Lundblad provides perspective on the recent work by Wright and colleagues (this issue) that reveals a tightly choreographed sequence of events that occur during telomere replication and end processing.
Keywords: telomere overhangs, DNA replication, Okazaki fragment RNA primer, aging, cancer
Abstract
Most human cells lack telomerase, the enzyme that elongates telomeres. The resulting telomere erosion eventually limits cell proliferation and tissue renewal, thereby impacting age-dependent pathologies. In this issue of Genes & Development, a technical tour-de-force by Chow and colleagues (pp. 1167–1178) reveals a highly choreographed sequence of events that processes newly replicated chromosome ends into mature telomeres. This sheds new light on an underappreciated contribution to telomere dynamics that may be as important as telomerase in dictating the correlation between life span and telomere length.
Telomeres occupy a small but very intense piece of chromosomal real estate. These natural chromosome termini must accomplish a crucial task in every cell cycle, which is to convince the exquisitely sensitive DNA repair machinery to leave these particular DNA ends alone. In human cells, this complex task is achieved by a suite of proteins aptly titled the shelterin complex (de Lange 2005). Thousands of repeats of a 6-base-pair (bp) sequence (5′-TTAGGG-3′ on the so-called G-rich strand) comprise the duplex region of human telomeres, which provides a specialized landing pad for shelterin. Protruding from the duplex DNA is a G-rich single-stranded extension (Henderson and Blackburn 1989; McElligott and Wellinger 1997) that is also bound by ssDNA-binding subunits of shelterin. When telomeres are unprotected—for example, in response to experimental depletion of components of the shelterin complex—the loss of this critical G-rich overhang and the resulting end-to-end fusions can be catastrophic for genome stability. Thus, cells make a substantial investment in both maintaining and shielding the last several hundred nucleotides of every chromosome end (de Lange 2009).
This single-strand extension is also the substrate for telomerase, which replenishes chromosome ends by synthesizing additional telomeric repeats (Collins 2011). However, telomeres only enjoy the benefits of telomerase-mediated elongation in human cells that rely on continuous proliferation, such as those found in stem cell compartments (Artandi and DePinho 2010). In most tissues, telomerase is down-regulated, resulting in slow erosion of chromosomal termini with each round of cell division. Telomeres that are too short are no longer afforded the protection of the shelterin complex, with the ensuing DNA damage response leading to either cell death through apoptosis or a halt to cell division referred to as cellular senescence. Although the cellular response to severe telomere attrition may provide an important barrier to the development of cancer (de Lange and DePinho 1999), there is a substantial downside. Tissue renewal is critical to organ maintenance and regeneration; telomere length reservoirs that are too short to sustain organ function throughout a normal life span can be a substantial contributing factor to age-dependent degenerative organ failure in the lung, liver, or bone marrow (Armanios 2009).
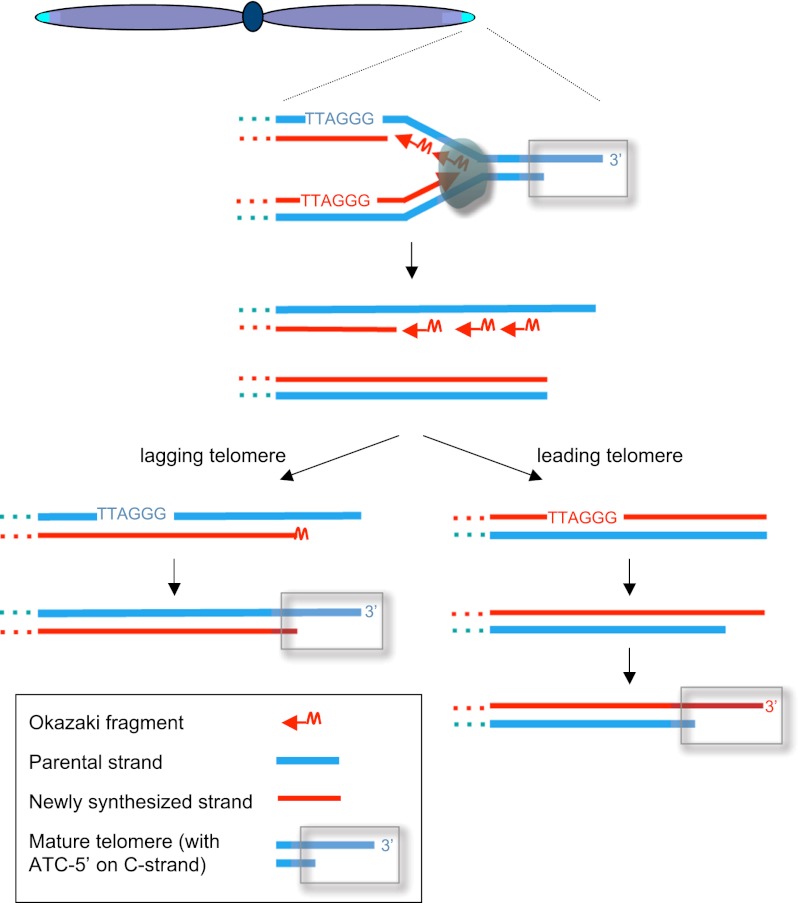
