenges systems biology will face over the next decade is in breaking the divide between classical and high-throughput methods. Its role is not to replace any of the classical techniques from biochemistry or genetics but to provide a set of organizing principles that integrate these methods (Figure 2). The way forward is undoubtedly through close integration of multiple disciplines to crack the biological system using every means possible.
Supplementary Material
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adler AS, Lin M, Horlings H, Nuyten DSA, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat. Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100073. 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asyali MH, Colak D, Demirkaya O, Inan MS. Gene expression profile classification: a review. Curr. Bioinforma. 2006;1:55–73. [Google Scholar]
- Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C, Linding R, Llense F, Heffern E, Martin-Blanco E, et al. Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science. 2008;322:453–456. doi: 10.1126/science.1158739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Computat. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000065. e1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Mitchell-Olds T. From genotype to phenotype: systems biology meets natural variation. Science. 2008;320:495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Sismour AM. Synthetic biology. Nat. Rev. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Laurent LC, Ren B, Loring JF, Fan JB. Unraveling epigenetic regulation in embryonic stem cells. Cell Stem Cell. 2008;2:123–134. doi: 10.1016/j.stem.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proc. Natl. Acad. Sci. USA. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Breslin T, Krogh M, Peterson C, Troein C. Signal transduction pathway profiling of individual tumor samples. BMC Bioinformatics. 2005;6:163. doi: 10.1186/1471-2105-6-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, et al. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics. 2009;25:3327–3329. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Cheang MCU, van de Rijn M, Nielsen TO. Gene expression profiling of breast cancer. Annu. Rev. Pathol. Mech. Dis. 2008;3:67–97. doi: 10.1146/annurev.pathmechdis.3.121806.151505. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M, Smoot M, Cerami E, Kuchinsky A, Landys N, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R, Suarez B, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am. J. Hum. Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu. Rev. Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JLY, Prasad TSK, Chahwan C, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. USA. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Efroni S, Schaefer CF, Buetow KH. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS One. 2007;2:e425. doi: 10.1371/journal.pone.0000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- Ergün A, Lawrence C, Kohanski M, Brennan T, Collins J. A network biology approach to prostate cancer. Mol. Syst. Biol. 2007;3:82. doi: 10.1038/msb4100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord C, Tutt A, Johnson D, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke L, Bakel HV, Fokkens L, de Jong ED, Egmont-Petersen M, Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE. 2008;96:1254–1265. [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang T, Li X, Wang Q, Xu J, et al. Towards precise classification of cancers based on robust gene functional expression profiles. BMC Bioinformatics. 2005;6:58. doi: 10.1186/1471-2105-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Srivas R, Guénolé A, van Attikum H, Krogan N, Kerr K. Genome-wide association data reveal a global map of genetic interactions. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000782. e1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding A. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;321:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer M, Fung E, Wildenhain J, Pierce S, Hoon S, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Snitkin E, Delisi C. VisANT: an integrative framework for networks in systems biology. Brief Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- Ingenuity. Ingenuity Pathways Analysis Software. http://www.ingenuity.com/ [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotech. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Lee E, Chuang H-Y, Kim J-W, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000217. e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang J, Chen T, Wang Y, Xin S, et al. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 2008;18:1177–1189. doi: 10.1038/cr.2008.309. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Lee H, Wang L, Sun F. CGI: a new approach for prioritizing genes by combining gene expression and protein-protein interaction data. Bioinformatics. 2007;23:215–221. doi: 10.1093/bioinformatics/btl569. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. Mol. Cell Biol. 2009;10:672–681. doi: 10.1038/nrm2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol. Syst. Biol. 2008a;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R, St.Onge RP, Hartman JL, Giaever G, Roth FP. Defining genetic interaction. Proc. Natl. Acad. Sci. USA. 2008b;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod D, Widmann C. DNA-damage sensitizers: potential new therapeutical tools to improve chemotherapy. Crit. Rev. Oncol. Hematol. 2007;63:160–171. doi: 10.1016/j.critrevonc.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70(12):4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Myers RH. Huntington’s disease genetics. NeuroRx. 2004;1:255–262. doi: 10.1602/neurorx.1.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol. Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Ekins S, Nikolskaya T, Bugrim A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 2005;158:20–29. doi: 10.1016/j.toxlet.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- Oti M, Brunner H. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein–protein interactions. J. Med. Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem. Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- Peng G, Luo L, Siu H, Zhu Y, Hu P, et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2009;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Ronen K, Vlasblom J, Greenblatt J, Wodak SJ. Local coherence in genetic interaction patterns reveals prevalent functional versatility. Bioinformatics. 2008;24:2376–2383. doi: 10.1093/bioinformatics/btn440. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray analysis and tumor classification. New Engl. J. Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic V, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140(5):744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Meth. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Rommens J, Iannuzzi M, Kerem B, Drumm M, Melmer G, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn I, Schinzel A, Barbie D, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga N, Wang J, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum. Mol. Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat. Rev. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- Spooncer E, Brouard N, Nilsson SK, Williams B, Liu MC, et al. Developmental fate determination and marker discovery in hematopoietic stem cell biology using proteomic fingerprinting. Mol. Cell Proteomics. 2008;7:573–581. doi: 10.1074/mcp.M700292-MCP200. [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson JP, Stalpers LJ, Esveldt-van Lange RE, Franken NA, Haveman J, et al. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS Med. 2006;3:e422. doi: 10.1371/journal.pmed.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck DP, Kluger HM, Kluger Y. Characterizing disease states from topological properties of transcriptional regulatory networks. BMC Bioinforma. 2006;7:236. doi: 10.1186/1471-2105-7-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Karp RM, Shamir R. Detecting disease-specific dysregulated pathways via analysis of clinical expression profiles. Lect. Notes Comput. Sci. 2008a;4955:347. [Google Scholar]
- Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shlomi T, Kupiec M, Shamir R. From E-MAPs to module maps: dissecting quantitative genetic interactions using physical interactions. Mol. Syst. Biol. 2008b;4:209. doi: 10.1038/msb.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert JP, Kanehisa M. Extracting active pathways from gene expression data. Bioinformatics. 2003;19(Suppl. 2):ii238–ii244. doi: 10.1093/bioinformatics/btg1084. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, et al. Genome-wide identification of posttranslational modulators of transcription factor activity in human B cells. Nat. Biotech. 2009;27:829–837. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23:1537–1544. doi: 10.1093/bioinformatics/btm129. [DOI] [PubMed] [Google Scholar]
- Yang WS, Stockwell B. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9:R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- Yeo S, Jeong S, Kim J, Han JS, Han YM, Kang YK. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2007;359:536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.