Thus, insights about the events that contribute to telomere erosion in the absence of telomerase should have great relevance to a fundamental aspect of human aging. The susceptibility of chromosome ends to sequence loss was recognized even before the discovery of telomerase, as the mechanisms that govern semiconservative DNA replication were elucidated. With a new appreciation of the complexities of this process, it became apparent that replication of linear chromosomes presents a particular dilemma (Watson 1972; Olovnikov 1973). The “end replication problem,” as it came to be known, stems from the inherent asymmetry of how linear duplex DNA is copied during each cell division (Okazaki et al. 1968; Sugimoto et al. 1968). As the replisome—a complex DNA replication machinery composed of multiple polymerases—moves along the chromosome, each strand is copied by one or more polymerases dedicated to that strand. One newly synthesized strand is produced continuously (the “leading” strand), whereas the opposing (“lagging”) strand is replicated in short discontinuous segments of DNA called Okazaki fragments (Fig. 1). These discontinuous strands are initiated by a very short stretch of RNA, which primes each round of DNA synthesis. Subsequent maturation of Okazaki fragments (replacement of RNA primers by DNA synthesis, followed by ligation of each segment) (Burgers 2009) ultimately generates a continuous product on the lagging strand as well. However, at the extreme termini of chromosomes replicated by lagging strand synthesis, removal of the terminal RNA primer results in a potentially irreparable loss of a very small portion of chromosomal DNA (Fig. 1). With multiple rounds of cell division, the cumulative loss due to the removal of a succession of terminal RNA primers would be incompatible with long-term maintenance of the genome. This end replication problem, postulated >40 years ago, predicted the existence of a counterbalancing mechanism to replenish lost sequences, which is, of course, accomplished by the enzyme telomerase.
Figure 1.
The completion of semiconservative DNA replication results in daughter chromosomes with asymmetric termini that undergo processing to produce mature telomeres. (Left) The newly replicated telomere resulting from lagging strand synthesis retains the terminal RNA primer, which is removed approximately an hour later in a process that also specifies the terminal nucleotides on the C-strand (—ATC-5′) to generate a mature telomere. (Right) The initial product of leading strand DNA replication is a terminus that is blunt; resection of the C-strand to create the mature G-rich overhang occurs in two temporally discrete stages.
But in human cells that lack telomerase, just how serious a problem is loss of the terminal RNA primer? If the last Okazaki fragment initiates at the very terminus of the templating parental strand of DNA, potentially less than two telomeric repeats would be lost at each cell division on the lagging strand, since the primer itself is only 10 nucleotides (nt). Although telomere length shows substantial variation among individual humans (and even among individual telomeres) (Baird et al. 2003), a starting telomere length of just a thousand repeats would allow an impressive number of population doublings in telomerase-deficient tissues during a human life span. Furthermore, experimental observations in ciliates and yeast suggest that the terminal Okazaki fragment does in fact initiate at the very end of the chromosomal DNA molecule (Klobutcher et al. 1981; Marcand et al. 1999; Larrivee et al. 2004). One could argue that it makes teleological sense (no pun intended) that the cellular DNA replication machinery would use whatever mechanisms necessary to ensure close to complete duplication of the genome, particularly in single-cell organisms. Given other parallels in telomere biology between single-cell and multicellular eukaryotes, these assumptions about the placement of the terminal Okazaki fragment have been extended to human cells. This extrapolation has not been challenged, however, largely due to the potential technical impasse: Assessing the status of ∼100 nt at the termini of 46 chromosomes spanning a genome composed of >3.4 billion bp is a nontrivial endeavor.
But as Wright and colleagues (Chow et al. 2012) show in this issue of Genes & Development, they are up to the task. Building on an elegant series of prior publications that have used ever more sophisticated assays to monitor the status of the G-rich single-strand overhang (Sfeir et al. 2005, Chai et al. 2006; Zhao et al. 2008), they have provided the most detailed picture to date of the events that occur at chromosomal ends following the completion of conventional semiconservative DNA replication. Their analysis reveals a complex progression of events that are necessary to process the newly replicated DNA ends into “mature” telomeres that exhibit the vital G-rich overhang, with apparently distinct activities dedicated to the leading and lagging termini. In particular, they overthrow prior assumptions about the position of the terminal RNA primer during replication of the telomere lagging strand, showing instead that the terminal Okazaki fragment is situated ∼70–100 nt from the end. Stunningly, this means that each round of DNA replication leaves up to 100 nt of the genome unreplicated on each lagging terminus. This has substantial implications when considering mechanisms that drive telomere shortening, as this placement of the terminal primer means that as much as ∼1000 nt can be lost in just ∼20 population doublings.
The critical technical hurdle in these experiments was the ability to separate termini synthesized by leading strand synthesis from those produced by lagging strand processes in sufficient quantities to allow subsequent high-resolution detection of the length of the G-rich overhang in each population. To do so, Wright and colleagues (Chow et al. 2012) turned to the classic Meselson and Stahl (1958) experiment, in which newly replicated DNAs of different densities were separated on a cesium chloride gradient. In their updated version, incorporation of the nucleoside analog 5-bromodeoxyuridine into newly synthesized DNA resulted in telomeres generated by leading strand synthesis with a heavier density than those telomeres produced by lagging strand processes (which will be called leading telomeres and lagging telomeres, for simplicity). Leading and lagging strand telomere fractions were collected at various time points from synchronized cells progressing through the cell cycle, with two technically distinct assays used to assess the terminal structure in these two populations of telomeres.
These assays were sensitive enough to detect an obvious (and expected) difference between leading and lagging telomeres at the earliest possible time point following DNA replication. At leading telomeres, there was no detectable G-rich overhang, consistent with the prediction that the initial product of semiconservative DNA replication should be a blunt DNA end. In contrast, G-rich overhangs that were close to their mature size were observed at lagging strand telomeres, even though the terminal RNA primers had not yet been removed from the C-strand. Initial detection of these 10-nt RNA primers relied less on technical prowess and more on old-school knowledge by using an exonuclease from a bacteriophage that cannot digest DNA/RNA hybrids. Whereas the C-strand of leading strand telomeres was readily digested by this exonuclease, the lagging telomere fraction was completely resistant immediately following semiconservative replication, arguing that the C-strand of lagging telomeres terminated in RNA. Direct evidence came from an elegantly designed experiment in which C-strands from both leading and lagging telomeres were ligated to a biotinylated oligomer and purified by binding to streptavidin-conjugated beads. RNase digestion released the lagging strand C-strands from the beads, whereas leading strand C-strands remained bound, directly implicating the presence of RNA on the lagging C-strand.
A somewhat surprising feature of these experiments was the stability of this terminal RNA primer on the C-strand, which was retained for several hours following completion of lagging strand DNA synthesis. Whether this delay is a regulated aspect of telomere end processing is only one of several questions raised by this study. Equally interesting will be the identification of the enzyme responsible for removal of this 10-nt RNA primer. Although it is formally possible that the same nucleases that promote Okazaki fragment maturation will be used, our current understanding of this process argues against this premise. The task of removal of RNA primers from Okazaki fragments is assigned to structure-specific nucleases that recognize a “flap” structure generated when the RNA primer is displaced by DNA synthesis (Burgers 2009). With no mechanism to displace the terminal RNA, this invokes other enzymatic activities. Given the investment that the cell makes in ensuring telomere function, one speculative possibility is that there is a telomere-specific RNase designed for this task.
Left unaddressed in the above discussion is the fate of the newly replicated leading strand termini. As pointed out ∼20 years after the end replication postulate (Lingner et al. 1995), leading strand telomeres face their own predicament every time a linear chromosome is replicated (Fig. 1). Unlike lagging strands, which have the ability to regenerate the crucial G-rich overhang simply as a default consequence of Okazaki fragment placement, the immediate result of leading strand synthesis is a blunt terminus (or nearly blunt, as the final 1–2 nt are often missing) (Sfeir et al. 2005). This necessitates another set of operations, distinct from the events occurring at lagging termini, to ensure that the C-strand of this potentially genotoxic blunt terminus is resected to produce the necessary G-rich overhang. Solutions to this long-standing problem have often invoked parallels with double-strand break repair, in which a similar 5′-to-3′ resection of one strand of a newly generated break exposes a single-stranded substrate for homologous recombination. However, at least in the case of human telomeres, a shelterin-associated nuclease (fittingly named Apollo, based on similarities with a related nuclease called Artemis) is responsible for resection of leading strand telomeres (van Overbeek and de Lange 2006). When Apollo is absent, leading strand telomeres lose their protective G-rich overhang, leading to chromosome fusions and activation of the ATM-dependent DNA damage response (Wu et al. 2010). Temporally, nucleolytic resection of the C-strand of leading strand termini (presumably by Apollo) does not occur immediately following completion of semiconservation DNA replication (Chow et al. 2012). Instead, a delay of roughly an hour occurs before G-rich overhangs can be detected on leading strand telomeres, well behind the appearance of overhangs on lagging strand telomeres. This raises an intriguing question: What ensures that this hour-long period of blunt DNA ends does not engage the DNA damage machinery?
One final processing step is required before these two termini can be considered fully mature telomeres. At the majority of human telomeres, the C-rich strand terminates with the sequence CCAATC-5′ (Sfeir et al. 2005); specificity for the sequence of the final 5′ nucleotide of the C-strand is also observed at ciliate telomeres (Fan and Price 1997). Thus, telomere maturation is not complete until the 5′-terminal sequence of the recessed C-strand has been determined. On lagging telomeres, specification of this 5′-terminal nucleotide on the C-strand is concomitant with removal of the RNA primer (Chow et al. 2012). In contrast, the nuclease-mediated conversion of the blunt leading telomeres into mature telomeres occurs in two stages; somewhat surprisingly, there is a delay between the initial C-strand resection and determination of the final nucleotide of the C-strand. Does this mean two discrete enzymatic activities, or is Apollo (and possibly other nucleases) specific enough to fulfill both tasks? If the latter, the pause might imply some sort of proofreading or fidelity check. However, the functional significance of this C-strand sequence specificity is unclear. Determination of the final nucleotide of the C-strand might simply be a passive consequence of binding of the G-rich overhang by subunits of the shelterin complex, with constraints on resection due to proteins bound in register with the sequence of the telomeric repeats (Sfeir et al. 2005).
Thus, complete processing of newly replicated termini into mature telomeres occurs in several steps (Fig. 1), with events at lagging strand termini preceding those at leading termini. The significance of this study goes well beyond the revelation that the position of the terminal RNA primer on lagging strands makes a more substantial contribution to telomere shortening than had been previously appreciated. The identification of temporally discrete steps helps define questions about potential points of regulation as well as enzymatic activities yet to be discovered. Since most human cells lack appreciable levels of telomerase activity, studies such as this help focus our collective attention on aspects of telomere dynamics that have the potential to modulate when cells reach the end of their replicative life span.
One final consideration about telomere end processing centers around a small subpopulation of chromosomal ends that are generated through incomplete replication of the duplex telomeric tract. In the analysis reported by Chow et al. (2012), the method used to fractionate leading and lagging strand termini ensured that only those telomeres in which the replisome had fully traversed the duplex telomere were analyzed. Partially replicated telomeres with intermediate densities, which resolved midway between the peaks of fully replicated leading versus lagging telomeres, were specifically excluded. However, an increasing body of evidence indicates that replication does not always successfully proceed all the way through the entire telomeric duplex tract, resulting in dissociation of the replisome due to replication fork collapse. As might be expected when the replisome encounters highly repetitive DNA, telomeric DNA presents a challenge for the DNA replication machinery (Makovets et al. 2004; Ohki and Ishikawa 2004). In fact, one of the tasks of the shelterin complex is to facilitate replication of duplex telomeric DNA, as telomeres display the characteristics of fragile sites when the TRF1 subunit of the shelterin complex is absent (Martínez et al. 2009; Sfeir et al. 2009). Defects in other factors that are not necessarily telomere-specific can also confer lagging strand-specific replication defects at telomeres (Crabbe et al. 2004; Saharia et al. 2008). One of these factors, FEN1 (Flap endonuclease I), has long been studied for its role in Okazaki fragment processing, as discussed above; however, Stewart's group (Saharia et al. 2010) has instead implicated the FEN1 nuclease in promoting replication restart at telomeres, potentially by processing stalled replication forks.
Although most telomeres escape the consequences of a replication default, the outcome of a stalled replication fork and subsequent collapse of the replisome is an abruptly shortened telomere. Such ultrashort telomeres can, in fact, be detected in senescent human cells, using a PCR-based assay designed to detect single telomere ends as a subpopulation that is distinct from telomeres that have eroded more gradually (Baird et al. 2003). DNA termini that are created as the result of replication fork collapse are also presumably structurally dissimilar from termini generated by the DNA replication machinery running off the end, with the potential to invoke a whole different cast of characters (Petermann and Helleday 2010). Therefore, one additional intriguing question for the future will be whether chromosome termini generated by replication mishaps will follow the choreography described by Chow et al. (2012) or whether a distinctly different dance routine will be required to process this particular subpopulation into mature telomeres.
Acknowledgments
Research in my laboratory is supported by R37 AG11728 from the National Institutes of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.195339.112.
References
- Armanios M 2009. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet 10: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA 2010. Telomeres and telomerase in cancer. Carcinogenesis 31: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D 2003. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 [DOI] [PubMed] [Google Scholar]
- Burgers PM 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284: 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE 2006. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE 2012. Early and late steps in telomere overhang processing in normal human cells: The position of the final RNA primer drives telomere shortening. Genes Dev (this issue). doi: 10.1101/gad.187211.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K 2011. Single-stranded DNA repeat synthesis by telomerase. Curr Opin Chem Biol 15: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- de Lange T 2005. Shelterin: The protein complex that shapes and safe-guards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, DePinho RA 1999. Unlimited mileage from telomerase? Science 283: 947–949 [DOI] [PubMed] [Google Scholar]
- Fan X, Price CM 1997. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol Biol Cell 8: 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ER, Blackburn EH 1989. An overhanging 3′ terminus is a conserved feature of telomeres. Mol Cell Biol 9: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci 78: 3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR 1995. Telomerase and DNA end replication: No longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA 2009. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ 1997. The terminal DNA structure of mammalian chromosomes. EMBO J 16: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M, Stahl FW 1958. The replication of DNA in Escherichia coli. Proc Natl Acad Sci 44: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Ishikawa F 2004. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res 32: 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci 59: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41: 181–190 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA 2008. Flap endonuclease 1 contributes to telomere stability. Curr Biol 18: 496–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA 2010. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J Biol Chem 285: 27057–27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE 2005. Telomere-end processing: The terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE 1994. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Okazaki T, Okazaki R 1968. Mechanism of DNA chain growth, II. Accumulation of newly synthesized short chains in E. coli infected with ligase-defective T4 phages. Proc Natl Acad Sci 60: 1356–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T 2006. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol 16: 1295–1302 [DOI] [PubMed] [Google Scholar]
- Watson JD 1972. Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE 2008. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res 36: e14 doi: 10.1093/nar/gkm1063 [DOI] [PMC free article] [PubMed] [Google Scholar]